Abstract
HIV cure research is increasingly focused on anatomical tissues as sites for residual HIV replication during combined antiretroviral therapy (cART). Tissue-based HIV could contribute to low-level immune activation and viral rebound over the course of infection and could also influence the development of diseases, such as atherosclerosis, neurological disorders and cancers. cART-treated subjects have a decreased and irregular presence of HIV among tissues, which has resulted in a paucity of actual evidence concerning how or if HIV persists, replicates and evolves in various anatomical sites during therapy. In this study, we pooled 1806 HIV envelope V3 loop sequences from twenty-six tissue types (seventy-one total tissues) of six pre-cART subjects, four subjects with an unknown cART history who died with profound AIDS, and five subjects who died while on cART with an undetectable plasma viral load. A computational approach was used to assess sequences for their ability to utilize specific cellular coreceptors (R5, R5 and X4, or X4). We found that autopsied tissues obtained from virally suppressed cART+ subjects harbored both integrated and expressed viruses with similar coreceptor usage profiles to subjects with no or ineffective cART therapy (i.e., significant plasma viral load at death). The study suggests that tissue microenvironments provide a sanctuary for the continued evolution of HIV despite cART.
Keywords: HIV, Coreceptor, Anatomical tissues, Combined antiretroviral therapy, HIV sequence data, Bioinformatics
For those infected with the human immunodeficiency virus (HIV+), carefully monitored cART therapy will reduce plasma viral loads (pVL) to undetectable levels and increase CD4+ T-cell levels, thus allowing HIV+ persons to live longer, healthier lives. Still, some cART-treated patients develop potentially fatal diseases such as atherosclerosis, neurological deficits and cancers at a higher rate than HIV seronegative (HIV−) people, indicating that pathogenic pathways can remain active during cART (Cesarman, 2013; Huysentruyt et al., 2012; Brew and Chan, 2014; Hsue et al., 2009; Crowe et al., 2010).
Prior to cART, the initial AIDS pandemic resulted in a wealth of autopsy material, which permitted a thorough study of HIV evolution in tissues. These studies found that, with some exceptions (e.g., viral compartments such as brain and breast milk), HIV proviral DNA in organs were often related to viral RNA species obtained from plasma (Ball et al., 1994). In the cART era, sequencing HIV from autopsy tissues derived from cART+ subjects remains challenging due to the paucity of appropriate, well-documented samples and the reduced amount of amplifiable HIV in a given set of tissues (Rose et al., 2016). Moreover, a single-genome sequencing approach is required in these studies to ensure that the limited HIV species in a tissue are not resampled. Despite these hurdles, recent studies have confirmed that both expressed and integrated proviral HIV can be present in some tissues during prolonged cART treatment (Rose et al., 2016; Lamers et al., 2016a), highlighting the importance of HIV tissue sanctuaries as a major challenge in curing HIV infection. These studies raise a serious question: how and why are these viruses maintained in tissues during cART despite what appears to be effective clearance of virus from blood? In a recent study, we found that the evolutionary rate of HIV sampled from tissues was not significantly different than that of pre-ART virus sampled from blood (Rose et al., 2016). Further, tissues harboring HIV were frequently associated with pathologic changes (Lamers et al., 2016a). These findings directed the current study to examine another HIV evolutionary metric, predicted viral coreceptor usage, in pre-cART vs. cART-treated vs. virally suppressed subjects' autopsy tissues.
HIV enters immune cells via binding a cellular coreceptor, usually CCR5 (and thus is denoted an R5 virus) and more rarely CXCR4 (denoted as an X4 virus) (Goodenow and Collman, 2006). Mixed viral populations (R5 and X4), of viruses that have evolved to use both coreceptors (R5X4), usually emerge prior to X4 species (Tasca et al., 2008). These coreceptor transitions can be attributed in part to structural changes in HIV gp120, the target for neutralizing antibodies. The interplay of gp120 with the immune system drives selection against least fit variants during successive rounds of replication and results in genetic variability. During clinical latency, in the years prior to AIDS, blood samples from HIV+ subjects primarily harbor R5 viruses, because X4 variants are more easily recognized and neutralized by a healthier immune system (Tasca et al., 2008). In advanced disease, when the immune system has exhausted its resources, a reduction in selective pressure allows R5X4 or X4 variants to emerge. This process also explains why R5 viruses are the most common founder phenotype in newly infected individuals, as transmission is most likely to occur between healthier individuals (Tasca et al., 2008). While coreceptor studies have predominantly focused on blood-derived or molecularly cloned HIV, few studies have considered coreceptor usage in tissue-based HIV populations, especially those derived from patients on cART. Here, we make use of the availability of tissues from three sets of HIV+ subjects (untreated, treated but not virologically suppressed, and treated with successful viral suppression) to address the novel hypothesis that HIV coreceptor usage in tissues is independent of cART.
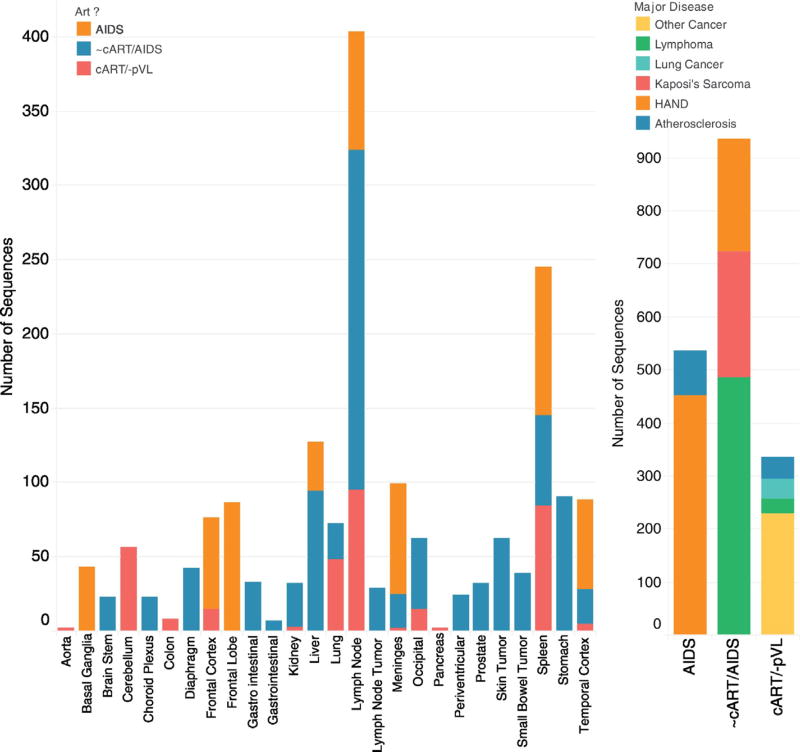
Genetic determinants of HIV-1 coreceptor usage are primarily concentrated within the 35-amino acid hypervariable V3 loop of the envelope protein gp120 (Cann et al., 1992; Stamatatos and Cheng-Mayer, 1993;Milich et al., 1993). We analyzed envelope V3 loop sequences collected from 71 autopsy specimens from 15 US patients infected with HIV subtype B sampled from 1994 to 2009 who died due to a variety of AIDS and non-AIDS pathologies (Table 1) (Rose et al., 2016; Salemi et al., 2009; Lamers et al., 2010). A total of 1806 sequences (DNA and RNA) were evaluated from 26 distinct tissue types (Fig. 1). Each subject varied in cART history: six had no prior cART experience (Lamers et al., 2009; Lamers et al., 2016b), four were prescribed cART with unknown adherence (Lamers et al., 2009), a detectable pVL, and profound AIDS noted at autopsy; five had well-documented, suppressive cART and no detectable pVL at death (Rose et al., 2016; Lamers et al., 2016a); extensive histopathology for the tissues derived from these five patients as well as disease progression and therapy has been previously published (Lamers et al., 2016a).
Table 1.
Subject information.
| Subject ID | HIV status | Primary disease | Secondary disease | cART | NADIR CD4 | Last plasma CD4 | Last plasma VL |
|---|---|---|---|---|---|---|---|
| AM | AIDS | AIDS lymphoma | Hodgkin's disease | No | NKc | NK | NK |
| IV | AIDS lymphoma | Pneumonia | NK | NK | NK | ||
| CX | HAD | none | NK | NK | NK | ||
| HCK1 | MACa/PCPb/Wasting | Kaposi's sarcoma | NK | 5 | NK | ||
| HCK2 | AIDS lymphoma | Kaposi's sarcoma | NK | NK | NK | ||
| HCK3 | Kaposi's sarcoma | None | NK | NK | NK | ||
| BW | AIDS lymphoma | Dementia | Unknown adherence | NK | 4 | NK | |
| AZ | Atherosclerosis | Polypharmacy | NK | 110 | 8020 | ||
| GA | HAD | Atherosclerosis/PCP | 50 | <299 | NK | ||
| DY | HAD | MAC | 2 | NK | 7,180,000 | ||
| HC02 | HIV+ | Plasmacytoma | HAND | Yes | 20 | 20 | <40 |
| HC03 | Anal carcinoma | Hodgkin's disease | 21 | 21 | <40 | ||
| HC05 | Prostate cancer | Anal carcinoma | 58 | 154 | <40 | ||
| HC08 | Lung cancer | None | 17 | 112 | <40 | ||
| HC09 | Alzheimer's | Atherosclerosis | 220 | 268 | <40 |
MAC = Mycobacterium avian complex.
PCP = pneumocystis pneumonia.
NK = not known.
Fig. 1.
Number of sequences, tissues, and subject pathology used for study. Left, the number of sequences for each tissue type is colored by three treatment categories (AIDS subjects without ART, subjects with varied ART exposure who died from AIDS and sequences derived from cART+ subjects who died with no viral load). Right, the number of sequences studied for each treatment category is colored by primary disease pathology of the nine subjects studied. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Coreceptor usage can be predicted computationally with reasonable accuracy using a variety of algorithms. Here, two algorithms were used for prediction: WebPSSM Sinsi (Jensen et al., 2003) and ZAPP (Lamers et al., 2016c; Fogel et al., 2015). WebPSSM is a commonly used web-based approach that is based on the positional frequency of amino acids along the V3 loop. For sequences with scores that are intermediate between R5 (x < −6.96) or X4 (x > −2.88), WebPSSM uses the “11/25 rule,” which proposes that a positively charged amino acid at positions 11 or 25 is likely an X4 isolate. The program classifies X4 species at 84% and R5 species at 96% accuracy. ZAPP (Zoetic Amino Acid Protein Profiler) is a newer approach that was used because of its ability to predict dual tropic (R5X4) as well as R5 and X4 sequences. Along with positional information, ZAPP uses 77 amino acid physicochemical properties at each position in an aligned set of sequences as input to an evolved neural network for a coreceptor usage prediction (Lamers et al., 2016c; Fogel et al., 2015). ZAPP scores on a continuous scale from R5 (x < 0.378) to R5X4 (0.378 ≤ x < 1.13) to X4 (x ≥ 1.13). ZAPP classification accuracy is 94.5% for R5, 83.2% for R5X4 and 79.6% for X4.
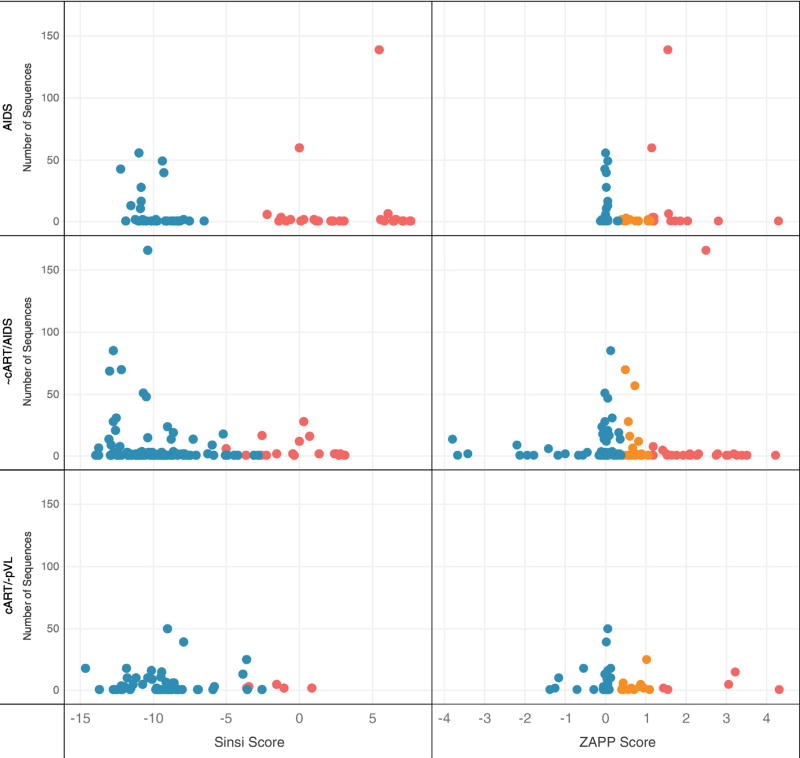
Fig. 2 shows the number of sequences, algorithm scores and predicted coreceptor usage for the three patient data sets (subjects prior to cART with AIDS, subjects with variable cART with AIDS, and virally suppressed/ cART+ subjects). WebPSSM results agreed with ZAPP predictions 75% of the time. Of the remaining discordant predictions, 10% were classified by ZAPP as dual tropic (R5X4), which would have been scored as either R5 or X4 using WebPSSM. X4 viruses were predicted within tissues for all 3 groups of subjects by both algorithms. In the virally suppressed patients, WebPSSM identified 4.2% of the isolates as X4, while ZAPP predicted 7.2% as X4 and 15.5% as R5X4. Both algorithms result in the same general conclusion: there is no evidence that cART halted the evolution of R5X4 and X4 HIV derived from tissues.
Fig. 2.
Coreceptor usage prediction for WebPSSM sinsi (left) and ZAPP (right) algorithms. 556 brain and 1249 non-brain sequences were evaluated from 26 different tissue types (71 tissues in total) collected from 15 subjects. Sequences from subjects were grouped into three categories (y-axis): No cART/AIDS (934 sequences), variable cART/AIDS (536 sequences), and cART+ subjects who were virally suppressed (335 sequences). Predicted X4 variants (red), predicted dual-tropic variants (R5X4) (orange) and predicted R5 variants (blue) are indicated. WebPSSM predicts envelope sequences as R5 (x < −6.96) or X4 (x > −2.88); for sequences with scores that are intermediate between R5 or X4, the program uses the “11/25 rule,” which proposes that a positively charged amino acid at positions 11 or 25 is likely an X4 isolate. ZAPP predicts over a continuous range using thresholds for coreceptor assignment as R5 (x < 0.378), R5X4 (0.378 ≤ x < 1.13), and X4 (x ≥ 1.13). The results demonstrate no remarkable differences among coreceptor affinity pre- and post-ART. Many of the autopsy samples derived from cART+/virally suppressed subjects revealed various tissue-based pathologies (Brew and Chan, 2014). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Numerous studies have focused on resting HIV+ CD4+ T-cells in blood during cART; however, we are just learning how HIV evolves in anatomical tissues during long term cART therapy. The presence of HIV in tissues with mixed coreceptor affinity, especially R5X4 and X4, suggests at least two scenarios that deserve further study: 1) despite cART, there can be an inability in tissue microenvironments to neutralize HIV, and, 2) similar to the emergence of X4 variants in the blood of AIDS patients, there may be an association between the pathology identified in these tissues and R5X4/X4 evolution. Of note, in an earlier study, we identified increased rates of HIV recombination in tissues with abnormal pathology (Lamers et al., 2009). Since increased rates of viral recombination are associated with increased rates of evolution, coreceptor usage is yet another metric suggesting that localized tissue pathology may coincide with the ability of HIV to evolve in microenvironments. Unfortunately, due to the small sample size, any association with co-receptor usage and any of the primary/secondary disease presented by the donors was not discernable, highlighting the need for future studies using large cross-sectional tissue-based resources, which may have important implications for future therapies aimed at an HIV cure or in treating HIV-associated comorbidities.
Acknowledgments
This work was supported by the U.S. National Institutes of Health under Grants NIMHR01-MH100984 and NCI UM1-CA181255 to Michael S. McGrath and the NIMH U24-MH100929 to Elyse J. Singer.
Footnotes
GenBank accession numbers
KU708874-KU709342; HM002482-HM001362; new sequence data KY270561-KY270659.
Disclosures
Susanna L. Lamers, Rebecca Rose, David J. Nolan and Andrew Barbier are employed by Bioinfoexperts, LLC. Gary B. Fogel and Enoch Liu are employed by Natural Selection, Inc.
References
- Ball JK, Holmes EC, Whitwell H, Desselberger U. Genomic variation of human immunodeficiency virus type 1 (HIV-1): molecular analyses of HIV-1 in sequential blood samples and various organs obtained at autopsy. J. Gen. Virol. 1994;75(Pt 4):67–79. doi: 10.1099/0022-1317-75-4-867. [DOI] [PubMed] [Google Scholar]
- Brew BJ, Chan P. Update on HIV dementia and HIV-associated neurocognitive disorders. Curr. Neurol. Neurosci. Rep. 2014;14:468. doi: 10.1007/s11910-014-0468-2. [DOI] [PubMed] [Google Scholar]
- Cann AJ, Churcher MJ, Boyd M, O'Brien W, Zhao JQ, Zack J, et al. The region of the envelope gene of human immunodeficiency virus type 1 responsible for determination of cell tropism. J. Virol. 1992;66:305–309. doi: 10.1128/jvi.66.1.305-309.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E. Pathology of lymphoma in HIV. Curr. Opin. Oncol. 2013;25:487–494. doi: 10.1097/01.cco.0000432525.70099.a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe SM, Westhorpe CL, Mukhamedova N, Jaworowski A, Sviridov D, Bukrinsky M. The macrophage: the intersection between HIV infection and atherosclerosis. J. Leukoc. Biol. 2010;87:589–598. doi: 10.1189/jlb.0809580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel GB, Lamers SL, Liu ES, Salemi M, McGrath MS. Identification of dual-tropic HIV-1 using evolved neural networks. Biosystems. 2015;137 doi: 10.1016/j.biosystems.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenow MM, Collman RG. HIV-1 coreceptor preference is distinct from target cell tropism: a dual-parameter nomenclature to define viral phenotypes. J. Leukoc. Biol. 2006;80:965–972. doi: 10.1189/jlb.0306148. [DOI] [PubMed] [Google Scholar]
- Hsue PY, Hunt PW, Schnell A, Kalapus SC, Hoh R, Ganz P, et al. Role of viral replication, antiretroviral therapy, and immunodeficiency in HIV-associated atherosclerosis. AIDS. 2009;23:1059–1067. doi: 10.1097/QAD.0b013e32832b514b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysentruyt LC, Lamers SL, McGrath MS. The Prevelence of HIV-1 DNA in AIDS-related lymphoma and Kaposi's Sarcoma throughout the AIDS epidemic. Infect. Agents Cancer. 2012;7(Suppl. 1):P43. [Google Scholar]
- Jensen MA, Li FS, van 't Wout AB, Nickle DC, Shriner D, He HX, et al. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 2003;77:13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Salemi M, Galligan DC, de Oliveira T, Fogel GB, Granier SC, et al. Extensive HIV-1 intra-host recombination is common in tissues with abnormal histopathology. PLoS One. 2009;4:e5065. doi: 10.1371/journal.pone.0005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Salemi M, Galligan DC, Morris A, Gray R, Fogel G, et al. Human immunodeficiency virus-1 evolutionary patterns associated with pathogenic processes in the brain. J. Neuro-Oncol. 2010;16:230–241. doi: 10.3109/13550281003735709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Rose R, Maidji E, Agsalda-Garcia M, Nolan DJ, Fogel GB, et al. HIV DNA is frequently present within pathologic tissues evaluated at autopsy from cART-treated patients with undetectable viral load. J. Virol. 2016a doi: 10.1128/JVI.00674-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Rose R, Nolan DJ, Fogel GB, Barbier AE, Salemi M, et al. HIV-1 evolutionary patterns associated with metastatic Kaposi's Sarcoma during AIDS. Sarcoma. 2016b;2016:4510483. doi: 10.1155/2016/4510483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Fogel GB, Liu ES, Salemi M, McGrath MS. On the physicochemical and structural modifications associated with HIV-1 subtype B tropism transition. AIDS Res. Hum. Retrovir. 2016c;32:829–840. doi: 10.1089/aid.2015.0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J. Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R, Lamers SL, Nolan DJ, Maidji E, Faria NR, Pybus OG, et al. HIV maintains an evolving and dispersed population among multiple tissues during suppressive cART with periods of rapid expansion corresponding to the onset of cancer. J. Virol. 2016 doi: 10.1128/JVI.00684-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M, Lamers SL, Huysentruyt LC, Galligan D, Gray RR, Morris A, et al. Distinct patterns of HIV-1 evolution within metastatic tissues in patients with non-Hodgkins lymphoma. PLoS One. 2009;4:e8153. doi: 10.1371/journal.pone.0008153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Cheng-Mayer C. Evidence that the structural conformation of envelope gp120 affects human immunodeficiency virus type 1 infectivity, host range, and syncytium-forming ability. J. Virol. 1993;67:5635–5639. doi: 10.1128/jvi.67.9.5635-5639.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasca S, Ho SH, Cheng-Mayer C. R5X4 viruses are evolutionary, functional, and antigenic intermediates in the pathway of a simian-human immunodeficiency virus coreceptor switch. J. Virol. 2008;82:7089–7099. doi: 10.1128/JVI.00570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]