Abstract
Adeno-associated virus (AAV) is the most common vector for clinical gene therapy of the CNS. This popularity originates from a high safety record and the longevity of transgene expression in neurons. Nevertheless, clinical efficacy for CNS indications is lacking, and one reason for this is the relatively limited spread and transduction efficacy in large regions of the human brain. Using rationally designed modifications of the capsid, novel AAV capsids have been generated that improve intracellular processing and result in increased transgene expression. Here, we sought to improve AAV-mediated neuronal transduction to minimize the existing limitations of CNS gene therapy. We investigated the efficacy of CNS transduction using a variety of tyrosine and threonine capsid mutants based on AAV2, AAV5, and AAV8 capsids, as well as AAV2 mutants incapable of binding heparan sulfate (HS). We found that mutating several tyrosine residues on the AAV2 capsid significantly enhanced neuronal transduction in the striatum and hippocampus, and the ablation of HS binding also increased the volumetric spread of the vector. Interestingly, the analogous tyrosine substitutions on AAV5 and AAV8 capsids did not improve the efficacy of these serotypes. Our results demonstrate that the efficacy of CNS gene transfer can be significantly improved with minor changes to the AAV capsid and that the effect is serotype specific.
Keywords: AAV, CNS, capsid, mutant
Introduction
The use of recombinant adeno-associated virus (rAAV) in pre-clinical and clinical gene therapy treatments of neurological disorders has surpassed the use of other vectors such as lentivirus.1, 2 The preferential use of rAAV is mainly due to the strong safety profile, efficacious neuronal transduction, and longevity of expression, particularly in non-dividing cells such as neurons.3 Nevertheless, clinical efficacy is largely absent, and concerns related to the immunogenicity of the viral capsids still remain. The limited clinical success has been attributed to the inability to efficiently scale transduction between species (i.e., from mice to non-human primates [NHPs] to humans),4 differential properties of the healthy and the diseased brain,4 and the differential ability of rAAV to infect the aged versus the young brain,5 among others. To address such shortcomings, researchers have engaged in a veritable arms race to enhance the efficacy of rAAV using both rational design methods based on the known structure of rAAV6 and molecular evolution techniques to select for capsids with various desirable properties.7, 8, 9
One of the first major advancements in enhancing transduction was the finding that protein tyrosine kinases phosphorylate specific residues on the AAV2 capsid surface,10 resulting in ubiquitination and degradation of the virion before it reaches the nucleus. To counteract this process, Srivastava and colleagues6, 10, 11 mutated specific, surface-exposed AAV2 capsid tyrosine (Y) residues to phenylalanine (F) because these residues cannot be phosphorylated. These capsid mutants exhibit improved intracellular transport with the resultant increase of virions arriving in the nucleus, ultimately vastly improving transduction both in vitro6, 10 and in vivo.11
A second rate-limiting step of AAV transduction is nuclear entry of the viral particle. Recent work demonstrated that AAV enters the nucleus via the nuclear pore complex, a process facilitated via an interaction with importin-α and -β.12 This step appears to be relatively inefficient because particles remain outside the nuclear membrane for long times following infection.13, 14 As for intracellular transport, nuclear translocation may be negatively regulated by the phosphorylation of exposed threonine (T) amino acids. Indeed, mutation of an exposed threonine residue to a valine (T491V or T492V) results in improved nuclear import and a commensurate improvement in transduction of AAV2 and AAV6, respectively, because valines cannot be phosphorylated.15, 16 However, whether this phenomenon was entirely mediated via a direct effect on nuclear import is not entirely clear.
Heparan sulfate proteoglycan (HSPG) was originally viewed as the sole canonical receptor for AAV2 infection.17 However, it is now evident that AAV2 can undergo HSPG-independent attachment and internalization,18, 19, 20 and that the dependency on HSPG for infection varies between cell types.18 On the contrary, because of the high degree of normal HSPG expression, both in the extracellular matrix and on the surface of a multitude of cells throughout the body, deletion of the HSPG binding motif is thought to be a viable means to retarget, and improve, rAAV2 transduction by avoiding sequestration in non-target tissue or cells.19
Herein, we sought to improve rAAV transduction in the CNS using a rational capsid design approach. We generated capsid amino acid mutations that were predicted to enhance the overall efficacy of internalized viral particles to reach the nucleus (Y-F and T-V mutations) and AAV2 HSPG null capsid mutants predicted to improve extracellular spread of virions.18 Several of these capsid alterations exhibit significantly increased transduction efficacy in non-CNS organs such as the eye11, 21 or the liver.15 However, this is the first study to examine the potential utility of these engineered capsid mutants in the CNS. We compared the transduction efficacy following intraparenchymal injections into the striatum or hippocampus of a variety of capsid mutants based on rAAV2, rAAV5, and rAAV8, all serotypes that have garnered a significant interest in the treatment of CNS disorders. Transduction efficiency was assessed via quantification of transduced cells, transgene expression levels, and volumetric spread of transgene expression. Moreover, we evaluated whether these capsid alterations modulated the tropism of rAAV. Our results demonstrate that the inclusion of various mutations into the rAAV capsid significantly alters the number of transduced cells, magnitude of transgene expression, and volumetric spread, but only the mutant rAAV2 incapable of binding HSPG exhibited altered cell type tropism to some extent.
Results
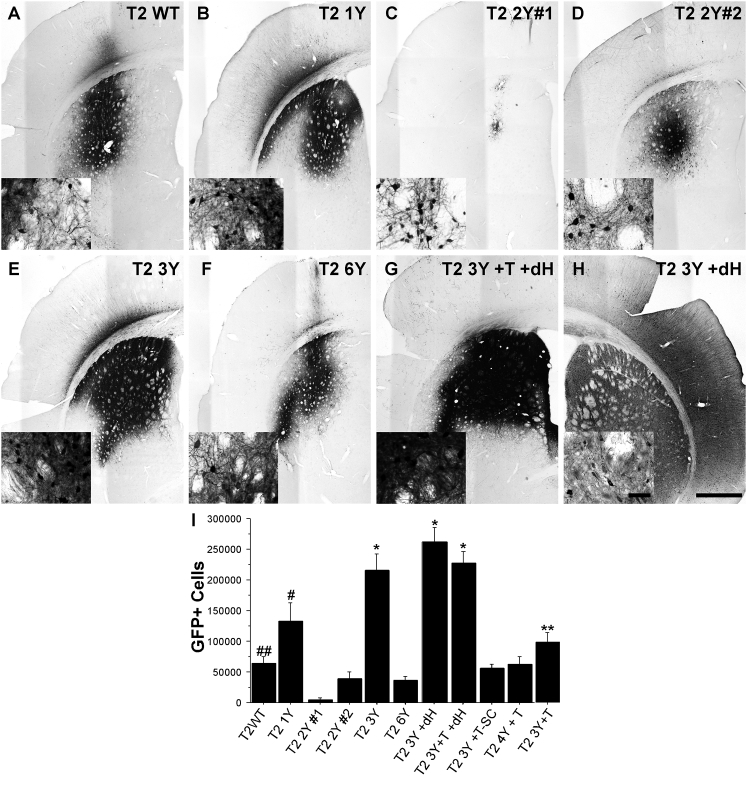
AAV2 Capsid Mutants Enhance Transduction Efficacy following Intrastriatal Delivery
The striatum (caudate/putamen) has been targeted clinically in the treatment of Parkinson’s disease (PD) using various growth factors, and targeting this brain region is being investigated for other neurological indications such as Huntington’s disease.22 To evaluate the transduction efficacy of capsid mutant rAAVs in this clinically relevant brain region, we injected adult Sprague-Dawley rats in the striatum with 2 μL (normalized to 1.2 × 1012 vector genomes/mL [vg/mL]) of either wild-type (WT) AAV2 or various AAV2 capsid mutants (see Table 1) carrying a GFP reporter construct. Animals were sacrificed 1 month following the injection, a time at which peak expression is typically achieved,23 and brains were processed for immunohistochemistry to quantify number of transduced cells. Qualitative observations revealed that all the rAAV vectors used in this study transduced striatal neurons, but to widely varying degrees (Figures 1A–1H; Figure S1). Stereological cell counting of GFP+ striatal neurons revealed that the T2 3Y +dH (260,952 GFP+ cells; Figures 1H and 1I), T2 3Y +T +dH (226,648 GFP+ cells; Figures 1G and 1I), and T2 3Y (216,137 GFP+ cells; Figures 1E and 1I) mutant capsids produced the most robust transduction of striatal neurons compared with all other capsid types (p < 0.001 versus T2 1Y, p < 0.0001 versus all other capsids). The T2 1Y capsid infected significantly more cells (133,000; Figures 1B and 1I) than T2 2Y#1 (4,184; Figures 1C and 1I), T2 2Y#2 (39,835; Figures 1D and 1I), T2 6Y (36,327; Figures 1F and 1I), T2 4Y +T (62,467; Figure S1B; Figure 1I), and T2 WT (64,656; Figures 1A and 1I) (p < 0.004 versus all groups). T2 3Y +T (99,172; Figure S1C; Figure 1I) exhibited significantly greater transduction as compared with T2 2Y#1 (Figures 1C and 1I), T2 2Y#2 (Figures 1D and 1I), and T2 6Y (Figures 1F and 1I; p < 0.01). Interestingly, several capsids (T2 Y#2 [Figures 1D and 1I], T2 6Y [Figures 1F and 1I], T2 4Y +T [Figure S1B; Figure 1I], T2 3Y +T [Figure S1C; Figure 1I]) did not provide improved transduction over T2 WT (Figures 1A and 1I). Notably, T2 2Y#1 (Figures 1C and 1I) exhibited a deficiency in neuronal transduction (p < 0.03) compared with T2 WT (Figures 1A and 1I). The utilization of a self-complementary genome (T2 3Y +T -SC; 56,635; Figures S1A and S1C; Figure 1I) did not improve overall transduction; rather, there was a trend toward a reduction in the overall number of GFP+ cells (p = 0.09). This is not entirely surprising, however, because the benefit with a self-complementary genome is to overcome the rate-limiting step of double-stranded genome formation, which occurs early following nuclear translocation.24 Thus, the lack of an improvement seen in this study is likely the result of the later time point (4 weeks) chosen for analysis.
Table 1.
List of Capsid Amino Acid Substitutions Utilized in This Study
| Mutant Name | Amino Acid Mutations |
|---|---|
| AAV2 | |
| T2 WT | N/A |
| T2 1Y | Y444F |
| T2 3Y | Y444F, Y500F, Y730F |
| T2 2Y #1 | Y500F, Y730F |
| T2 2Y #2 | Y444F, Y730F |
| T2 6Y | Y252F, Y272F, Y444F, Y500F, Y704F, Y730F |
| T2 3Y +T | Y444F, Y500F, Y730F, T491V |
| T2 4Y + T | Y272F, Y444F, Y500F, Y730F, T491V |
| T2 3Y +T +dH | Y444F, Y500F, Y730F, T491V, R585S, R588T, R487G |
| T2 3Y +dH | Y444F, Y500F, Y730F, R585S, R588T, R487G |
| AAV5 | |
| T5 WT | N/A |
| T5 1Y | Y719F |
| T5 2Y | Y263F, Y719F |
| T5 5Y | Y242F, Y263, Y689F, Y693F, Y719F |
| AAV8 | |
| T8 WT | N/A |
| T8 1Y | Y733F |
| T8 2Y | Y447F, Y733F |
| T8 2Y +T | Y447F, Y733F, T494V |
dH, abolished heparin sulfate binding domain; F, phenylalanine; G, glycine; S, serine; T, threonine; V, valine; WT, wild-type; Y, tyrosine.
Figure 1.
Rationally Designed AAV2 Capsid Mutants Significantly Enhance Striatal Transduction
Adult Sprague-Dawley rats received intrastriatal injections of an AAV2 vector (2 μL of 1.2 × 1012 vg/μL for all viruses) as defined in Table 1. One month later, the animals were sacrificed and processed for transgene (GFP) immunoreactivity. (A–H) Representative images of GFP immunoreactivity in the striatum following the injection of (A) T2 WT (n = 8), (B) T2 1Y (n = 7), (C) T2 2Y#1 (n = 6), (D) T2 2Y#2 (n = 8), (E) T2 3Y (n = 7), (F) T2 6Y (n = 8), (G) T2 3Y +T +dH (n = 12), and (H) T2 3Y +dH. Insets are higher magnification images taken at the periphery of the transduction area. (I) Stereological quantification of striatal GFP+ neurons revealed that T2 3Y, T2 3Y +T +dH, and T2 3Y +dH transduced significantly more striatal neurons than any other capsid (*p < 0.001) followed by T2 1Y (#p < 0.004 versus remaining groups except T2 3Y +T). Several capsid serotypes did not exhibit an improvement over WT (T2 2Y#2, T2 6Y, T2 4Y +T, T2 3Y +T), and T2 2Y#1 exhibited significant impairment in transduction as compared with the T2 WT control (##p < 0.03). The T2 3Y +T capsid showed higher transduction versus T2 2Y#1, T2 2Y#2, and T2 6Y (**p < 0.01). (I) Error bars represent mean + SD. (H) Scale bar, 1 mm (inset scale bar, 50 μm); scale bar applies to all micrographs.
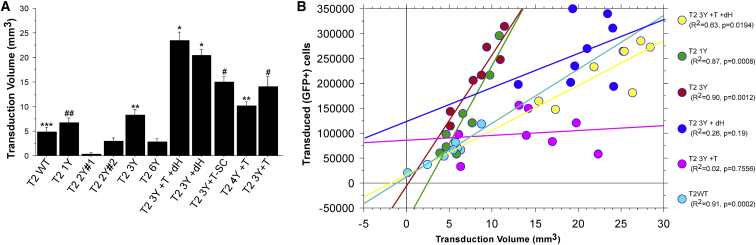
AAV2 Capsid Mutants Exhibit a Significantly Increased Volume of Transduction following Intrastriatal Injections Compared with WT AAV2
The ability to transduce large structures such as the striatum is not only dependent on the infectivity of the virion, but also on the ability of the viral capsid to diffuse throughout the extracellular environment. In order to determine the volume of transduction, we outlined the area of transduction (i.e., the area containing GFP+ cells) of serial sections throughout the striatum and utilized the Cavalieri method to estimate the volume (Figure 2A). The effects of the capsid mutations were remarkable, particularly for the T2 3Y +dH and T2 3Y +T +dH capsids, which transduced approximately 20.3 mm3 and 23.4 mm3, respectively. This volumetric spread was nearly five times greater than that observed with T2 WT (4.86 mm3) and was significantly greater than all other mutants (p < 0.0001). However, virions also exhibited improved diffusion without the removal of the canonical HSPG binding site, although less so than dH mutants. T2 3Y +T (14.1 mm3) transduced a greater volume than all capsid variants (except for HSPG mutants; p < 0.02 versus T2 4Y +T; p < 0.001 versus others). In addition, the T2 4Y +T (10.22 mm3) and T2 3Y (8.35 mm3) capsids facilitated greater diffusion than T2 WT (p < 0.04). Again, T2 2Y#1 exhibited an impairment in transduction (as assessed by volume) compared with WT (0.32 mm3; p < 0.01). No other capsid (T2 1Y [6.69 mm3], T2 2Y#2 [3.03 mm3], or T2 6Y [2.88 mm3]) exhibited different transduction volumes from the WT AAV2 capsid. As expected, no effect on diffusion was seen with the use of a self-complementary genome (T2 3Y +T -SC [14.94 mm3] versus T2 3Y +T [14.1 mm3]).
Figure 2.
Removal of the Canonical HSPG Binding Site Significantly Improves Viral Vector Spread
The volume of transduction from animals shown in Figure 1 and Figure S1 was estimated using the Cavalieri method. (A) Volumetric measurements of striatal transduction revealed that the vector spread was significantly enhanced with the T2 3Y +dH and T2 3Y +T +dH capsids as compared with any other capsid (*p < 0.0001). Moreover, all T-V mutants exhibited significantly enhanced transduction volume (#p < 0.0001 versus T2 WT, T2 1Y, T2 2Y#1, T2 2Y#2, T2 3Y, T2 6Y) compared with other capsids, with the exception of T2 4Y +T, which was not different from T2 3Y. **p < 0.005 versus T2 WT, T2 2Y#1, and T2 2Y#2, T2 6Y; ##p < 0.05 versus T2 2Y#1, T2 2Y#2, and T2 6Y; ***p < 0.01 versus T2 2Y#1. Error bars represent the mean + SD. (B) Plot of GFP+ cells versus transduction volume. A clear pattern emerged for several of the capsid mutants tested. For instance, both T2 3Y and T2 1Y infected an increased number of neurons within a limited volume. In contrast, T2 3Y +dH and T2 3Y +T +dH (which transduced a similar number of neurons as T2 3Y) transduced neurons over a much greater volume.
Next, a regression analysis was performed comparing the number of transduced cells with the transduction volume (Figure 2B) where a clear pattern emerged for several of the mutants. For instance, it was apparent that the HSPG null mutants T2 3Y +dH (Figure 2B, dark blue circles) and T2 3Y +T +dH (Figure 2B, yellow circles) transduced less cells per volume (R2 = 0.26 and 0.63, respectively). This was in stark contrast with injection of T2 3Y (Figure 2B, red circles) and T2 1Y (Figure 2B, green circles), which resulted in an increase in the number of transduced cells across a smaller volume in closer proximity to the injection site (R2 = 0.90 and 0.87, respectively). In contrast, several of the other mutants did not exhibit a correlation between transduction volume and number of cells transduced (e.g., T2 3Y +T; Figure 2B, purple circles; R2 = 0.017).
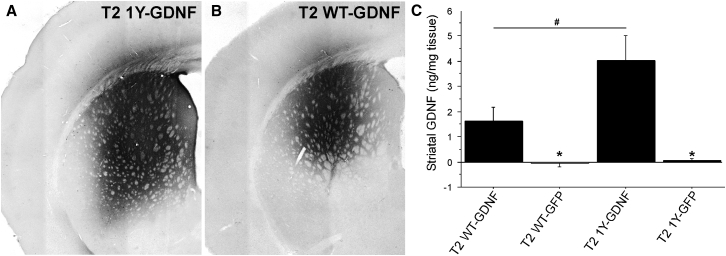
AAV Capsid Tyrosine Mutant Enhances Striatal Levels of the Therapeutic Transgene GDNF
Quantification of GFP+ neurons as a metric of transduction efficacy is binary (yes or no) and does not provide information as to how efficiently GFP+ cells are transduced (i.e., how many genome copies have made it to the nucleus in each GFP+ cell) beyond that which is required to detect a positive GFP signal. Moreover, these measurements do not provide information on the efficiency of transduction using a mammalian protein normally expressed in the brain that may represent a potentially therapeutic factor. Thus, we measured the level of striatal transgene expression of a therapeutic gene (i.e., glial cell line-derived neurotrophic factor [GDNF]) using an AAV2 capsid mutant that exhibited dramatically improved transduction of the striatum to further investigate the potential clinical utility of these capsid mutants. We chose a capsid with a focal transduction pattern (T2 1Y) because GDNF readily diffuses away from the injection site,25 and limiting the spread of the vector would allow us to ensure accurate measurements of transgene levels. Using the same injection paradigms as described above, animals received intrastriatal injections of T2 WT or T2 1Y expressing either GFP or GDNF. One month later, animals were sacrificed and striatal GDNF protein content was measured via ELISA. As expected, T2 1Y-mediated GDNF expression (4.0 ng/mg tissue; Figures 3A and 3C) far exceeded that achieved with T2 WT (1.6 ng/mg tissue; Figures 3B and 3C). Expression of GDNF was not detectable with either of the GFP control viruses (Figure 3C).
Figure 3.
Increase in Transduction Efficacy Is Reflected by Increased GDNF Transgene Levels
An AAV genome containing an expression cassette encoding human glial cell line-derived neurotrophic factor (GDNF) was packaged into T2 WT or T2 1Y, and the same capsid mutants with a GFP expression cassette were utilized as controls. Adult Sprague-Dawley rats received unilateral intrastriatal injections of either vector (2 μL of 1.2 × 1012 vector genomes/μL for all viruses); 1 month later, the striatum was collected for ELISA protein measurements and histology. (A and B) Representative images of striatal GDNF immunoreactivity from animals injected with T2 1Y-GDNF (A) or T2 WT-GDNF (B). (C) ELISA protein measurements of the human GDNF transgene levels in the AAV-injected striatal samples from animals injected with either T2 WT-GDNF (n = 8), T2 WT-GFP (n = 4), T2 1Y-GDNF (n = 7), or T2 1Y-GFP (n = 4). Transduction with T2 1Y-GDNF resulted in significantly higher striatal GDNF levels than that produced by T2 WT-GDNF (#p = 0.01). No GDNF was detected in either GFP control-treated subject. *p = 0.01 versus T2 WT-GDNF and p = 0.001 versus T2 1Y-GDNF. Error bars represent the mean + SD.
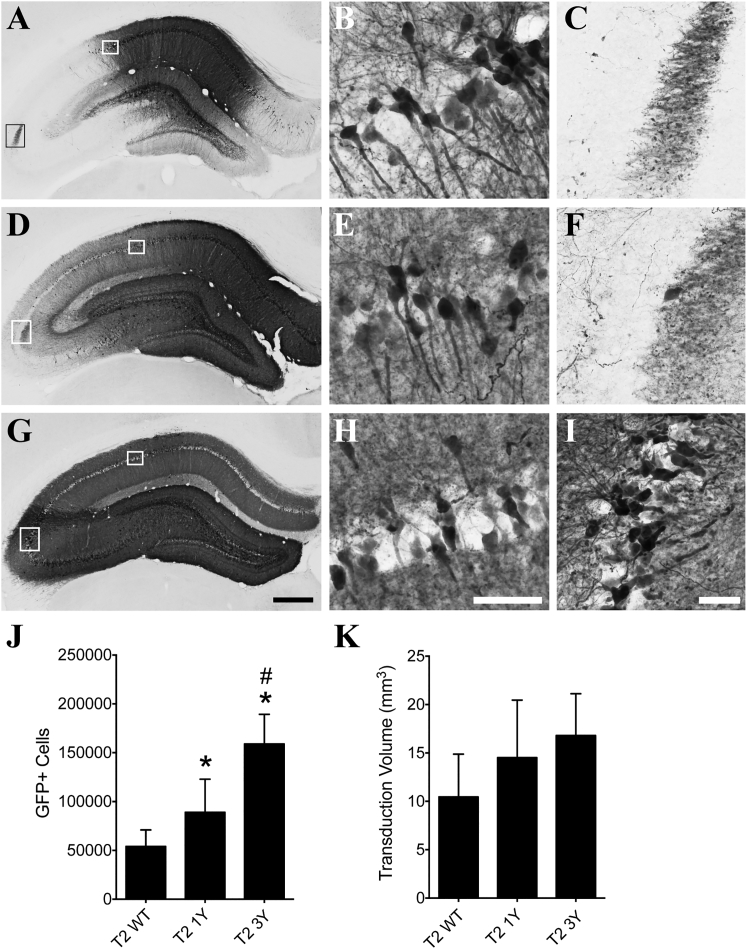
Efficacy of Transduction following Hippocampal Injections of AAV2 Capsids
We injected a subset of the AAV2 capsid mutants carrying a GFP reporter into the hippocampus to determine whether targeting of a different structure within the CNS would confer similar improvements in transduction to that seen in the striatum. We injected 2 μL of the same vector batches used in the striatum experiments into the center of dorsal hippocampus and sacrificed the animals 1 month later. All of the viruses tested effectively transduced hippocampal neurons, but to varying degrees (Figures 4A–4I). The T2 3Y virus clearly transduced cells throughout the entire dorsal hippocampus (Figures 4G–4I), while the T2 1Y virus did not appear to spread as readily (Figures 4D–4F) and the T2 WT virus showed the least spread to regions farther from the injection site (e.g., the CA3 pyramidal cell layer; Figures 4A–4C). Stereological counts of GFP+ cells showed that the T2 3Y virus (158,891 GFP+ cells) transduced significantly more hippocampal neurons when compared with both T2 WT (54,066 GFP+ cells) and T2 1Y viruses (89,095 GFP+ cells), and T2 1Y infected significantly more cells than the T2 WT virus (p < 0.001; Figure 4J). Stereological measurement of transduction volume showed a similar relationship between the viruses (i.e., T2 WT < T2 1Y < T2 3Y) but did not reach statistical significance (p = 0.065; Figure 4K).
Figure 4.
AAV2 Capsid Mutations Significantly Improve Transduction of the Hippocampus
Adult Sprague-Dawley rats received unilateral intrahippocampal injections of either T2 WT, T2 1Y, or T2 3Y virus (2 μL of 1.2 × 1012 vg/μL). One month later, the animals were sacrificed and processed for transgene (GFP) immunoreactivity. (A–C) Representative micrographs of T2 WT transduction in the hippocampus; white boxes in (A) are regions shown in (B) and (C). (D–F) Representative micrographs of T2 1Y mutant AAV transduction throughout the hippocampus; white boxes in (D) are regions shown in (E) and (F). (G–I) Representative micrographs of T2 3Y transduction throughout the entire hippocampus; white boxes in (G) indicate regions shown in (H) and (I). Note that all viruses transduce dentate granule cells (A, D, and G). (J) Unbiased stereological quantitation of GFP+ neurons showed that the T2 1Y virus (n = 9) transduced significantly more cells than the T2 WT virus (n = 8), and the T2 3Y virus (n = 7) transduced significantly more than T2 WT and T2 1Y in the hippocampus (*p < 0.05 versus T2 WT and #p < 0.05 versus T2 1Y, one-way ANOVA, Tukey’s post hoc). (K) Unbiased stereological quantitation of the transduction volume showed a trend toward a difference among the T2 WT (n = 8), T2 1Y (n = 9), and T2 3Y (n = 7) viruses (p = 0.065, one-way ANOVA) in the hippocampus. Scale bars, 500 μm (G, also applies to A and D); 100 μm (H, also applies to B and E); 50 μm (I, also applies to C and F).
Efficacy of Transduction following Intrastriatal Injections Using AAV5 Mutant Capsids
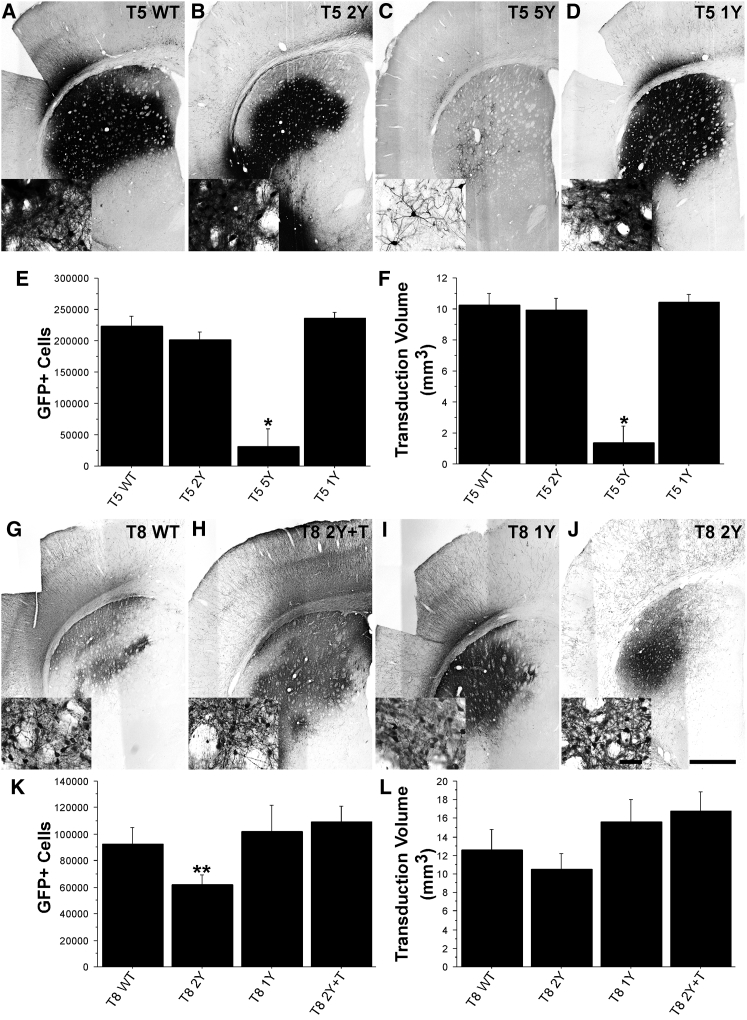
AAV5 is a popular serotype of choice for CNS applications and has been successfully used as a vector to correct lysosomal storage disease when delivered by intrastriatal injection.26 Moreover, AAV5 is considered a viable alternative in clinical trials for use in patients with a pre-existing immunity to rAAV2/2.27 Hence, we evaluated whether mutations to tyrosine residues (Figures 5C and 5D; Figure S2) on the AAV5 capsid would enhance transduction akin to that seen with AAV2. Unexpectedly, Y-F mutations in the AAV5 capsid (T5 1Y: 236,093 GFP+ cells [Figures 5D and 5E]; T5 2Y: 201,127 GFP+ cells [Figures 5B and 5E]) did not increase the number of transduced cells above the T5 WT capsid (223,658 cells; Figures 5A and 5E), and in fact, the mutation of five distinct tyrosine residues resulted in significantly less infectivity of the vector (T5 5Y; 30,561 GFP+ cells; p < 0.0001; Figures 5C and 5E). Similarly, these AAV5 mutations did not improve the transduction volume following a single intrastriatal injection (T5 WT: 10.22 mm3, T5 2Y: 9.95 mm3, T5 1Y: 10.46 mm3; Figure 5F), and the quintuple mutant showed a smaller transduction volume (T5 5Y: 1.35 mm3) compared with WT AAV5 capsid.
Figure 5.
Incorporation of Tyrosine Mutants into AAV5 and AAV8 Capsids Does Not Improve Transduction Properties following Intrastriatal Injections
Adult Sprague-Dawley rats received intrastriatal injections of either AAV5 or AAV8 vectors (2 μL of 1.2 × 1012 vg/μL) as defined in Table 1. One month later, the animals were sacrificed and processed for transgene (GFP) immunoreactivity. (A–D) Representative micrographs of striatal GFP immunoreactivity following injection with AAV5 capsid mutants: (A) T5 WT (n = 9), (B) T5 2Y (n = 8), (C) T5 5Y (n = 8), and (D) T5 1Y (n = 8). (E and F) Stereological cell counts of transduced neurons (E) or transduction volume (F) suggests that incorporation of these mutations does not improve transduction compared with WT AAV5. In contrast, the incorporation of five mutations (T2 5Y) resulted in a dramatic reduction in transduction (*p < 0.0001). (G–J) Representative micrographs of striatal GFP immunoreactivity following transduction with AAV8-based capsids: (G) T8 WT (n = 8), (H) T8 2Y +T (n = 7), (I) T8 1Y (n = 8), and (J) T8 2Y (n = 7). (K and L) As indicated by the stereological quantification (K) and volumetric analysis (L), there were no significant differences between the various mutants except a modest reduction in the number of transduced cells with the T8 2Y virus as compared with T8 2Y +T (**p = 0.02). (E, F, K, and L) Error bars represent the mean + SD.
AAV8 Capsid Mutants Do Not Improve Overall Transduction
AAV8 is considered another prime candidate for CNS applications.28 Accordingly, we evaluated the impact of several mutations in the AAV8 capsid (Figure S2). The incorporation of these capsid changes (T8 2Y: 61,772 GFP+ cells [Figures 5J and 5K]; T8 1Y: 101,923 GFP+ cells [Figures 5G and 5K]; T8 2Y +T: 109,270 GFP+ cells [Figures 5H and 5K]) did not improve overall transduction compared with the T8 WT capsid (92,722 GFP+ cells; Figures 5G and 5K). On the contrary, the incorporation of Y4447F+Y733F (T8 2Y; Figures 5J and 5K) resulted in significantly fewer transduced cells (p = 0.02 versus T8 2Y +T) and smaller transduction volume compared with T8 WT capsids (Figure 5L; 12.59 mm3; T8 2Y: 10.50 mm3; T8 1Y: 15.67 mm3; T8 2Y +T: 16.75 mm3). Moreover, the spread of transduction throughout the striatum correlated poorly with the number of transduced cells, and no pattern emerged for either capsid (data not shown).
Comparison across Serotypes
When comparing all capsids and/or serotypes utilized in this study it becomes clear that in terms of the number of transduced cells, the HSPG null capsids T2 3Y +dH and T2 3Y +T +dH, as well as T2 3Y, performed as well as WT AAV5 and the AAV5 mutants. Remarkably, T2 3Y +dH and T2 3Y +T +dH exhibit a much greater volume of transduction than any other capsid type analyzed herein (p < 0.001 versus all groups).
AAV Capsid Modifications Do Not Alter Tropism of the Viral Vectors
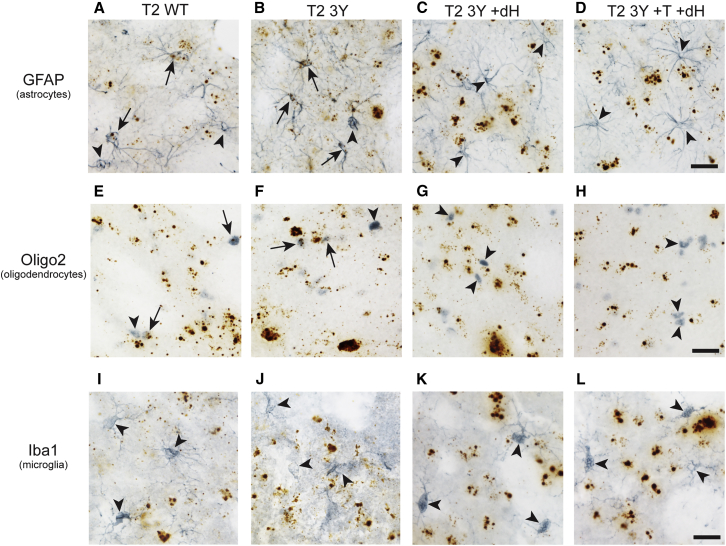
AAV2, AAV5, and AAV8 are known to show neuronal, astrocyte, and oligodendrocyte tropism.29, 30 In contrast, WT AAV capsids typically do not facilitate microglial transduction, but a recent report described limited infection of microglia using a modified AAV6 capsid.31 It was unclear whether the new mutant capsid rAAVs used here alter the tropism for astrocytes, oligodendrocytes, and/or microglia. We focused primarily on AAV2 capsids to determine whether these capsid modifications altered glial tropism using a combination of RNAscope in situ hybridization to detect rAAV genomes and either glial fibrillary acidic protein (GFAP; an astrocyte marker), oligodendrocyte transcription factor 2 (Olig2; an oligodendrocyte marker), or ionized calcium binding adaptor molecule 1 (Iba1, a microglial marker) immunohistochemistry (IHC) to identify glial cells in brain sections. The T2 WT and T2 3Y capsids displayed clear tropism for astrocytes (Figures 6A and 6B) and oligodendrocytes (Figure 6E and 6F), whereas T2 Y3 +dH and T2 Y3 +T +dH did not appear to transduce astrocytes (Figures 6C and 6D) or oligodendrocytes (Figures 6G and 6H). All of the other AAV2 mutant capsids tested displayed tropism for astrocytes (Figure S3) and oligodendrocytes (Figure S4), and qualitatively the extent of astrocyte and oligodendrocyte transduction was associated with the overall level of transduction for each virus. None of the AAV capsids used here showed an apparent tropism for microglia, as indicated by the lack of in situ hybridization puncta within microglia in the striatum of animals injected with T2 WT, T2 3Y, T2 3Y +dH, or T2 3Y +T +dH (Figures 6I and 6L). There was no apparent transduction of microglia by the other AAV2 mutants (Figure S5), and we confirmed that microglia were not transduced by the AAV5 or AAV8 capsids tested here (Figure S6). Collectively, these data suggest the tropism for glial cells is largely unchanged by the mutant AAV capsids tested with the exception of a lack of astrocytic and oligodendrocytic transduction with T2 Y3 +dH and T2 Y3 +T +dH capsids.
Figure 6.
AAV2 Capsid Tyrosine Mutations Do Not Alter the Tropism of the Viral Vector, while Deletion of Heparan Sulfate Binding Eliminates Glial Tropism
Adult Sprague-Dawley rats received intrastriatal injections of AAV2 vectors (2 μL of 1.2 × 1012 vg/μL) as defined in Table 1. One month later, the animals were sacrificed and processed for dual-label viral genome in situ hybridization (brown punctate staining) and glial cell IHC (blue-gray staining). (A–D) Representative sections from animals injected with T2 WT (A) and T2 3Y (B) indicate that both viruses display tropism for astrocytes, as indicated by GFAP+ cells containing brown in situ hybridization puncta (arrows), whereas T2 3Y +dH (C) and T2 3Y +T +dH (D) do not appear to contain viral genomes, indicating a loss of astrocyte tropism (arrowheads). (E–H) Representative sections from animals injected with T2 WT (E) and T2 3Y (F) indicate that both viruses display tropism for oligodendrocytes, as indicated by Oligo2+ cells containing brown in situ hybridization puncta (arrows), whereas T2 3Y +dH (G) and T2 3Y +T +dH (H) do not appear to contain viral genomes, indicating a loss of oligodendrocyte tropism (arrowheads). (I–L) Representative sections from animals injected with T2 WT (I), T2 3Y (J), T2 3Y +dH (K), and T2 3Y +T +dH (L) show that Iba1+ cells do not appear to contain viral genomes, indicating a lack of microglial tropism (arrowheads). Additional in situ hybridization-IHC results with other AAV mutant capsids are in Figures S3–S6. Scale bars, 25 μm (D, H, and L, and apply to all images in each row).
Discussion
Incorporation of AAV2 Capsid Surface Y-F Substitutions Significantly Enhances Striatal and Hippocampal Transduction
Here, we demonstrate that the rational reengineering of the AAV capsid can improve transduction efficacy and volumetric spread following stereotaxic delivery to the CNS. Previous work demonstrated that the mutation of specific surface tyrosine residues on the AAV2 capsid results in significantly augmented transduction efficiency of similar capsid mutants in cultured cells10 and other organ systems in vivo such as the eye.11, 18 This improvement in transduction is presumably mediated through an increased number of particles escaping degradation while traversing the endosomal pathway, ultimately resulting in an increase in genomes that enter the nucleus.
Several studies in different cell and tissue types suggest that the effects of capsid mutations on transduction efficacy may be dependent upon target cell phenotypes. Here, we found that increased transduction was not mediated via a single amino acid substitution per se. Some of the single amino acid changes tested here, such as the Y444F substitution, enhanced transduction compared with the WT capsid, but combination with other substitutions, such as Y730F, eliminated the enhanced transduction. In contrast, adding the Y500F mutation to the Y444F and Y730F mutations further enhanced the efficacy of the viral vector infectivity over Y444F alone. Interestingly, the Y730F mutation alone significantly improves transduction properties in the retina.11 It was unexpected that triple, but not double, Y to F substitutions produced an enhancement over a single mutation. In other words, our results suggest that mutations that increase transduction alone show differential effects when added in certain combinations, which is in stark contrast with that seen from in vitro studies.32, 33 The AAV2 Y444, Y500, and Y730 residues are not located adjacent to each other on the capsid surface (Figure S2A). The in vitro data thus suggest that there is no requirement for a (functional) interaction between the various surface tyrosine residues in order for transduction to occur. Rather, a possible explanation for our discrepant findings may be that this phenomenon is target cell specific. In a previous study, we tested the same capsid tyrosine mutants in neurons of the enteric nervous system, and no differences in transduction efficacy were observed.34 Indeed, the Y730 residue is in close proximity to the HS binding motif in AAV2 (Figure S2A), and it is possible that these mutations have an effect on the binding of specific co-receptors that may be differentially expressed or regulated in different cells and/or tissues.35 Moreover, it is possible that mutations in certain tyrosine residues can affect the affinity of the capsid for HS.18 Together, the divergent effects of various capsid mutants in different cell types and tissue types suggest that the effects of these mutations on transduction efficacy are dependent on tissue or target cell phenotypes.
Incorporation of an Additional T-V Substitution in AAV2 Did Not Further Enhance Transduction
The AAV2 T491V mutation is associated with improved intracellular trafficking and nuclear translocation of the viral capsid,15 although it is unclear whether this threonine is directly involved in nuclear import of the viral particle. However, transduction remained the same, or was impaired, following intrastriatal delivery of viral mutants containing the T491V mutation. Our findings are discordant with observations made in cell culture.15 In addition to the inherent differences between cell culture and in vivo model systems, there are several potential explanations for these discrepancies. For instance, as further discussed below, the T-V mutants exhibited improved diffusion properties as compared with non-T491V mutants (Figure 2). Thus, it is possible that the dilution of viral particles due to diffusion away from the injection site reduced the overall MOI in the region of transduction, effectively reducing the ability to detect transduced cells.
Diffusion Properties in the Parenchymal Space Are Improved with Altered Receptor Binding Properties
It is evident that certain capsid mutations drastically alter the diffusion of viral particles through the extracellular milieu. In particular, substitutions of the amino acids that abolish HS binding (Figures 2A and 2B)18 lead to a dramatic increase in transduction spread with AAV2. This increased spread is not surprising because the extracellular matrix contains high levels of HSPG;36 thus, it is likely that the removal of HS binding capacity prevented extracellular sequestration of viral particles. In addition, removal of the HS binding motif also may reduce the overall neuronal affinity for this capsid, providing greater ability to diffuse from the injection sphere. Interestingly, we found that AAV capsids without HS binding ability did not effectively transduce glial cells, suggesting an alteration in the tropism, and the subsequent reduction in glial sequestration may contribute to enhanced spread as well. In contrast, the Y-F mutations only have a modest effect on HS binding.18 Accordingly, our data show that these mutants exhibit a more limited spread that was only slightly improved over the WT capsid. Finally, vectors with the added T-V mutations also exhibit overall improvements in diffusivity. As is the case with Y-F substituted amino acids, T491 does not directly interact with the HS binding residues,18 but instead resides in close proximity to the HS receptor binding site (R484, R487, R532, R585, R588) (Figures 2A and 2B). Thus, it is possible that mutation of this amino acid alters the affinity for HS and reduces extracellular capture of viral particles. Indeed, we did not see an improvement in diffusion when the T-V mutation was combined with the mutation of HS binding residues, lending further support to the idea that the T-V mutation increases spread through the same mechanism (i.e., disruption of HSPG binding).
Early studies in CHO cells found that HSPG is a crucial receptor for AAV2, and several studies support a prominent role for this mechanism of entry into multiple cell types.17 However, we found here that capsids with impaired or abolished HS binding also effectively transduce neurons in the CNS, which mirrors recent findings in the retina18 and other tissue types.19, 20, 37, 38 Together, these studies suggest that a still unknown receptor may mediate strong neuronal transduction of AAV2. For instance, the recently identified type I transmembrane protein KIAA0319L appears to be crucial for transduction for a number of AAV serotypes in a variety of cell types.39 Nevertheless, further studies are needed to identify these non-HSPG-mediated pathways of AAV2 entry into neurons. Interestingly, infection of astrocytes and oligodendrocytes was virtually abolished with the removal of the HS binding site, supporting the idea that HSPG binding is required for the infection of certain types of cells.
Comparison Identifies Serotype-Specific Transduction Property Differences
WT AAV2 is associated with being a poor gene therapy vector for the CNS compared with other AAV serotypes.40 Our findings with the HS binding-site-modified vectors suggest that this, at least in part, is due to an overabundance of available HSPG in the brain parenchyma, acting to sequester virions. In agreement with previous studies, our data show that WT AAV5 is more efficient in transducing striatal neurons when compared with WT AAV2, but when the AAV2 capsid is altered by site-specific mutations it produces similar, or significantly improved, transduction. Interestingly, neither WT nor mutant AAV8 capsid variants exhibited improved transduction in neurons of the striatum when compared with AAV2 or AAV5, which contrasts with previous results in rodent studies.24, 41, 42, 43 One reason for these discrepancies may be the titers at which the viruses were used. In this study, we utilized a relatively low dose of virus (total of 2.4 × 109 genome copies injected) to avoid a ceiling effect, whereas the other studies typically used higher titers. Importantly, work by Baekelandt and colleagues42 showed that AAV8 is disproportionately inefficient at lower titers as compared with AAV5 and AAV7, which may account for the discrepant findings in our study. Alternatively, the poor relative infectivity observed with AAV8 may also be explained by the finding that AAV8 exhibited a rather large overall volume of transduction as compared with AAV5; this increase in diffusion may have diluted the effective concentration of AAV8 throughout the transduction area to that below the limits of detection. These studies highlight the importance of viral serotypes and titers in evaluating transduction in experimental studies, and ultimately, this information will guide the most efficacious utilization of AAVs in clinical applications.
Y-F and T-V Mutations Do Not Alter Tropism, But Ablation of HSPG Binding Restricts Tropism to Neuronal Cells
Following intraparenchymal delivery, AAV primarily infects neurons; however, a small number of astrocytes and oligodendrocytes are also transduced.24, 29, 30, 44 In contrast, microglia have proven to be refractory to AAV transduction. However, the incorporation of different Y-F and T-V mutations in the AAV6 capsid results in modest microglia transduction,31 suggesting that capsid modifications such as those utilized here can alter the tropism of the capsid. To that end, we evaluated whether any of our mutant capsids were capable of infecting microglia, and whether there were qualitative changes in the level of astrocyte or oligodendrocyte transduction with the AAV2 capsid mutants.
The ability to detect transduction in various cell types depends on detecting transgene production. However, the activity of ectopic promoters differs between different cell populations in the brain,44 and prior studies have utilized cell-specific promoters to detect transduction of non-neuronal cells. We took a different approach to analyze the tropism of our vectors by using a combination of in situ hybridization against a non-transcribed portion of the viral genome45 combined with IHC for markers specific to each type of glial cell (GFAP for astrocytes, Oligo2 for oligodendrocytes, and Iba1 for microglia). This approach allows us to assess transduction of the various cell types independent of viral transgene expression. We were unable to find evidence that viral genomes co-localize with Iba1. Thus, the AAV capsid mutants tested here are not capable of infecting microglia. It remains to be seen whether strong microglial transduction is at all achievable using AAV, or whether there are significant basic biological hurdles preventing this from occurring. Moreover, there were no obvious changes in astrocyte or oligodendrocyte infection for a majority of the AAV2 mutants. Interestingly, we did not observe viral genomes in either oligodendrocytes or astrocytes in tissue injected with the HS binding mutants. This suggests that whereas HSPG binding is not required for neuronal transduction, it is required for infection of neuronal support cells.
Translational and Clinical Implications of Observed Improved Transduction
Gene therapy of the CNS is increasingly of interest to the medical community. Currently, numerous trials have utilized rAAV in the treatment of disorders such as Parkinson’s disease, Alzheimer’s disease, and Canavan disease, among others.46, 47, 48 Nevertheless, transduction efficacy has not always translated well from preclinical studies to clinical trials, and efficacy and spread of the viral vector has often been disappointing.4 Regardless of the biological origin of this deficiency, it must be overcome to increase the potential of rAAV therapies. Indeed, a more comprehensive coverage of the CNS will be required in diseases where large regions of the brain are affected, such as lysosomal storage disorders. Numerous considerations, in addition to interspecies scaling, have to be made when translating rodent studies into the NHP and, ultimately, humans. For instance, we have established that age is an important covariate in transduction efficiency, where certain serotypes exhibit deficiencies in distinct anatomical locations.5, 49 Thus, age-matching experimental subjects in preclinical testing is increasingly important. Moreover, anatomical differences between species must be considered. For instance, in this study, we targeted the rodent striatum, which is one continuous structure enclosed within the corpus callosum and the lateral ventricles. In NHPs and humans, however, the analogous structure (the caudate and putamen) is physically separated by the internal capsule. Thus, future preclinical studies using gene therapy approaches should include multiple injections sites in the target region to better replicate the approaches required to translate these therapies to humans. Finally, the fundamental biology and behavior of AAV in the brain does not change to a significant extent when comparing rodents with NHPs; rather, the changes observed in such direct comparisons are related to differential promoter activities between species;44 thus, when doing translational work, it is important to optimize expression for human applications and to consider regulatory components within the viral genome.
Our observations suggest that the rational engineering of vector capsids is a relevant component of future therapies and a viable mechanism to improve rAAV infectivity and spread. However, one question one may ask is, why not simply use a more effective naturally occurring serotype? Translation of AAV-based therapies requires a complex decision making process to achieve the therapeutic goals that this work and others will help facilitate. We did not detect an appreciable difference in the number of cells transduced by the strongest AAV2 mutants and WT AAV5, but there were substantial differences in other transduction parameters (i.e., spread of the virus). Thus, if the clinical goal is to transduce a discrete area at high efficiency, then either AAV5 or Y-F AAV2 mutants are reasonable choices; however, mutant AAV2 capsids lacking HS binding are a superior choice if large areas must be transduced at high efficiency. Furthermore, the current findings and previous studies24, 40 clearly indicate differences in cell tropism with different serotypes and mutant capsids. Removal of HS binding prohibited astrocyte and oligodendrocyte tropism; thus, applications that may need to avoid transduction of these cell populations would benefit from the HS binding mutants. Additional considerations that may warrant the use of one serotype over another is whether a patient has pre-existing immunity to a specific serotype. Moreover, various serotypes are differentially impaired in the aged brain.5, 49 Finally, the increased efficacy of these vectors would also allow the use of lower titers, thereby lowering the risk of an immune response. Currently, the altered properties of these Y-F mutant capsids are largely believed to be based on reduced degradation during intracellular trafficking, making the kinetics of capsid breakdown different from WT AAVs. Consequently, additional work is needed to determine the immunogenicity of these vectors, particularly using a re-administration paradigm, because this immune response is specifically guided against the WT capsid and capsid proteins.50 Nevertheless, the penultimate test of the translational value of these engineered vectors will be in preclinical models of neurological disease such as Parkinson’s disease and Huntington’s disease, among others.
Conclusions
In conclusion, we have shown that the incorporation of rationally chosen mutations in the AAV2 capsid confers improved infectivity and spread following stereotaxic delivery into the CNS. The combination of capsid serotype and various versions of mutant capsid proteins appears to provide a remarkable level of versatility in altering the transduction profile of the viruses, which may help to tailor applications of AAVs in different ways. Importantly, the use of such capsid mutants results in an increased production of a therapeutic transgene, thus lending itself to further clinical development for disorders where the transduction of large structures is required.
Materials and Methods
Capsid Clones
AAV2
The packaging plasmid pACG2 for AAV2 (T2 WT) was mutated using site-directed mutagenesis to generate the capsid mutants as previously described.6, 18, 21 Capsid tyrosine mutants (Table 1; Figures 2A and 2B) were generated by mutating Y444F (T2 1Y), Y444F+Y500F+Y730F (T2 3Y), Y500F+Y730F (T2 2Y#1), Y444F+Y730F (T2 2Y#2), Y252F+Y272F+Y444F+Y500F+Y704F+Y730F (T2 6Y), T2 3Y and T491V (T2 3Y +T), and T2 3Y +T + Y272F (T2 4Y +T), and the heparin sulfate binding mutants were generated by mutating R585S, R588T, and R487G on the T2 3Y capsid (T2 3Y +dH) or the T2 3Y +T capsid (T2 3Y +T +dH).
AAV5
To generate AAV5 capsid mutants (Table 1; Figures 2C and 2D), AAV5 capsid containing plasmid pACG2r5c (T5 WT) was mutated at Y719F (T5 1Y), Y263F+Y719F (T5 2Y), and Y242F+Y263+Y689F+Y693F+Y719F (T5 5Y) as previously described.21
AAV8
Similarly, the plasmid pE518-VD2/8 encoding the AAV8 cap gene (T8 WT) was mutated as follows: Y733F (T8 1Y), Y447F+Y733F (T8 2Y), Y447F+Y733F+T494V (T8 2Y +T) as previously described21 (Table 1; Figures S2E and S2F). All mutations were verified using standard sequencing of the DNA plasmids containing the cap gene.
Viral Genomes and Vector Packaging
All vectors contained an identical viral genome encoding humanized GFP under the control of the hybrid chicken β-actin/cytomegalovirus promoter (CBA). In addition, to test the contribution of a self-complementary genome to the efficacy of transduction, we packaged a self-complementary genome encoding a truncated CBA promoter18 together with GFP into the T2 3Y +T capsid (referred to as T2 3Y +T -SC). The GDNF genome contained the same CBA promoter followed by the human GDNF coding sequence with an N-terminal signal peptide.25 All viral genomes were flanked by AAV2 inverted terminal repeats. Viral particles were generated as previously described.51, 52 In brief, the viral genome was co-transfected with a helper plasmid encoding either cap mutant described above together with plasmids encoding rep and helper functions. Viral particles were purified using an iodixanol gradient followed by column chromatography. Viral titers were determined using a qPCR assay, and all vector preparations utilized in this study were normalized to 1.2 × 1012 vg/mL using a balanced salt solution (Alcon Laboratories).
Animals and Surgery
Experiments were conducted on young adult (220 g) male Sprague-Dawley rats (Harlan) in accordance with guidelines of the Michigan State University Institutional Animal Care and Use Committee (AUF 10/12-196-00). Rats were housed two per cage, maintained in a light-controlled (12 hr light/dark cycle, lights on at 6:00 a.m.) and temperature-controlled (22 ± 1°C) room, and provided with food and water ad libitum.
In the striatal injection groups, animals received bilateral injections of the various vectors.23 The animals in the hippocampal injection groups received unilateral injections because the decussating fibers of the hippocampus cause bilateral transduction from a unilateral injection.40 Similarly, animals in the GDNF experiment were injected unilaterally in the striatum in order to retain the naive hemisphere as an internal control. All surgery was performed under 2% isoflurane anesthesia. Rats were placed in a stereotaxic frame and received a 2 μL injection of either vector. Injection coordinates for the striatum were AP ± 0.0 mm, ML ± 3.0 mm, DV (from dura) −4.0 mm; injection coordinates for the hippocampus were AP −3.3 mm, ML +2.0 mm, DV (from dura) −2.6 mm. A Hamilton syringe fitted with a glass capillary needle (Hamilton Gas Tight syringe 80,000, 26 s/2” needle [Hamilton], coated in SigmaCote53) was used for the injection. The needle was lowered to the site and vector injection began immediately at a rate of 0.5 μL/min. For striatal injections, the needle remained in place for 1 min following the injection and then retracted 1 mm, where it remained in place for an additional 4 min before being fully retracted. For hippocampal injections, the needle remained in place after the injection for the full 5 min before retraction.
Euthanasia and Tissue Preparation
Euthanasia was performed as approved by the Michigan State University (MSU) Institutional Animal Care and Use Committee (IACUC), and all euthanasia procedures were consistent with the recommendations of the American Veterinary Medical Association. All rats were sacrificed 1 month after the vector delivery. Rats were deeply anesthetized (60 mg/kg pentobarbital, intraperitoneally [i.p.]) and perfused intracardially with heparinized (10 U/mL) 0.9% saline, followed by ice-cold 4% paraformaldehyde (PFA). Rat brains were immediately removed and placed in 4% paraformaldehyde for an additional 24 hr (4°C) and thereafter transferred to 30% sucrose (in 0.1 M PO4 buffer) until saturated. Brains in the GDNF experiments were removed immediately following the saline perfusion and hemisected coronally at the level of the cerebral peduncles. The anterior striatum (used for GDNF ELISA) was dissected from the portion anterior to the cut,54 weighed, and immediately frozen at −80°C. The region posterior to the cut, containing the posterior striatum, was post-fixed in 4% PFA for 72 hr and thereafter treated with sucrose as described above. Brains were thereafter cut coronally into a series of 40-μm-thick (for histology) and 20-μm-thick (for RNAscope in situ hybridization) sections using a freezing stage sliding microtome.
Immunohistochemistry
Immunohistochemistry (IHC) was performed on free-floating sections as previously described.54 Primary antibodies used were rabbit anti-GFP (Abcam ab290, 1:20,000). Following the primary incubation, sections destined for stereology (GFP) were incubated in biotinylated secondary antibodies against rabbit IgG (Millipore AP132b 1:500; Merck Millipore) for 2 hr at room temperature, treated with Vector ABC detection kit using horseradish peroxidase (Vector Laboratories). GFP expression was visualized by treatment of 0.5 mg/mL 3,3′ diaminobenzidine and 0.03% hydrogen peroxide in Tris buffer. Sections were mounted on subbed slides, dehydrated with increasing concentrations of ethanol followed by xylene, and coverslipped with Cytoseal (Thermo Fisher Scientific).
Images were taken on a Nikon Eclipse 90i microscope with a QICAM camera (QImaging). Images were taken from subjects that displayed mean transduction as measured by stereological cell counts and created in Photoshop 7.0, with brightness, saturation, and sharpness adjusted only as needed to best represent the immunostaining as viewed directly in the microscope.
Stereology
Striatal and hippocampal GFP+ cells were quantified using unbiased stereology using StereoInvestigator software (Version 4.03; MBF Bioscience). Brain sections were visualized using 4× magnification (Olympus BX53 microscope equipped with a motorized stage [Olympus] and a QImaging 2000R camera [QImaging]), and the transduction area was outlined. GFP+ cells from every sixth section, with six to nine sections per animal, were counted using the optical fractionator method with a 60× oil objective. The coefficient of error for each estimate was calculated according to Gundersen and Jensen,55 and was less than 0.1 (Gundersen, m = 1). Transduction volume was calculated using the Cavalieri estimator in StereoInvestigator using a grid spacing of 100 μm. The outlined transduction areas of GFP+ cells from the optical fractionator probe were filled using the Paint Cavalieri mode.
GDNF ELISA
Dissected striata from AAV-GDNF-treated (or GFP control) animals were analyzed using a GDNF Emax kit (Promega). Samples were homogenized in 100 μL of buffer according to the kit protocol using a 300 V/T Ultrasonic homogenizer (BioLogics). Striatal samples were assayed at 1:50 or 1:100 dilutions and quantified with a Multiskan spectrophotometer (Thermo Fisher Scientific). Raw plate values were adjusted for tissue weight, and striatal GDNF protein content was calculated using a standard curve.
Dual-Label RNAscope In Situ Hybridization and Immunohistochemistry
To evaluate whether the viruses infected microglial cells, we double-labeled tissue sections with in situ hybridization using a custom-ordered probe against a non-transcribed portion of the viral genome (probe 408321; Advanced Cell Diagnostics)45 and IHC with either GFAP antibody (Z0334; Dako), Oligo2 antibody (ab109186; Abcam), or Iba1 antibody (019-19741; Wako) to detect astrocytes, oligodendrocytes, or microglia, respectively. In brief, sections (20 μm thick) from animals injected with WT and mutant AAVs in the striatum were first labeled with in situ hybridization following a previously established protocol,45 which produces brown punctate signal. After developing the in situ hybridization labeling, the sections were stained for IHC using the GFAP (1:20,000 dilution), Oligo2 (1:50 dilution), or Iba1 (1:1,000 dilution) using the same protocol as described previously,45 which produces a blue-gray reaction product using the VectorSG peroxidase substrate according to the manufacturer’s instructions (SK-4700; Vector Labs). Images were captured and processed for publication as described above.
Visualizing AAV Capsid Surface Residues
The coordinates for the VP3 monomers of AAV2 (accession number Research Collaboratory for Structural Bioinformatics [RCSB] PDB: 1LP3), AAV5 (accession number RCSB PDB: 3NTT), and AAV8 (accession number RCSB PDB: 2QA0) were used to generate a capsid (60-mer) by icosahedral symmetry matrix manipulation using the Oligomer generator subroutine in the VIPERdb online server (http://viperdb.scripps.edu/oligomer_multi.php).56 The capsid coordinates were used to generate surface density images on which the positions of the tyrosine (red), threonine (yellow), and basic (blue) residues mutated in this study (Table 1) were displayed. The surface image was generated using the PyMOL program (The PyMOL Molecular Graphics System version 1.3; Schrödinger). A two-dimensional “roadmap” image, showing the positions of the mutated residues on the capsid surfaces of AAV2, AAV5, and AAV8, was generated using the RIVEM program57 (Figure S2).
Statistical Analysis
Experimenters were blind to all vector group assignments during data collection and analysis. All statistical tests and graphs were done using Statview (version 5.0). All comparisons between different vector constructs were performed using one-way ANOVA tests, and p ≤ 0.05 was considered statistically significant. When overall significance was achieved, Tukey’s post hoc test was used for pairwise comparisons between groups. A simple linear regression analysis was performed to detect the relationship between striatal transduction volume and the number of GFP+ cells. All AAVs used here showed clear transduction of neurons; thus, any subject that displayed no obvious transduction was excluded from all analyses.
Author Contributions
F.P.M. conceived the project, designed the experiments, and wrote the manuscript. N.M.K. and C.E.S. assisted with experimental design, execution of the study, and data analysis. R.C.S. and B.C. assisted with stereology. W.W.H., S.L.B., and S.E.B. assisted with mutant design and capsid mutagenesis. A.B. and M.A.-M. assisted with AAV capsid structure analysis. All authors critically read the manuscript.
Conflicts of Interest
W.W.H. and the University of Florida have a financial interest in the use of AAV therapies and own equity in a company (AGTC) that might, in the future, commercialize some aspects of this work.
Acknowledgments
We would like to acknowledge Nathan Kuhn and Tessa Grabinski (Michigan State University) for their assistance with histological processing. This work was supported by NIH grants R56AG052328 (to F.P.M. and C.E.S.), P30EY021721 (to W.W.H.), and RO1EY024280 (S.E.B. and M.A.M.), Mercy Health Saint Mary’s (to N.M.K. and F.P.M.), the Foundation Fighting Blindness (to S.E.B.), and an unrestricted grant from Research to Prevent Blindness to the University of Florida Department of Ophthalmology (to S.E.B. and W.W.H.).
Footnotes
Supplemental Information includes six figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.06.011.
Supplemental Information
References
- 1.Kantor B., Bailey R.M., Wimberly K., Kalburgi S.N., Gray S.J. Methods for gene transfer to the central nervous system. Adv. Genet. 2014;87:125–197. doi: 10.1016/B978-0-12-800149-3.00003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manfredsson F.P., Mandel R.J. Development of gene therapy for neurological disorders. Discov. Med. 2010;9:204–211. [PubMed] [Google Scholar]
- 3.Marks W.J., Jr., Baumann T.L., Bartus R.T. Long-term safety of patients with Parkinson’s disease receiving rAAV2-neurturin (CERE-120) gene transfer. Hum. Gene Ther. 2016;27:522–527. doi: 10.1089/hum.2015.134. [DOI] [PubMed] [Google Scholar]
- 4.Bartus R.T., Herzog C.D., Chu Y., Wilson A., Brown L., Siffert J., Johnson E.M., Jr., Olanow C.W., Mufson E.J., Kordower J.H. Bioactivity of AAV2-neurturin gene therapy (CERE-120): differences between Parkinson’s disease and nonhuman primate brains. Mov. Disord. 2011;26:27–36. doi: 10.1002/mds.23442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polinski N.K., Gombash S.E., Manfredsson F.P., Lipton J.W., Kemp C.J., Cole-Strauss A., Kanaan N.M., Steece-Collier K., Kuhn N.C., Wohlgenant S.L., Sortwell C.E. Recombinant adenoassociated virus 2/5-mediated gene transfer is reduced in the aged rat midbrain. Neurobiol. Aging. 2015;36:1110–1120. doi: 10.1016/j.neurobiolaging.2014.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong L., Li B., Mah C.S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R.W., Zolotukhin I., Warrington K.H., Jr., Weigel-Van Aken K.A. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maheshri N., Koerber J.T., Kaspar B.K., Schaffer D.V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 8.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowles D.E., McPhee S.W., Li C., Gray S.J., Samulski J.J., Camp A.S., Li J., Wang B., Monahan P.E., Rabinowitz J.E. Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol. Ther. 2012;20:443–455. doi: 10.1038/mt.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong L., Li B., Jayandharan G., Mah C.S., Govindasamy L., Agbandje-McKenna M., Herzog R.W., Weigel-Van Aken K.A., Hobbs J.A., Zolotukhin S. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology. 2008;381:194–202. doi: 10.1016/j.virol.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrs-Silva H., Dinculescu A., Li Q., Min S.H., Chiodo V., Pang J.J., Zhong L., Zolotukhin S., Srivastava A., Lewin A.S., Hauswirth W.W. High-efficiency transduction of the mouse retina by tyrosine-mutant AAV serotype vectors. Mol. Ther. 2009;17:463–471. doi: 10.1038/mt.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicolson S.C., Samulski R.J. Recombinant adeno-associated virus utilizes host cell nuclear import machinery to enter the nucleus. J. Virol. 2014;88:4132–4144. doi: 10.1128/JVI.02660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao P.J., Li C., Neumann A., Samulski R.J. Quantitative 3D tracing of gene-delivery viral vectors in human cells and animal tissues. Mol. Ther. 2012;20:317–328. doi: 10.1038/mt.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao P.J., Samulski R.J. Cytoplasmic trafficking, endosomal escape, and perinuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J. Virol. 2012;86:10462–10473. doi: 10.1128/JVI.00935-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aslanidi G.V., Rivers A.E., Ortiz L., Song L., Ling C., Govindasamy L., Van Vliet K., Tan M., Agbandje-McKenna M., Srivastava A. Optimization of the capsid of recombinant adeno-associated virus 2 (AAV2) vectors: the final threshold? PLoS ONE. 2013;8:e59142. doi: 10.1371/journal.pone.0059142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandya J., Ortiz L., Ling C., Rivers A.E., Aslanidi G. Rationally designed capsid and transgene cassette of AAV6 vectors for dendritic cell-based cancer immunotherapy. Immunol. Cell Biol. 2014;92:116–123. doi: 10.1038/icb.2013.74. [DOI] [PubMed] [Google Scholar]
- 17.Summerford C., Samulski R.J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boye S.L., Bennett A., Scalabrino M.L., McCullough K.T., Van Vliet K., Choudhury S., Ruan Q., Peterson J., Agbandje-McKenna M., Boye S.E. Impact of heparan sulfate binding on transduction of retina by recombinant adeno-associated virus vectors. J. Virol. 2016;90:4215–4231. doi: 10.1128/JVI.00200-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perabo L., Goldnau D., White K., Endell J., Boucas J., Humme S., Work L.M., Janicki H., Hallek M., Baker A.H., Büning H. Heparan sulfate proteoglycan binding properties of adeno-associated virus retargeting mutants and consequences for their in vivo tropism. J. Virol. 2006;80:7265–7269. doi: 10.1128/JVI.00076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi X., Fang G., Shi W., Bartlett J.S. Insertional mutagenesis at positions 520 and 584 of adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors with eliminated heparin- binding ability and introduced novel tropism. Hum. Gene Ther. 2006;17:353–361. doi: 10.1089/hum.2006.17.353. [DOI] [PubMed] [Google Scholar]
- 21.Kay C.N., Ryals R.C., Aslanidi G.V., Min S.H., Ruan Q., Sun J., Dyka F.M., Kasuga D., Ayala A.E., Van Vliet K. Targeting photoreceptors via intravitreal delivery using novel, capsid-mutated AAV vectors. PLoS ONE. 2013;8:e62097. doi: 10.1371/journal.pone.0062097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhury S.R., Hudry E., Maguire C.A., Sena-Esteves M., Breakefield X.O., Grandi P. Viral vectors for therapy of neurologic diseases. Neuropharmacology. 2017;120:63–80. doi: 10.1016/j.neuropharm.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reimsnider S., Manfredsson F.P., Muzyczka N., Mandel R.J. Time course of transgene expression after intrastriatal pseudotyped rAAV2/1, rAAV2/2, rAAV2/5, and rAAV2/8 transduction in the rat. Mol. Ther. 2007;15:1504–1511. doi: 10.1038/sj.mt.6300227. [DOI] [PubMed] [Google Scholar]
- 24.Aschauer D.F., Kreuz S., Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS ONE. 2013;8:e76310. doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manfredsson F.P., Tumer N., Erdos B., Landa T., Broxson. C.S., Sullivan L.F., Rising A.C., Foust K.D., Zhang Y., Muzyczka N. Nigrostriatal rAAV-mediated GDNF overexpression induces robust weight loss in a rat model of age-related obesity. Mol. Ther. 2009;17:980–991. doi: 10.1038/mt.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G., Chen Y.H., He X., Martins I., Heth J.A., Chiorini J.A., Davidson B.L. Adeno-associated virus type 5 reduces learning deficits and restores glutamate receptor subunit levels in MPS VII mice CNS. Mol. Ther. 2007;15:242–247. doi: 10.1038/sj.mt.6300016. [DOI] [PubMed] [Google Scholar]
- 27.Hildinger M., Auricchio A., Gao G., Wang L., Chirmule N., Wilson J.M. Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J. Virol. 2001;75:6199–6203. doi: 10.1128/JVI.75.13.6199-6203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asokan A., Schaffer D.V., Samulski R.J. The AAV vector toolkit: poised at the clinical crossroads. Mol. Ther. 2012;20:699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H., McCarty D.M., Bruce A.T., Suzuki K. Oligodendrocyte-specific gene expression in mouse brain: use of a myelin-forming cell type-specific promoter in an adeno-associated virus. J. Neurosci. Res. 1999;55:504–513. doi: 10.1002/(SICI)1097-4547(19990215)55:4<504::AID-JNR10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 30.Feng X., Eide F.F., Jiang H., Reder A.T. Adeno-associated viral vector-mediated ApoE expression in Alzheimer’s disease mice: low CNS immune response, long-term expression, and astrocyte specificity. Front. Biosci. 2004;9:1540–1546. doi: 10.2741/1323. [DOI] [PubMed] [Google Scholar]
- 31.Rosario A.M., Cruz P.E., Ceballos-Diaz C., Strickland M.R., Siemienski Z., Pardo M., Schob K.L., Li A., Aslanidi G.V., Srivastava A. Microglia-specific targeting by novel capsid-modified AAV6 vectors. Mol. Ther. Methods Clin. Dev. 2016;3:16026. doi: 10.1038/mtm.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryals R.C., Boye S.L., Dinculescu A., Hauswirth W.W., Boye S.E. Quantifying transduction efficiencies of unmodified and tyrosine capsid mutant AAV vectors in vitro using two ocular cell lines. Mol. Vis. 2011;17:1090–1102. [PMC free article] [PubMed] [Google Scholar]
- 33.Li M., Jayandharan G.R., Li B., Ling C., Ma W., Srivastava A., Zhong L. High-efficiency transduction of fibroblasts and mesenchymal stem cells by tyrosine-mutant AAV2 vectors for their potential use in cellular therapy. Hum. Gene Ther. 2010;21:1527–1543. doi: 10.1089/hum.2010.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benskey M.J., Kuhn N.C., Galligan J.J., Garcia J., Boye S.E., Hauswirth W.W., Mueller C., Boye S.L., Manfredsson F.P. Targeted gene delivery to the enteric nervous system using AAV: a comparison across serotypes and capsid mutants. Mol. Ther. 2015;23:488–500. doi: 10.1038/mt.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie Q., Bu W., Bhatia S., Hare J., Somasundaram T., Azzi A., Chapman M.S. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brightman M.W. The brain’s interstitial clefts and their glial walls. J. Neurocytol. 2002;31:595–603. doi: 10.1023/a:1025783326667. [DOI] [PubMed] [Google Scholar]
- 37.Shi W., Bartlett J.S. RGD inclusion in VP3 provides adeno-associated virus type 2 (AAV2)-based vectors with a heparan sulfate-independent cell entry mechanism. Mol. Ther. 2003;7:515–525. doi: 10.1016/s1525-0016(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 38.Xing G., Zhang L., Zhang L., Heynen T., Li X.L., Smith M.A., Weiss S.R., Feldman A.N., Detera-Wadleigh S., Chuang D.M., Post R.M. Rat nurr1 is prominently expressed in perirhinal cortex, and differentially induced in the hippocampal dentate gyrus by electroconvulsive vs. kindled seizures. Brain Res. Mol. Brain Res. 1997;47:251–261. doi: 10.1016/s0169-328x(97)00056-9. [DOI] [PubMed] [Google Scholar]
- 39.Pillay S., Meyer N.L., Puschnik A.S., Davulcu O., Diep J., Ishikawa Y., Jae L.T., Wosen J.E., Nagamine C.M., Chapman M.S., Carette J.E. An essential receptor for adeno-associated virus infection. Nature. 2016;530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burger C., Gorbatyuk O.S., Velardo M.J., Peden C.S., Williams P., Zolotukhin S., Reier P.J., Mandel R.J., Muzyczka N. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol. Ther. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 41.McFarland N.R., Lee J.S., Hyman B.T., McLean P.J. Comparison of transduction efficiency of recombinant AAV serotypes 1, 2, 5, and 8 in the rat nigrostriatal system. J. Neurochem. 2009;109:838–845. doi: 10.1111/j.1471-4159.2009.06010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taymans J.M., Vandenberghe L.H., Haute C.V., Thiry I., Deroose C.M., Mortelmans L., Wilson J.M., Debyser Z., Baekelandt V. Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Hum. Gene Ther. 2007;18:195–206. doi: 10.1089/hum.2006.178. [DOI] [PubMed] [Google Scholar]
- 43.Klein R.L., Dayton R.D., Leidenheimer N.J., Jansen K., Golde T.E., Zweig R.M. Efficient neuronal gene transfer with AAV8 leads to neurotoxic levels of tau or green fluorescent proteins. Mol. Ther. 2006;13:517–527. doi: 10.1016/j.ymthe.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watakabe A., Ohtsuka M., Kinoshita M., Takaji M., Isa K., Mizukami H., Ozawa K., Isa T., Yamamori T. Comparative analyses of adeno-associated viral vector serotypes 1, 2, 5, 8 and 9 in marmoset, mouse and macaque cerebral cortex. Neurosci. Res. 2015;93:144–157. doi: 10.1016/j.neures.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Grabinski T.M., Kneynsberg A., Manfredsson F.P., Kanaan N.M. A method for combining RNAscope in situ hybridization with immunohistochemistry in thick free-floating brain sections and primary neuronal cultures. PLoS ONE. 2015;10:e0120120. doi: 10.1371/journal.pone.0120120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leone P., Shera D., McPhee S.W., Francis J.S., Kolodny E.H., Bilaniuk L.T., Wang D.J., Assadi M., Goldfarb O., Goldman H.W. Long-term follow-up after gene therapy for canavan disease. Sci. Transl. Med. 2012;4:165ra163. doi: 10.1126/scitranslmed.3003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rafii M.S., Baumann T.L., Bakay R.A., Ostrove J.M., Siffert J., Fleisher A.S., Herzog C.D., Barba D., Pay M., Salmon D.P. A phase 1 study of stereotactic gene delivery of AAV2-NGF for Alzheimer’s disease. Alzheimers Dement. 2014;10:571–581. doi: 10.1016/j.jalz.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Marks W.J., Jr., Bartus R.T., Siffert J., Davis C.S., Lozano A., Boulis N., Vitek J., Stacy M., Turner D., Verhagen L. Gene delivery of AAV2-neurturin for Parkinson’s disease: a double-blind, randomised, controlled trial. Lancet Neurol. 2010;9:1164–1172. doi: 10.1016/S1474-4422(10)70254-4. [DOI] [PubMed] [Google Scholar]
- 49.Polinski N.K., Manfredsson F.P., Benskey M.J., Fischer D.L., Kemp C.J., Steece-Collier K., Sandoval I.M., Paumier K.L., Sortwell C.E. Impact of age and vector construct on striatal and nigral transgene expression. Mol. Ther. Methods Clin. Dev. 2016;3:16082. doi: 10.1038/mtm.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peden C.S., Manfredsson F.P., Reimsnider S.K., Poirier A.E., Burger C., Muzyczka N., Mandel R.J. Striatal readministration of rAAV vectors reveals an immune response against AAV2 capsids that can be circumvented. Mol. Ther. 2009;17:524–537. doi: 10.1038/mt.2008.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benskey M.J., Sandoval I.M., Manfredsson F.P. Continuous collection of adeno-associated virus from producer cell medium significantly increases total viral yield. Hum. Gene Ther. Methods. 2016;27:32–45. doi: 10.1089/hgtb.2015.117. [DOI] [PubMed] [Google Scholar]
- 52.Zolotukhin S., Byrne B.J., Mason E., Zolotukhin I., Potter M., Chesnut K., Summerford C., Samulski R.J., Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 53.Benskey M.J., Manfredsson F.P. Intraparenchymal stereotaxic delivery of rAAV and special considerations in vector handling. Methods Mol. Biol. 2016;1382:199–215. doi: 10.1007/978-1-4939-3271-9_14. [DOI] [PubMed] [Google Scholar]
- 54.Manfredsson F.P., Burger C., Rising A.C., Zuobi-Hasona K., Sullivan L.F., Lewin A.S., Huang J., Piercefield E., Muzyczka N., Mandel R.J. Tight Long-term dynamic doxycycline responsive nigrostriatal GDNF using a single rAAV vector. Mol. Ther. 2009;17:1857–1867. doi: 10.1038/mt.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gundersen H.J., Jensen E.B. The efficiency of systematic sampling in stereology and its prediction. J. Microsc. 1987;147:229–263. doi: 10.1111/j.1365-2818.1987.tb02837.x. [DOI] [PubMed] [Google Scholar]
- 56.Carrillo-Tripp M., Shepherd C.M., Borelli I.A., Venkataraman S., Lander G., Natarajan P., Johnson J.E., Brooks C.L., 3rd, Reddy V.S. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao C., Rossmann M.G. Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 2007;158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.