Abstract
Tumour heterogeneity in cancers has been observed at the histological and genetic levels, and increased levels of intra-tumour genetic heterogeneity have been reported to be associated with adverse clinical outcomes. This review provides an overview of radiomics, radiogenomics, and habitat imaging, and examines the use of these newly emergent fields in assessing tumour heterogeneity and its implications. It reviews the potential value of radiomics and radiogenomics in assisting in the diagnosis of cancer disease and determining cancer aggressiveness. This review discusses how radiogenomic analysis can be further used to guide treatment therapy for individual tumours by predicting drug response and potential therapy resistance and examines its role in developing radiomics as biomarkers of oncological outcomes. Lastly, it provides an overview of the obstacles in these emergent fields today including reproducibility, need for validation, imaging analysis standardisation, data sharing and clinical translatability and offers potential solutions to these challenges towards the realisation of precision oncology.
INTRODUCTION
Tumour heterogeneity in cancers has been observed at the histological and genetic levels (1–3), and increased levels of intra-tumour genetic heterogeneity (4–8) have been reported to be associated with adverse clinical outcomes (9, 10). In oncological imaging, phenotypic heterogeneity between and within tumours of a given patient is readily apparent and various imaging features are routinely described subjectively in radiology reports (11). Recently, however, imaging research has focused increasingly on the newly emergent field of radiomics, which is defined as a high-throughput process in which a large number of shape, edge, and texture metrics are extracted and quantified objectively and in a reproducible form (12–14) (Fig. 1). These quantitative metrics can provide important insights into tumour phenotype and as well as the interaction of the tumour with its microenvironment, defined as “habitat imaging” (11, 15). In the effort to delineate the biological and clinical implications of these new quantitative metrics, radiomic metrics obtained from magnetic resonance imaging (MRI), including diffusion-weighted (DW) and dynamic-contrast-enhanced (DCE) MRI sequences, computed tomography (CT), and combined 2-[18F]-fluoro-2-deoxy-D-glucose (FDG) positron-emission tomography (PET)/CT have been further correlated with genomics data, a process defined as radiogenomics (16). Radiogenomics and outcome data can be meaningfully mined with the goal of developing robust biomarkers that may potentially aid cancer diagnosis, improve assessment of treatment response, and better predict patient outcome.
Figure 1.
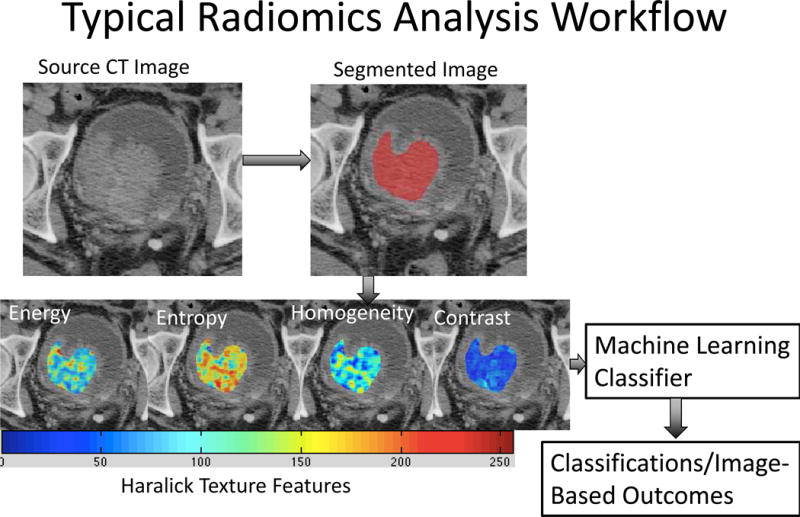
Radiomics analysis workflow. Radiomics-based analysis starts with segmentation of the structure(s) of interest, in this case, bladder cancer. Various texture features including Haralick textures are generated. A machine-learning classifier is trained using features generated from several images. Classification of various measures is then performed on never-seen-before images.
POTENTIAL VALUE OF RADIOMICS AND RADIOGENOMICS
To date, several studies have focused on the correlation and integration of radiomics with genomics (defined as the systematic study of the complete DNA sequences [genome] of organisms (17)) and proteomics (defined as the systematic study of the complete complement of proteins [proteome] of organisms (18)) data, and their results continue to support the notion that radiomic metrics may perform relatively well as surrogates of molecular alterations and protein expression found in tissue samples.
Radiomics approaches for assisting in diagnosis and assessment of cancer aggressiveness
Studies using radiomic analyses have shown that radiomic metrics are capable of distinguishing between benign and malignant tissue and aiding in the assessment of cancer aggressiveness in a variety of clinical settings. Analysis of heterogeneity in enhancement patterns, founded on perfusion deficits, which ultimately lead to different micro-environmental selection pressures (11), has been performed with promising findings. For example, grey level co-occurrence matrix analyses of DCE images distinguished between benign and malignant breast lesions with very high diagnostic accuracy (19). Texture analysis of DCE images using a structured fractal-based approach improved differentiation between low- and high-grade gliomas by orders of magnitude (20); however, texture analysis is not limited to enhancement patterns. Measures of heterogeneity in T1-weighted (W), T2W, and DW MRI images can reveal differences in cellular density in tumours, which in turn can be matched to histological findings and aid in distinguishing malignant versus benign soft-tissue masses (21). Similarly, analysis of prostate MRI using T2W and DWI sequences has been successful in discriminating prostate cancer from benign prostate tissue and in providing information on prostate cancer aggressiveness using Gleason scores. (5). In this study, five textural features (entropy, inertia, energy, correlation, and homogeneity) on apparent diffusion coefficient (ADC) maps and two textural features (inertia and correlation) on T2W imaging were significantly different between prostate tumours and benign prostate tissue (5). Using textural features, the same group also reported 90% accuracy for discriminating Gleason score of six tumours (which are usually considered “low risk”) from those with a Gleason score of ≥7 (5). Similarly, a study by Fehr et al.22 used texture features derived from ADC and T2W MRI to obtain classification of Gleason patterns with >92% accuracy (Fig. 2).
Figure 2.
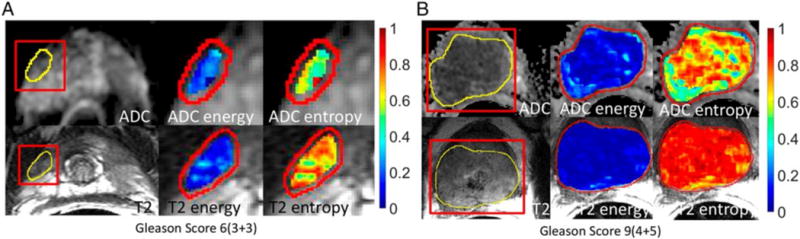
Example of texture analysis on ADC map and T2W image of prostate cancer. Energy and entropy values are overlaid on the tumour on ADC map (the top row) and T2W image (the bottom row). The texture features differ between a tumour of Gleason score (GS) 6 (3+3) (a) and a tumour of GS 9 (4+5) (b). Fehr et al.22 reported that texture analysis together with machine learning had distinguished GS6 (3+3) versus GS ≥7 and GS 7(3+4) versus GS (4+3) with high accuracy: 93% and 92%, respectively. Reprinted with permission from “Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images,” by Fehr et al.,22 Proc Natl Acad Sci U.S.A, 2015 Nov 17; 112(46):E6265-E6273.
Associations between radiomics, radiogenomics, and patient outcomes
Radiomics has the potential to identify imaging phenotypes of prognostic value by exploring intra-tumour heterogeneity features. For example, Grove et al.8 assessed speculation and entropy gradients in patients with lung cancer to find that these measures were strong prognostic indicators in patients with early stage lung cancer. Another study by Aerts et al.23 demonstrated that a radiomic signature (size, shape, texture, and wavelets) was predictive of outcome in independent cohorts of patients with lung cancer. Additionally, this same signature could be applied to patients with head and neck cancer with equivalent prognostic power (23). The presence of different habitats can be a source of radiomic features due to their distinct volumes, each with a specific combination of flow, cell density, necrosis, and oedema (12) (Fig. 3). Habitat distribution in patients with glioblastoma multiforme has enabled us to discriminate between cancers that progress quickly and those that are more indolent (24).
Figure 3.
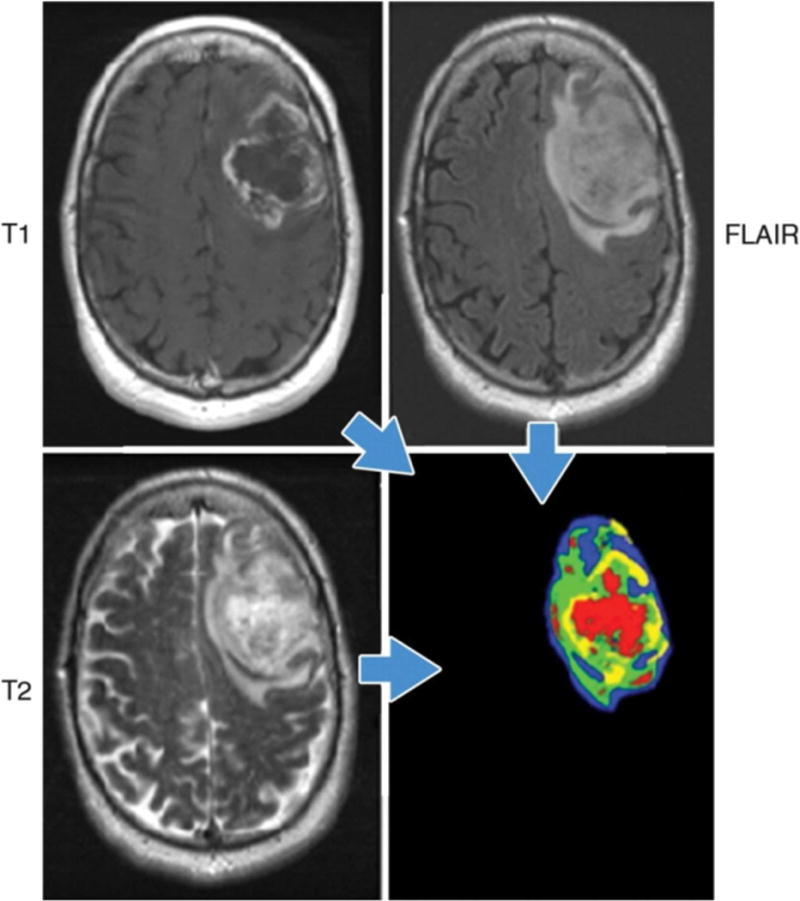
Example of habitat imaging in a patient with glioblastoma multiforme. Habitat imaging (lower right) is obtained by the combination of contrast-enhanced T1W, T2W, and fluid-attenuated inversion recovery (FLAIR) images. Each voxel of a tumour is assigned a specific colour depending on the combination of signal intensity (high/low) of these sequences, e.g., the red voxel is low on T1W images, and high on T2W and FLAIR images in this case. The clusters of voxels with specific colours yield regions that reflect different physiologic microenvironments, called habitats. This regional analysis would help the deeper understanding of tumour heterogeneity. Figure provided courtesy of R. A. Getenby.
Texture analysis can also be applied to PET. A study by Nair et al.25 demonstrated correlation of PET imaging features with gene expression, which led to an association of metagene clusters to imaging features and yielded good prognostic models in patients with resected non-small cell lung cancer.
Finally, correlation of radiogenomic data may enable us to make a decision about where to biopsy, when necessary. Quantitative analysis of regionally distinct radiomic features has the potential to inform precisely about the best biopsy sites by identifying the locations within complex tumours that are most likely to contain important diagnostic, prognostic, or predictive information. In fact PET, after overlaying functional information on CT or MRI images, has been employed to guide biopsies in the abdomen and in patients with bone disease, demonstrating the potential of radiomics to enable better-informed decisions about ideal biopsy sites. (26, 27).
Improving evaluation of treatment response
Radiogenomic analysis can be used to guide therapy for treatment of individual tumours by predicting drug response and potential therapy resistance. While Kuo et al.28 were the first to explore specific hepatocellular carcinoma imaging phenotypes that correlated with doxorubicin drug response in 2007 (28), more recently, a study in women who completed treatment for locally advanced breast cancer suggested that texture analysis of DCE MRI can enable us to predict response to neoadjuvant chemotherapy before its initiation (29). Understanding the spatial and temporal phenotypic, physiological, and genetic heterogeneity of most solid tumours has led to the realisation that most chemotherapy responses are not durable and that targeted therapies are necessary to improve outcomes (6, 30). Furthermore, inter- and intra-tumour phenotypic heterogeneity are associated with treatment failure and therapy resistance (11). For example, in a study of malignant gliomas using fused MRI sequences, the regions of tumour that were poorly perfused on post-T1W contrast-enhanced images may exhibit areas of low or high water content on T2W images and low or high diffusion on DWI. Thus, high or low densities can coexist in poorly perfused volumes, creating perfusion–diffusion mismatches. Poorly perfused regions with high cell density are concerning because they can represent cell populations that are adapted to live in micro-environmental conditions associated with poor perfusion. The associated hypoxia, acidosis, and nutrient deprivation has been suggested to select for cells that are resistant to apoptosis, and subsequently, are likely to be resistant to therapy (31, 32).
Developing radiomics as biomarkers of oncological outcomes
The field of biomarker discovery and validation has evolved rapidly over the past few years with the emergence of precision medicine, aiming to tailor medical care to the individual by taking into account the variability in genes, environment, and lifestyle. Biomarkers must accurately reflect the underlying molecular cancerous machinery (33, 34) and are measured by assays often requiring tissue analysis obtained through surgery or biopsy. The limitations of this approach include its invasive nature and the fact that samples are often obtained from only a portion of a generally heterogeneous lesion and cannot completely represent the lesion’s anatomical, functional, and pathological properties (35). Various groups have proposed that image-derived indices may reliably identify important subregions non-invasively and create predictive imaging biomarkers (36). These imaging biomarkers could then act as non-invasive measurements of functional and physiological processes in vivo and provide information on treatment planning, prognosis, and therapy response (35). For example, O’Connor et al.37 predicted colorectal cancer liver metastasis shrinkage following anti-angiogenic and cytotoxic therapy by using heterogeneity of tumour vascular enhancement as a prognostic/predictive biomarker; however, such an endeavour has proven to be difficult in radiogenomics, making implementation of predictive biomarkers more difficult than expected (38). In fact, only a few single genes, transcriptomic, epigenetic or structural genomic alterations in tumours have been discovered that could serve as clinically implementable biomarkers. For example, in breast cancer, only a few prognostic gene signatures and three traditional single gene predictive markers, namely oestrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2), are in routine clinical use (13, 39). The reasons behind the limited prognostic/predictive value of many proposed biomarkers are manifold, and include intra- and inter-tumour genetic heterogeneity; technical issues including reproducibility and reliability of the biomarker assay; and retrospective, small and often clinically heterogeneous patient cohorts (34).
Despite these challenges, given the potential usefulness of imaging biomarkers, the National Cancer Institute (NCI) in the United States has established the Quantitative Imaging Network, which promotes the development of imaging methods, protocols, and software tools (19). Although a lot of work remains to be done, a multidisciplinary approach that combines phenotype and genetics is an important strategy for advancing precision medicine.
CHALLENGES AND FUTURE DIRECTIONS
Radiogenomics is still in its early phases and its implementation requires completion of a series of steps to make it usable in daily clinical practice. First, it requires standardisation of imaging protocols, including image acquisition and post-processing, as well as robust segmentation algorithms that require minimal operator input (13). As with any study, a radiogenomics study must be validated against a set of independent data, to ensure its reproducibility. Radiomic analyses require large image datasets with the expectation that large numbers may be able to overcome some of the inherent heterogeneities in clinical imaging. Hence, the need for informatics databases that allow for image data sharing along with medical and genetic data across sites becomes very important. Incorporating detailed clinical and patient risk factor data into radiomics is essential in clinical translation and development of biomarkers. Currently, the most important goal is optimisation of each of these steps, with a future plan to gradually harmonise and standardise the entire process (13).
Reproducibility
Radiomics has remarkable potential to accelerate precision medicine; however, it faces challenges common to biomarker development including suboptimal study designs, high technical complexity, data overfitting, incomplete result reporting, and the presence of many confounding variables, especially when dealing with retrospective data, which makes it difficult to ensure reproducibility (12). For example, in radiomics, a large number of imaging features may be computed, which increases the risk of extracted features becoming higher than the number of samples in a study, reducing its power and increasing the probability of overfitting the data (12). Dimensionality reduction offers a solution to this particular issue by selection of task-specific features or by combining the original features to obtain a new set of features by using methods, such as principal component analysis (13). Furthermore, although some standardised tools for genomic profiling exist, they are not universally accepted and applied across different institutions (12).
The issue of reproducibility extends beyond radiomics to biomedical research in general. A 2009 analysis of biomedical research reports found that at least 50% of studies were too poor, insufficient, or incomplete to be usable (40). Furthermore, when scientists at Amgen (Thousand Oaks, CA, USA) tried to replicate 53 landmark studies in the basic science of cancer, they were able to reproduce the original results of just six (41). In order to address this issue, editors from more than 30 high-impact-factor biomedical journals have imposed common standards for statistical testing and to improve access to raw data (42). Furthermore, reporting guidelines have been developed by many organisations, such as the Equator network, which focus on improving health research quality and transparency (43). Development of such standard guidelines provides a roadmap to navigate the complex issues inherent to radiomics, particularly acquisition and analysis of high-dimensional data (12); however, at present, adherence to these guidelines is not mandatory.
Need for validation
Validation with prospectively collected independent cohorts, ideally in the setting of clinical trials, is the reference standard for verifying an identified statistical association or biomarker (34, 44). As with any biomarker study, a retrospective radiomics investigation must be validated against a completely independent dataset, preferably from a different institution (12). In a thorough review by Bai et al.,35 a validation dataset was used in only eight out of 27 studies. One problem that prevents retrospective validation studies is the availability of publicly available radiomics data, which creates the need for shared databases that can be used as validation sets. The Cancer Genome Atlas (TCGA) has generated comprehensive multidimensional genomic data of more than 30 types of cancer, which together with the clinical annotations are available to the public (45). The Cancer Imaging Archive (TCIA) (46) is another publicly available resource that contains imaging corresponding to the patients in the TCGA database and can be used as valuable sources for both hypothesis-generating and validation purposes.
Sample size
Sample size determines the power of predictive classifier models used in radiomics and other biomedical studies. According to Gillies et al.,12 10 patients are needed for each feature in a model based on binary classifiers; however, the need for accommodation of additional clinical or genomic covariates demands a large sample size. Additionally, as few as 100 patients can be used in radiomics studies; however, datasets with larger samples provide more power (12). Furthermore, the presence of heterogeneity in clinical imaging requires a large enough dataset to overcome such inherent differences (13). Acquiring a sufficiently sized sample set requires large databases and data sharing capability across sites, which already exists in form of various online (13).
Standardisation of imaging analysis
Image acquisition parameters vary widely in routine clinical practice. For example, patient positioning, pixel or matrix size, section thickness, variable reconstruction algorithms, and washout period in the case of PET imaging vary widely in different institutions (13). As such, comparing results obtained across different machines becomes challenging and allows room for error. Furthermore, varying image parameters makes it more difficult to identify large numbers of image data examples with similar clinical parameters such as disease stage (13). For example, Kumar et al.13 showed the effect of the large variation of section thickness and pixel size in a study that aimed to develop prediction models by using image features to classify non-small cell cancer tumours. The large variation in this scenario and when performing radiomic analysis in general affects the information being extracted by image feature algorithms and subsequently classifier performance (13). As a result, there is an indisputable need for standardisation of imaging analysis and further research remains to be done in this area.
Image segmentation challenges
One of the most critical steps in feature data analyses is the segmentation of images into volumes of interest (VOI), which presents a variety of challenges. Many tumours have indistinct borders, which make reproducibility of their delineation an important issue (47, 48). Manual segmentation is often performed; however, it suffers from high inter- and intra-reader variability and is labour intensive, and hence, it is not feasible for examining the large datasets required in radiomics (13). Automatic and semiautomatic segmentation algorithms have been developed; however, there are no universal segmentation algorithm that can work for all medical image applications (13). Furthermore, even when using automatic or semi-automatic segmentation, the VOI will be different even when using the same algorithm performed multiple times with different initialisations (13). Thus, it becomes essential to develop agreed-upon metrics to evaluate segmentation algorithms. Currently, a consensus is emerging that computer-aided edge detection followed by manual adjustment may result in optimal reproducible segmentation (12).
Adequacy of the reference standards
Histopathology and results of genomic testing are often used as standards of reference; however, only few studies have explored the spatial relationship between imaging, genomics, and histopathology (15). Although there is a need for large prospective studies, a few issues arise when attempting to integrate imaging, genomic, and histopathological data. These studies must evaluate large and complex data across a range of biologically different scales (19, 49). One problem is that CT, MRI, or PET voxels are usually non-isotropic, where section thickness exceeds in-plane resolution. Hence, compared with genomic and histopathology biomarkers, this represents many orders of magnitude difference in scale (49), making it challenging to validate image heterogeneity biomarkers against histopathology. Furthermore, it is unclear whether imaging, genomics, and histopathology show spatial correspondence because they measure the same biology in different ways or alternatively measure different biology, providing complimentary data (15). If the last statement is true, this would open up new multidisciplinary strategies for advancing precision medicine by bringing imaging phenotype and genotype together, rather than assessing genomics in isolation (15).
Clinical translatability
Radiogenomics is still a relatively new field, and its full potential for clinical translation is yet to be explored; however, many studies have shown early promise. For example, in the research of hepatocellular carcinoma, microscopic venous invasion (MVI) is a sign of poor prognosis (50). Imaging techniques have failed to predict MVI and the only way it is diagnosed is by explanted tissue histology (51). In a study by Segal et al.,52 91 genes in the “venous invasion signature” were associated with two predominant imaging traits on CT—the presence of “internal arteries” and absence of “hypodense halos”. Banerjee et al.53 again demonstrated that these two imaging biomarkers, along with “tumour–liver difference,” were able to predict histological MVI with high precision. Furthermore, these three features were associated with early disease recurrence and poor overall survival (53). Beyond diagnosis, radiogenomics can have a significant impact on treatment guidance and overall survival (35). In a study of clear cell renal cell carcinoma patients, Jamshidi et al.54 developed a prognostic multigene signature termed radiogenomic risk score consisting of four CT imaging features (tumour necrosis pattern, transition zone, tumour–parenchyma interaction, and tumour–parenchyma interface) to predict disease-specific survival, independent of disease stage, disease grade, and performance status. The ultimate goal of clinical translation of radiogenomics is to enhance the radiology report to beyond what is typically reported and include genomics data in addition to radiological data, which may have an impact on patient clinical management.
Data sharing
As mentioned above, there is need for the development of integrated publicly available databases where images and the extracted features are linked to clinical and molecular data (13) to ensure studies of sufficient size for statistical power. Such databases present their own challenges including de-identification, the need for large digital storage space, integration of clinical and molecular data to create a simple work stream, as well as reporting and exporting the data (13). Furthermore, as part of a larger network of quantitative imaging sites, we must also be able to exchange data according to an evolving set of standards. Various online repositories that host image data are currently available, such as the online CT image repository, the National Biomedical Image Archive (NBIA), hosted by NCI; however, in addition to images themselves, image annotations, outcome data, acquisition, scanner, and other information should be available. Currently, available clinical image data, which may be used for radiomics study, includes TCIA, TCGA, and others (12, 13, 45, 46). Ultimately, the goal is multi-institutional, national, or international consortia to agree to share data either through centralised or federated networks (12).
Need for well-designed prospective studies that account for spatial and temporal heterogeneity
Although initial retrospective studies linking imaging phenotype with genotype have shown high prognostic capabilities, they do not provide spatial information, as quantitative imaging features are generated and averaged over the entire tumour assuming that tumours are heterogeneous, but well mixed. This approach ignores the spatial heterogeneity readily apparent on imaging. Subregions of tumour that show distinct imaging features may have different biological behaviour, which may lead to different response to therapy or drive tumour progression and metastatic ability (15). Indeed, recent genomics work has highlighted the presence of intra-tumour genetic and transcriptomic variation (1, 3, 7, 38, 55, 56). Nevertheless, little effort is any has been put into integrating imaging, histopathology, and genomics (4, 57). In a retrospective study of 10 patients with breast cancer, heterogeneous enhancement correlated with genetic subtypes (4). Jenkinson et al.57 found significant differences in ADC values at tumour margins between oligodendroglial tumour genotypes that may reflect underlying tumour biology (57); however, there is a need for well-designed prospective studies focused on meaningful integration of imaging phenotype and genotype rather than genomics or imaging in isolation.
Important information can be gained by evaluating the quantitative parameters in discrete phenotypic clusters or regions of the tumour by combining perfusion, diffusion, and metabolic maps derived from multiparametric imaging, such as MRI and PET/CT, to create one set of phenotypic heterogeneity maps based on their similarities and differences. This in turn can be used to guide tissue sampling, from both primary tumours and metastatic disease, for detailed histological, immunohistochemical, genetic, epigenetic, and proteomic analysis (55) with the goal of assessing whether an imaging-based analysis of heterogeneity would be reflective of the underlying pathological and/or genomic heterogeneity of the tumour.
CONCLUSION
In conclusion, complementary innovations in massive parallel sequencing, proteomics, and imaging that allow spatial and temporal quantification of tumour heterogeneity and its changes during drug treatment will provide the basis for the realisation of precision oncology.
Acknowledgments
This research was funded in part through the NIH/NCI Cancer Center Support GrantP30 CA008748. The funding source had no involvement in the writing of the review and in the decision to submit the review for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The funding source had no involvement in the writing of the review and in the decision to submit the review for publication.
References
- 1.de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science (New York, NY) 2014;346(6206):251–6. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yates LR, Gerstung M, Knappskog S, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21(7):751–9. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz RF, Ng CK, Cooke SL, et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med. 2015;12(2):e1001789. doi: 10.1371/journal.pmed.1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. AJR Am J Roentgenol. 2012;199(3):654–63. doi: 10.2214/AJR.11.7824. [DOI] [PubMed] [Google Scholar]
- 5.Wibmer A, Hricak H, Gondo T, et al. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol. 2015;25(10):2840–50. doi: 10.1007/s00330-015-3701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–14. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grove O, Berglund AE, Schabath MB, et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS One. 2015;10(3):e0118261. doi: 10.1371/journal.pone.0118261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andor N, Graham TA, Jansen M, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22(1):105–13. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris LG, Riaz N, Desrichard A, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget. 2016;7(9):10051–63. doi: 10.18632/oncotarget.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269(1):8–15. doi: 10.1148/radiol.13122697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–77. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30(9):1234–48. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–6. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21(2):249–57. doi: 10.1158/1078-0432.CCR-14-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colen R, Foster I, Gatenby R, et al. NCI Workshop Report: clinical and computational requirements for correlating imaging phenotypes with genomics signatures. Translat Oncol. 2014;7(5):556–69. doi: 10.1016/j.tranon.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MeSH Browser. Genomics. Bethesda, MD: National Library of Medicine (US); 2016. Available at: http://www.ncbi.nlm.nih.gov/mesh/68023281. Accessed: 21 July 2016. [Google Scholar]
- 18.MeSH Browser. Proteomics. Bethesda, MD: National Library of Medicine (US); 2016. Available from: http://www.ncbi.nlm.nih.gov/mesh/?term=proteomics. Accessed: 21 July 2016. [Google Scholar]
- 19.Ahmed A, Gibbs P, Pickles M, Turnbull L. Texture analysis in assessment and prediction of chemotherapy response in breast cancer. J Magn Reson Imaging. 2013;38(1):89–101. doi: 10.1002/jmri.23971. [DOI] [PubMed] [Google Scholar]
- 20.Rose CJ, Mills SJ, O’Connor JP, et al. Quantifying spatial heterogeneity in dynamic contrast-enhanced MRI parameter maps. Magn Reson Med. 2009;62(2):488–99. doi: 10.1002/mrm.22003. [DOI] [PubMed] [Google Scholar]
- 21.Chen CK, Wu HT, Chiou HJ, et al. Differentiating benign and malignant soft tissue masses by magnetic resonance imaging: role of tissue component analysis. J Chin Med Assoc. 2009;72(4):194–201. doi: 10.1016/S1726-4901(09)70053-X. [DOI] [PubMed] [Google Scholar]
- 22.Fehr D, Veeraraghavan H, Wibmer A, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A. 2015;112(46):E6265–73. doi: 10.1073/pnas.1505935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature Comm. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou M, Hall L, Goldgof D, et al. Radiologically defined ecological dynamics and clinical outcomes in glioblastoma multiforme: preliminary results. Translat Oncol. 2014;7(1):5–13. doi: 10.1593/tlo.13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair VS, Gevaert O, Davidzon G, et al. Prognostic PET 18F-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non-small cell lung cancer. Cancer Res. 2012;72(15):3725–34. doi: 10.1158/0008-5472.CAN-11-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klaeser B, Wiskirchen J, Wartenberg J, et al. PET/CT-guided biopsies of metabolically active bone lesions: applications and clinical impact. Eur J Nucl Med Mol Imaging. 2010;37(11):2027–36. doi: 10.1007/s00259-010-1524-z. [DOI] [PubMed] [Google Scholar]
- 27.Tatli S, Gerbaudo VH, Mamede M, Tuncali K, Shyn PB, Silverman SG. Abdominal masses sampled at PET/CT-guided percutaneous biopsy: initial experience with registration of prior PET/CT images. Radiology. 2010;256(1):305–11. doi: 10.1148/radiol.10090931. [DOI] [PubMed] [Google Scholar]
- 28.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol. 2007;18(7):821–31. doi: 10.1016/j.jvir.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 29.Teruel JR, Heldahl MG, Goa PE, et al. Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed. 2014;27(8):887–96. doi: 10.1002/nbm.3132. [DOI] [PubMed] [Google Scholar]
- 30.Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467(7319):1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thews O, Nowak M, Sauvant C, Gekle M. Hypoxia-induced extracellular acidosis increases p-glycoprotein activity and chemoresistance in tumors in vivo via p38 signaling pathway. Adv Exp Med Biol. 2011;701:115–22. doi: 10.1007/978-1-4419-7756-4_16. [DOI] [PubMed] [Google Scholar]
- 32.Thews O, Dillenburg W, Rosch F, Fellner M. PET imaging of the impact of extracellular pH and MAP kinases on the p-glycoprotein (Pgp) activity. Adv Exp Med Biol. 2013;765:279–86. doi: 10.1007/978-1-4614-4989-8_39. [DOI] [PubMed] [Google Scholar]
- 33.Mehta S, Shelling A, Muthukaruppan A, et al. Predictive and prognostic molecular markers for cancer medicine. Ther Adv Med Oncol. 2010;2(2):125–48. doi: 10.1177/1758834009360519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayes DF. Biomarker validation and testing. Molecular oncology. 2015;9(5):960–6. doi: 10.1016/j.molonc.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai HX, Lee AM, Yang L, et al. Imaging genomics in cancer research: limitations and promises. Br J Radiol. 2016;89(1061):20151030. doi: 10.1259/bjr.20151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chalkidou A, O’Doherty MJ, Marsden PK. False discovery rates in PET and CT studies with texture features: a systematic review. PLoS One. 2015;10(5):e0124165. doi: 10.1371/journal.pone.0124165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Connor JP, Rose CJ, Jackson A, et al. DCE-MRI biomarkers of tumour heterogeneity predict CRC liver metastasis shrinkage following bevacizumab and FOLFOX-6. Br J Cancer. 2011;105(1):139–45. doi: 10.1038/bjc.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Translat Med. 2012;4(127):127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 39.Schultheis Ng CK, Bidard AM, Weigelt FC, Reis-Filho BJS. Breast cancer genomics from microarrays to massively parallel sequencing: paradigms and new insights. J Natl Cancer Inst. 2015;107(5) doi: 10.1093/jnci/djv015. [DOI] [PubMed] [Google Scholar]
- 40.Chalmers I, Glasziou P. Avoidable waste in the production and reporting of research evidence. Obstet Gynecol. 2009;114(6):1341–5. doi: 10.1097/AOG.0b013e3181c3020d. [DOI] [PubMed] [Google Scholar]
- 41.Begley CG, Ellis LM. Drug development: raise standards for preclinical cancer research. Nature. 2012;483(7391):531–3. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 42.McNutt M. Journals unite for reproducibility. Science (New York, NY) 2014;346(6210):679. doi: 10.1126/science.aaa1724. [DOI] [PubMed] [Google Scholar]
- 43.Simera I, Moher D, Hirst A, Hoey J, Schulz KF, Altman DG. Transparent and accurate reporting increases reliability, utility, and impact of your research: reporting guidelines and the EQUATOR Network. BMC Med. 2010;8:24. doi: 10.1186/1741-7015-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14(7):507–15. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The future of cancer genomics. Nat Med. 2015;21(2):99. doi: 10.1038/nm.3801. [DOI] [PubMed] [Google Scholar]
- 46.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–57. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rios Velazquez E, Aerts HJ, Gu Y, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists’ delineations and with the surgical specimen. Radiother Oncol. 2012;105(2):167–73. doi: 10.1016/j.radonc.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Dam IE, van Sornsen de Koste JR, Hanna GG, Muirhead R, Slotman BJ, Senan S. Improving target delineation on 4-dimensional CT scans in stage I NSCLC using a deformable registration tool. Radiother Oncol. 2010;96(1):67–72. doi: 10.1016/j.radonc.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Cebulla J, Kim E, Rhie K, Zhang J, Pathak AP. Multiscale and multi-modality visualization of angiogenesis in a human breast cancer model. Angiogenesis. 2014;17(3):695–709. doi: 10.1007/s10456-014-9429-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandarana H, Robinson E, Hajdu CH, Drozhinin L, Babb JS, Taouli B. Microvascular invasion in hepatocellular carcinoma: is it predictable with pretransplant MRI? AJR Am J Roentgenol. 2011;196(5):1083–9. doi: 10.2214/AJR.10.4720. [DOI] [PubMed] [Google Scholar]
- 51.Griffin N, Addley H, Sala E, et al. Vascular invasion in hepatocellular carcinoma: is there a correlation with MRI? Br J Radiol. 2012;85(1014):736–44. doi: 10.1259/bjr/94924398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25(6):675–80. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 53.Banerjee S, Wang DS, Kim HJ, et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62(3):792–800. doi: 10.1002/hep.27877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jamshidi N, Jonasch E, Zapala M, et al. The radiogenomic risk score stratifies outcomes in a renal cell cancer phase 2 clinical trial. Eur Radiol. 2016;26(8):2798–807. doi: 10.1007/s00330-015-4082-8. [DOI] [PubMed] [Google Scholar]
- 55.Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72(19):4875–82. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yates LR, Gerstung M. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. 2015;21(7):751–9. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jenkinson MD, Smith TS, Brodbelt AR, Joyce KA, Warnke PC, Walker C. Apparent diffusion coefficients in oligodendroglial tumors characterised by genotype. J Magn Reson Imaging. 2007;26(6):1405–12. doi: 10.1002/jmri.21062. [DOI] [PubMed] [Google Scholar]


