Abstract
While genome assembly projects have been successful in a number of haploid or inbred species, one of the main current challenges is assembling non-inbred or rearranged heterozygous genomes. To address this critical need, we introduce the open-source FALCON and FALCON-Unzip algorithms (https://github.com/PacificBiosciences/FALCON/) to assemble Single Molecule Real-Time (SMRT®) Sequencing data into highly accurate, contiguous, and correctly phased diploid genomes. We demonstrate the quality of this approach by assembling new reference sequences for three heterozygous samples, including an F1 hybrid of the model species Arabidopsis thaliana, the widely cultivated Vitis vinifera cv. Cabernet Sauvignon, and the coral fungus Clavicorona pyxidata that have challenged short-read assembly approaches. The FALCON-based assemblies were substantially more contiguous and complete than alternate short or long-read approaches. The phased diploid assembly enabled the study of haplotype structures and heterozygosities between the homologous chromosomes, including identifying widespread heterozygous structural variations within the coding sequences.
Introduction
De novo genome assembly is one of the most fundamental and important computations in genome research1–3. It has led to the creation of high quality reference genomes for many haploid or highly inbred species, and promoted gene discovery, comparative genomics, and other studies4–6. However, most currently available genome assemblies do not capture the heterozygosity present within a diploid or polyploid species7. Instead, most assemblers output a “mosaic” genome sequence that arbitrarily alternates between parental alleles8. Consequently, the variation between the homologous chromosomes will be lost, including allelic variations, structural variations (SVs) or even entire genes present in only one of the haplotypes. Furthermore, heterozygous genome assemblies are typically more fragmented, which has limited the identification and analysis of allele specific expression, long range eQTLs, or other haplotype-specific features9–11. These challenges are becoming more prominent as de novo sequencing projects are shifting towards more heterogeneous samples, such as outbred, wild type diploid, polyploid non-model organisms, or highly rearranged disease samples including human cancers.
While the problem of assembling diploid and polymorphic genomes is not new12, 13, it has not been solved with a universal and scalable solution. The computational methods for diploid assembly that have been proposed tend to produce highly fragmented results, often with contigs averaging just a few hundred bases to several kilobases12, 14, 15. Other approaches such as sequencing both parents and offspring (i.e. trios) 16, haploid sex cells17, clonal fosmid 18 or technologies such as “synthetic long read” 19, 20 are labor intense, costly and are often limited in assembly contiguity. Long-range scaffolding technologies (optical mapping, chromatin assays, etc.) are also often not possible for heterozygous short read genome assemblies as they demand well-assembled contig sequences (minimally contig N50 sizes 50 kbp to 100 kbp long) and can leave unresolved regions (N characters) inside the scaffolds.
SMRT® Sequencing has now become the leading method to finish bacterial genomes and provide high contiguity assemblies for mammalian scale genomes21, 22. The long read lengths, currently averaging ~10 kbp with some approaching 100 kbp, can span through many repetitive elements and assist to resolve more complicated diploid genomes. Nonetheless, currently available assemblers do not take advantage of the long reads to resolve haplotypes. In this paper, we present a new diploid-aware long-read assembler, FALCON, and an associated haplotype-resolving tool, FALCON-Unzip. They are designed to assemble haplotype contigs, “haplotigs”, representing the actual genome in its diploid state with homologous chromosomes independently represented and correctly phased (Fig. 1).
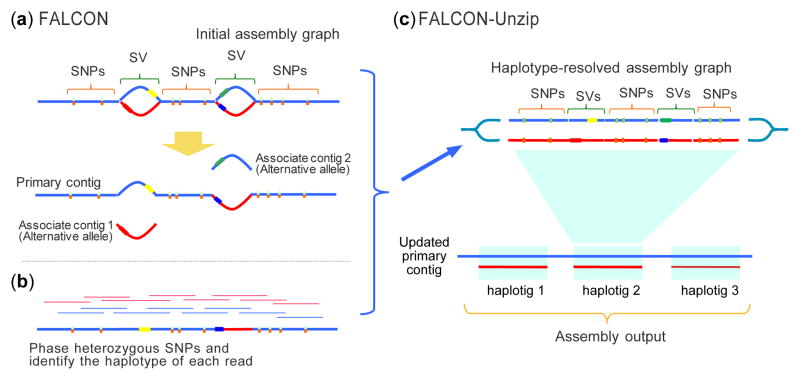
Figure 1. FALCON and FALCON-Unzip overview.
(a) The initial assembly is computed by FALCON, which error corrects the raw reads (not shown) and then assembles using a string graph of the read overlaps. The assembled contigs are further refined by FALCON-Unzip into the final set of contigs and haplotigs. (b) Phase heterozygous SNPs and group reads by haplotype (c) The phased reads are used to open up the haplotype-fused path and generate as output a set of primary contigs and associated haplotigs.
The FALCON assembler follows the design of the previously developed Hierarchical Genome Assembly Process (HGAP)23, although uses greatly optimized components (Supplementary Fig. 1a). FALCON begins by constructing the string graph from the sequencing reads, which contain sets of “haplotype-fused” contigs as well as “bubbles” representing divergent regions between the homologous sequences24 (Fig. 1a). Next, “FALCON-Unzip” finds heterozygous variants within the contigs, and identifies the haplotypes of the reads according to the phasing information among heterozygous positions (Fig. 1b). Phased reads are subsequently utilized for assembling haplotype-specific contigs “haplotigs” and primary contigs. (Fig. 1c, Supplementary Fig. 1b). The resultant haplotigs form the final diploid assembly with phased SNPs and SVs.
To evaluate FALCON-Unzip, we first apply it to a trio of Arabidopsis genomes (Col-0, Cvi-0 and the hybrid Col-0 x Cvi-0) and analyze the results with respect to each other and the TAIR10 genome 25. With the accuracy of FALCON-Unzip established, we assess the performance based on the genome of Vitis vinifera cv. Cabernet Sauvignon, a highly heterozygous outcrossed grape cultivar of major agricultural and economic importance. In the end, we apply FALCON-Unzip to a highly heterozygous wild-type diploid fungus, Clavicorona pyxidata, which has resisted previous short-read assembly approaches.
Results
Sequencing and assembly results of the Arabidopsis trio
We individually sequenced and assembled the inbred Col-0 and Cvi-0 genomes using FALCON (Supplementary Table 1). The contig N50 sizes were 7.4 Mb (Col-0) and 6.0 Mb (Cvi-0), about 10 to 100 times more contiguous than other recently published Arabidopsis assemblies26 (Table 1). Notably, the contiguity approached that of the highly curated TAIR10 assembly (10.9 Mbp contig N50), which had been assembled using expensive BAC-by-BAC sequencing25. The largest FALCON contigs spanned the length of entire chromosome arms (Fig. 2), creating a new high quality draft reference for Cvi-0.
Table 1.
Assembly Results.
| Species | Sample (Total coverage, Read length N50) | Assembler | Sequence | Assembly Size (Mb) | # contigs (scaffods) | N50 size (Mb) | N50 # | N90 size (Mb) | Max Contig Size (Mb) |
|---|---|---|---|---|---|---|---|---|---|
| A. thaliana |
Inbred Col-0 (130x, read N50 = 9 kbp) |
Canu | contigs | 131 | 1102 | 4.573 | 8 | 0.0069 | 11.186 |
|
| |||||||||
| FALCON | p-contigs | 120 | 377 | 7.353 | 7 | 1.278 | 12.197 | ||
|
| |||||||||
|
Inbred Cvi-0 (120x, read N50 = 9 kbp) |
Canu | contigs | 127 | 676 | 4.817 | 9 | 0.364 | 12.393 | |
|
| |||||||||
| FALCON | p-contigs | 120 | 260 | 6.073 | 7 | 1.993 | 14.370 | ||
|
| |||||||||
|
F1 Col-0 x CVI-0 (120x, read N50 = 17 kbp) |
Canu | contigs | 219 | 1897 | 1.554 | 17 | 0.042 | 15.379 | |
|
| |||||||||
| FALCON | p-contigs | 143 | 426 | 7.923 | 6 | 0.387 | 13.386 | ||
| a-contigs | 57 | 551 | 0.146 | 117 | 0.05 | 0.688 | |||
|
| |||||||||
| FALCON-Unzip | p-contigs | 140 | 172 | 7.961 | 7 | 0.504 | 13.319 | ||
| haplotigs | 105 | 248 | 6.920 | 6 | 0.571 | 11.648 | |||
|
| |||||||||
|
F1 Col-0 x CVI-0 (short reads) (60x, 250 bp reads) |
Platanus | scaffolds | 143 | 151779 | 0.0269 | 1290 | 0.00014 | 0.329 | |
|
| |||||||||
| SOAPdenovo, k=93 | scaffolds | 260 | 691629 | 0.00099 | 43570 | 0.00013 | 0.0825 | ||
|
| |||||||||
| V. vinifera |
Cabernet Sauvignon (140x, read N50 = 15 kbp) |
Canu | contigs | 1066 | 14489 | 0.139 | 1778 | 0.03 | 2.211 |
|
| |||||||||
| FALCON | p-contigs | 633 | 1314 | 2.392 | 72 | 0.362 | 14.114 | ||
| a-contigs | 184 | 1164 | 0.278 | 220 | 0.073 | 0.804 | |||
|
| |||||||||
| Falcon Unzip | p-contigs | 591 | 718 | 2.173 | 72 | 0.402 | 14.079 | ||
| haplotigs | 368 | 2037 | 0.779 | 127 | 0.075 | 3.926 | |||
|
| |||||||||
|
Cabernet Sauvignon (short reads) (46x, 100 bp reads) |
SOAPdenovo, k=33 | scaffolds | 1728 | 12879081 | 0.0001 | 791053 | 0.0001 | 0.0368 | |
|
| |||||||||
| SOAPdenovo, k=43 | scaffolds | 507 | 767707 | 0.0019 | 63857 | 0.0018 | 0.0310 | ||
|
| |||||||||
| C. pyxidata |
Clavicorona pyxidata (100x, read N50=16 kb) |
Canu | contigs | 60 | 432 | 0.646 | 16 | 0.045 | 4.390 |
|
| |||||||||
| Falcon | p-contigs | 43 | 133 | 1.49 | 8 | 0.218 | 4.829 | ||
| a-contigs | 12 | 172 | 0.0805 | 41 | 0.037 | 0.407 | |||
|
| |||||||||
| Falcon Unzip | p-contigs | 42 | 82 | 1.484 | 8 | 0.252 | 4.778 | ||
| haplotigs | 24 | 93 | 0.872 | 9 | 0.141 | 2.218 | |||
|
| |||||||||
|
Clavicorona pyxidata (short reads) ( 86x, 100 bp reads) |
Platanus | scaffolds | 39 | 26702 | 0.045 | 225 | 0.0013 | 0.489 | |
|
| |||||||||
| SOAPdenovo, k=19 | scaffolds | 52 | 157941 | 0.00055 | 15065 | 0.00013 | 0.070 | ||
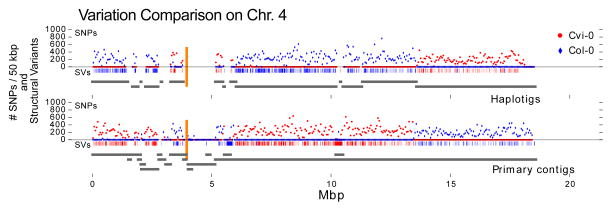
Figure 2. SNP density and Structural Variations in the FALCON-Unzip F1 Arabidopsis assembly.
The plot shows the primary contigs and haplotigs aligned to chromosome 4 of the TAIR reference assembly as grey line segments. Blue and Red colored dots show the number of Col-0 and Cvi-0 specific SNPs, respectively, per 50 kbp region of the assembled contig. The vertical orange lines indicate the centromere locations. The short vertical tick marks above the grey lines indicate the structural variations against Col-0 (blue) and Cvi-0 (red).
When comparing our Col-0 assembly to the TAIR10 assembly, the nucleotide sequence identity was greater than 99.98% (Supplementary Table 2). We applied BUSCO27 to evaluate the assembly completeness by identifying a set of highly conserved plant orthologs in the assembly (Supplementary Table 3). BUSCO identified 914 (95.6%) and 906 (94.8%) genes in the Col-0 and Cvi-0 assemblies, respectively, compared to 915 (95.7%) in the TAIR10 reference. The variations between Col-0 and Cvi-0 assemblies are summarized in Table 2.
Table 2.
Arabidopsis genome assembly comparisons
| HGAP inbreds, Col-0 vs. Cvi-0 | Falcon Unzip haplotigs vs primary contigs | |||
|---|---|---|---|---|
| Variant Type | events | Affected Bases | events | Affected Bases |
| SNP Count | 501,243 | 1,002,486 | 450,680 | 901,360 |
| indel > 50 bp | 1,051 | 882,736 | 966 | 798,438 |
| repeat contraction/expansion > 50 bp | 1,670 | 3,746,572 | 1,479 | 3,130,205 |
| tandem contraction/expansion > 50 bp | 73 | 97,319 | 65 | 85,495 |
| total SV > 50 bp detected | 2,794 | 4,726,627 | 2,510 | 4,014,138 |
| predicted CDS | Col-0:28,176, Cvi-0:27,797 | p:31,679, h:24,808 | ||
| Aligned CDS pairs | 27,424 | 24,808 | ||
| predicted coding sequence SNPs | 183,942 | 367,884 | 147,811 | 295,622 |
| other predicted coding sequence variants | 16,748 | 153,260 | 15,151 | 136,245 |
| local in-frame variants | 5,135 | 82,929 | 4,090 | 66,681 |
| local non in-frame variants | 11,613 | 70,331 | 11,061 | 69,564 |
To assess the performance of assembling a heterozygous genome, we generated and assembled short and long-read sequencing data of the F1 progeny with four leading assembly algorithms (Table 1). Canu (https://github.com/marbl/canu) is an updated genome assembler based on the MHAP overlapper and Celera® Assembler21, and was used to assemble long-read sequence data (Table 1, Supplementary Fig. 2) from the Col-0 x Cvi-0 F1 hybrid sample. The total size of the assembly was 219 Mb, slightly smaller than the expected diploid size of 238 Mb. The high level of polymorphisms, including a SNP rate of ~1/200bp and 1,051 SVs > 50 bp between the strains (Table 2), might cause fragmented assembly as the algorithm is not currently optimized for diploid genomes. Consequently, the contiguity of the F1 assembly was substantially worse (~3 fold less) than the Canu assembly of either inbred parents alone (Table 1).
We evaluated short-read assemblies with SOAPdenovo28 and Platanus29. SOAPdenovo is a widely used general-purpose genome assembler, and Platanus was specifically designed to assemble heterogeneous diploid genomes. The results for both assemblers were significantly less contiguous compared to Canu: SOAPdenovo assembled a total of 260 Mbp with a N50 = 990 bp even after k-mer optimization and error correction (Supplementary Fig. 3). Contigs assembled using Platanus were marginally improved, with an N50 = 26.9 kbp and a total assembly size of 143 Mbp, which is only slightly larger than the haploid genome size.
Unlike most genome assemblers that only generate a single set of contigs as the main assembly results, FALCON generates “primary contigs” (p-contigs) and “alternative contigs” (a-contig) that comprise the genome regions typified by SVs from the p-contigs (Methods). The a-contigs, representing local alternative sequences, spanned a total of 57 Mbp (~40% of the p-contigs) with a N50 = 146 kbp. Thus, FALCON alone produced 84% of the estimated 238 Mbp diploid genome. After the initial assembly, the FALCON-Unzip algorithm utilizes the heterozygosity information within the initial primary contigs for haplotype phasing (Fig. 1b, Supplementary Note). With the phasing information from the raw reads, FALCON-Unzip generates a subsequent set of p-contigs and the final haplotig set (h-contigs) that represents more contiguous haplotype specific sequence information than the a-contigs (Fig. 1c). After the “unzipping” process, the total size of the p-contigs was 140 Mbp (N50 = 7.96 Mbp) and the total size of the haplotigs was 105 Mbp (N50 = 6.92 Mbp). FALCON-Unzip restored the contiguity that was present in the assemblies of the individual inbred parental genomes (Table 1), but as a phased diploid genome.
Comparison of the F1 assembly of FALCON-Unzip, Platanus, and SOAPdenovo directly to the TAIR10 reference is detailed in the Supplementary Note (Supplementary Fig. 4, Supplementary Table 4). Overall, the variants from the FALCON-Unzip assembly captured 89% of the Platanus variants and 90% of the SOAP variants at a stringent requirement of the exact same variant type, size, and genomic location. However, the Platanus and SOAP assemblies captured only 37% and 1% of the FALCON-Unzip variants, respectively.
Col-0 x Cvi-0 F1 haplotig phasing quality analysis
We aligned the p-contigs and the haplotigs to the two parental inbred assemblies to evaluate the accuracy of haplotype separations. Ideally, each haplotig should be identical to one of the parental haplotypes and show variations against the other. We observe that most of the haplotigs only show SNPs or SVs to one of the parental genomes indicating that the phasing approach works accurately (Fig. 2, Supplementary Fig. 5). We assessed the accuracy by computing the ratio of differences (e.g. SNPs) to either of the parental assemblies within each haplotig (Supplementary Table 5). For the largest six haplotigs spanning 50% of the genome, the minority SNP percentages are all lower than 0.2%. The small minority SNP ratio represents either a small number of (1) local phasing errors, (2) incorrect SNP calls, and/or (3) assembly base errors, but demonstrates there are no significant segmental switching errors. Only 9 haplotigs (~2.5% of all haplotig bases) show a minority SNP ratio over 10%, and are generally associated with repetitive or low heterozygous regions. Finally, we aligned the haplotigs of the FALCON-Unzip assembly to analyze its ability to incorporate SNPs. We identified 450,680 SNPs among the haplotigs, compared to 501,243 found by aligning the Col-0 and Cvi-0 assemblies. Thus, FALCON-Unzip phased 85.7% of all SNPs and 91.9% of all SVs directly from the shotgun sequence assembly.
Col-0 x Cvi-0 F1 coding sequence prediction evaluation
In the F1 FALCON-Unzip assembly results, we estimated the overall base-to-base concordance rate at about 99.99% (QV40 in Phred scale). The insertion and deletion (indel) concordances to the parental lines were lower (about QV40) than the SNP concordance rate (about QV50), with most residual errors concentrated in long homopolymer sequences (Supplementary Table 6, Supplementary Fig. 6). We evaluated the impact of such errors on coding sequence prediction with AUGUSTUS (Supplementary Note, Supplementary Table 7). Interestingly, AUGUSTUS30 aligned 97% of all CDS of TAIR10 to our assembly without any indels, and the vast majority of BUSCO genes (877) were even found to be phased.
Vitis vinifera sequencing and diploid assembly results
We next assessed the performance of FALCON-Unzip on the genome of V. vinifera cv. Cabernet Sauvignon, which is an F1 of two very distinct cultivars, Cabernet Franc and Sauvignon Blanc and one of the world’s most widely cultivated red wine grape varieties. The long reads (Supplementary Table 1) were assembled using Canu, FALCON, and FALCON-Unzip (Table 1). FALCON-Unzip yielded the most contiguous assembly of 590 Mbp (N50 = 2.17 Mbp) and generated a total of 368 Mbp of associated haplotigs (N50 = 779 kbp). Both primary and associated contigs displayed overall high macro-synteny with the current V. vinifera genome reference (PN4002431; Supplementary Fig. 7). The total p-contig size was larger than the estimated genome size of V. vinifera (~500Mbp31). This suggests that in some cases FALCON-Unzip underestimated the alternative haplotype sequences, because of high heterozygosity between homologous regions. An analysis of synteny between different p-contigs to determine the extent of inclusion of redundant regions identified a total of 25 Mbp of syntenic blocks in the primary assembly (Supplementary Note).
Compared to Arabidopsis, the V. vinifera genome has more repeats and higher heterozygosity that makes it more challenging to assemble in general. Canu generated an assembly of 1,006 Mbp, which is roughly twice of the haploid genome size with a significantly smaller N50 = 139 kbp. Even with optimized k-mer sizes (33bp – 43bp), SOAPdenovo’s scaffold N50 size was smaller than 2 kbp and the contig N50 < 1 kbp (Supplementary Fig. 3). The Platanus results were unacceptably incomplete, with less than 1% of the expected genome size reported, most likely due to the limited available coverage. Nevertheless, even with high coverage levels (1,577 million reads) and multiple libraries, other published assemblies of different grape cultivars report contig N50 sizes of at most 41 kbp using Platanus32.
To assess completeness of the assemblies we used BUSCO as well as aligned the 29,971 mRNA sequences annotated from the current V. vinifera genome reference PN40024. Both approaches highlighted the completeness of the gene space in the FALCON-Unzip assembly (Supplementary Table 3 and 8). Furthermore, overall 80% of the 956 BUSCO genes and 16,981 of the 29,971 predicted complete genes from PN40024 were phased in the assembly. In contrast, less than 15% of the 956 BUSCO proteins were found within the most contiguous short-read assemblies suggesting that these assemblies are not only highly fragmented, but also markedly incomplete (Supplementary Table 3).
Clavicorona pyxidata sequencing and assembly results
To demonstrate the generality of the FALCON-Unzip approach to wild type heterozygous genomes, we apply the same assembly and analysis to C. pyxidata, a common coral fungus that grows on hardwoods across North America (haploid size ~42 Mbp). FALCON-Unzip produced the most contiguous assembly, followed by Canu (~2-fold less contiguous), and then followed distantly by the short-read assemblies (30 to >100 fold less contiguous) (Table 1). In lieu of a reference, we evaluated the assemblies using BUSCO and genomic sequencing data (SRA accession: SRR1800147, 86X, 150 bp reads). The results are summarized in Supplementary Note and Supplementary Table 3.
In contrast to the V. vinifera genome, the C. pyxidata genome has significantly skewed rates of heterozygosity, and about 50% is essentially homozygous. This suggests naturally occurring inbreeding or other selective pressures to limit variation in these regions. Different levels of heterozygosity between homologous chromosomes, seen in all three genomes, also affect the assembly sizes. We discuss such effect in detail in Supplementary Note and Supplementary Fig. 8–10.
For evaluating the phasing accuracy, we used the 150bp paired-end short-read data and called phased SNPs relative to the primary contigs with FreeBayes33 and HapCut34 (Supplementary Table 9). Due to the insert size limit of the short-read dataset, the phasing data only covered about 23% (9.72 Mbp) of the genome, but nearly all phased blocks, 96% to 98% depending on the variant call quality thresholds, are fully concordant with the FALCON-Unzip assembly (Supplementary Table 9). Comparison of homologous alleles within the genome with public available RNA Sequencing data (SRA accession SRR1589642) identified several candidate differentially expressed alleles (Supplementary Fig. 11).
Discussion
Genome sequencing projects aim to generate a high quality reference assembly that can serve as a foundation for various downstream analyses, e.g. gene finding, variant identification, or comparative and functional assays. While successful in a number of haploid or inbred species, one of the current main challenges for the genomics community is generating genome assemblies for non-inbred heterozygous genomes, which represent the vast majority of samples to be sequenced for biomedical, agricultural, or evolutionary studies. For heterozygous diploid genomes, we demonstrated FALCON and FALCON-Unzip can assemble PacBio SMRT Sequencing data into highly accurate, contiguous, and correctly phased primary contigs and haplotigs. Such haplotype specific assemblies present a true biological representation of the genome and empower study of haplotype structures and heterozygous variants, e.g. SVs and SNPs, between the homologous chromosomes not normally possible from other assemblers.
In all three genomes studied here, the FALCON/FALCON-Unzip assembly was significantly more contiguous (2 to 3 fold) than alternative long read assemblers of the same data, and much better (30 to >100 fold) than state-of-the-art short read assemblies. In the Arabidopsis F1-hybrid assembly, we evaluated the haplotype phasing accuracy by comparing the F1 assembly to the parental inbred genomes and determined that the haplotigs nearly perfectly matched one of their parental genomes with only ~2.5% of incorrectly phased sequences. While already accurate, in future work, we aim to further improve the phasing accuracy by analyzing the local assembly graph to predict hard-to-resolve regions and potential errors in the assembly. We showed that the small frequency of residual sequencing errors (<0.1%) had almost no effect on the identification of gene sequences. In the other two assemblies, we demonstrated greatly improved diploid representations of core genes, e.g. >90% in Arabidopsis F1 genome, from the FALCON/FALCON-Unzip assembly, and accurate phasing measured using orthogonal data (Supplementary Table 9).
Fundamentally both the raw sequencing read lengths and error rates may affect the haplotype and consensus accuracies. The genome complexity, especially the rate of heterozygous positions and the repetitive sequences, is also a major factor impacting the performances. Most haplotype-phasing algorithms utilize heterozygous SNPs and ignore any SVs. In contrast, FALCON-Unzip is designed to combine SNPs and SVs to separate haplotype information beyond what either alone provides to construct haplotype specific contigs. With long read lengths from SMRT Sequencing and increased levels of heterozygosity, this allows us to almost fully resolve both haplotype chromosomes for practically the entire Arabidopsis F1 genome with high contiguity. The other two genomes chosen for this study highlight some of the additional complexities that are possible for diploid genomes. In V. vinifera, we find homologous regions having very high rates of variations, likely from the out-crossing nature of the organism, while in C. pyxidata we discovered extended regions of unexpectedly low heterozygosity suggesting regions of increased selective pressures or complex naturally occurring inbreeding. While future increase of the read lengths will improve the separation of the haplotypes, we can already begin to utilize the assembly output to understand and represent the variations of heterozygosity within wide range of diploid genomes (Supplementary Table 10). The assembly results presented here were solely from PacBio SMRT Sequencing, but can in principle also be improved with other types of data, especially long range scaffolding data, and extended to higher ploidy genomes in the future.
The mosaic genome sequences that are commonly assembled today do not contain all of the genetic information of the variants between haplotypes. This makes it, among other things, difficult to probe the impact of epigenetic and differential gene expression and can exacerbate “reference-bias” when remapping sequencing data35. With FALCON-Unzip, however, almost the entire heterozygosity information is captured in the p-contigs and haplotigs, so the question of how haplotype specific variations affect gene expression, methylation patterns, or other regulatory interactions can be examined further. More systematic study of phased diploid references will expose the detailed cis-regulatory mechanisms of differential expression in diploid genomes to improve our general understanding of the biology beyond haploid genomes. Looking forward, with the advances of the SMRT sequencing technology, new algorithm and software development, we expect that there is a wide field of new opportunities for understanding diploid and polyploid genomic diversity and its impact on genome annotation, gene regulation and evolution.
Online Methods
DNA isolation and library preparation
For the Arabidopsis sample preparation, to minimize chloroplast DNA contamination, nuclei were isolated from leaf tissue as previous described36. Genomic DNA was isolated using standard purification columns and protocols (Qiagen®). For grapevine DNA extraction, young leaves (~1 cm diameter) were collected from Vitis vinifera cv. Cabernet Sauvignon clone 08 at Foundation Plant Services (UC Davis, Davis, CA). Plant tissue (1 g) was ground to a powder in a mortar containing liquid nitrogen. Ten mL of pre-warmed (65 °C) extraction buffer (300 mM Tris-HCl pH 8.0, 25 mM EDTA pH 8.0, 2 M NaCl, 2% (w/v) soluble PVP (MW 40000), 2% CTAB, 2% 2-mercaptoethanol) was added and the suspension was homogenized by inversion and incubated (65 °C) for 30 min in a water bath, mixing by inversion (every 5 min). Plant debris was removed by centrifugation (5000 rpm) for 5 min at room temperature and the supernatant was transferred into a new tube. Equal volume of chloroform:isoamyl alcohol (CIA, 24:1 v/v) was added and mixed by inversion for 5 min. Aqueous phase was segregated by 10 min centrifugation (5000 rpm) at room temperature and transferred gently into a new tube. RNase A was added to the sample (2 μg) and was incubated (37°C) for 30 min. After RNAse treatment, equal volume of CIA was added and centrifuged as above. 0.1 volume of 3 M NaOAc pH 5.2 and an equal volume of isopropanol were added for DNA precipitation, sample was mixed by inversion and then incubated (− 80 °C) for 30 min. DNA was collected by centrifugation (5000 rpm) for 30 min and the pellet was washed twice with 3 mL of 70 % ethanol. After 10 min centrifugation (5000 rpm), DNA pellet was air-dried at room temperature and resuspended in 500 μl of nuclease-free water. DNA quality was evaluated by pulse-gel electrophoresis, and quantity was determined using the Qubit fluorometer.
Shearing of the DNA was performed either with G-tubes (Covaris®) or by passage through a small bore needle37 to average size of 15 kbp to 40 kbp. The needle method was used during an evaluation of shearing techniques. However, both shearing methods produced libraries of comparable quality and sequencing performance. Sheared DNA was enzymatically repaired and converted into SMRTbell™ libraries prepared as described by the manufacturer (Pacific Biosciences). Non SMRTbell DNA was removed by exonuclease treatment. Finally, a BluePippin™ preparative electrophoresis purification step was performed (Sage Sciences) on the library to select insert sizes ranging from 7 to 50 kbp or from 15 to 50 kbp depending on the sequencing experiment. These size-selected libraries were used in subsequent sequencing steps.
Sequencing methods
Sequencing was performed on the PacBio RS II instrument as per the manufacturer’s recommendations. The Col-0 and Cvi-0 inbred Arabidopsis data sets were collected using P4-C2 chemistry with 4 hour movie lengths. The F1 Col-0 x Cvi-0 and the C. pyxidata and the V. vinifera cv Cabernet Sauvignon samples were run with P6 chemistry and 6 hour data collection movies.
Raw long-read error correction
All raw long-read sequences were aligned to each other using “daligner38” executed by the main script of the FALCON assembler. The overlap data and raw subreads are then processed to generate consensus sequences. The consensus-calling algorithm (FALCON-sense) was designed to preserve the information from heterozygous single nucleotide polymorphisms (SNP) and is described in detail in the Supplementary Note (Section “Updated FALCON consensus algorithm” and Supplementary Fig. 12).
Initial “haplotype-fused” assembly with a collapsed diploid-aware contig layout
After the error correction step, FALCON identifies the overlaps between all pairs of the pre-assembled error corrected reads. The read overlaps were used to construct a directed (in contrast to bi-directed) string graph following the Myers’ algorithm39. For diploid genomes with high heterozygosity, the string graph typically contains linear chains of “bubbles” (Supplementary Fig. 1b and Supplementary Fig. 13). We can decompose such linear chains into “simple” and “compound” paths where: a simple path is a path where there is no internal branching node and it also has unique source node and sink node, and a compound path is a collection of edges that represents a bubble with unique source and sink in the assembly graph. The algorithm for constructing such compound paths is described in the Supplementary Note. The non-branched collection of compound paths and simple paths are further combined to create unitigs. Genome repeats, sequencing errors or missing overlaps can introduce spurious unitigs. Empirically derived heuristic rules were applied to remove these artifacts and layout the primary contigs and the associated contigs. The graph reduction process is detailed in Supplementary Fig. 14. We call the final assembly graph the “haplotype-fused assembly graph G(f).”
Mapping and phasing the raw reads
In the draft assembly, each contig is simply a tiling sequence from the subsequences of a set of error corrected reads. Some of the raw reads have not yet been associated with any contigs. For example, if a read is “contained” within other reads (overlaps completely to a substring of another read), it is not used in constructing the first draft of the contigs. There are two strategies for identifying the raw-read to contig associations: (1) re-map all raw-reads to the contigs and find the best alignments; or (2) trace the read overlapping information to find out where a raw-read is most likely to be associated. FALCON-Unzip applies strategy (2), to avoid the time penalty for the re-mapping process, as the overlap information already exists. For each raw-read, FALCON-Unzip examines all overlapping reads. If a read is uniquely associated with one contig, then the raw-read is assigned to that contig. If there are multiple contigs associated with a read, it scores the matching contigs by the overlap lengths. In this case, a read is assigned to a target contig with the highest sum of overlap lengths.
For each primary contig, we collect all raw-reads associated with the primary contig and its associated contigs. We align the raw reads to the contigs with the BLASR aligner40 and call heterozygous SNPs (het-SNPs) by analyzing the base frequency of the detailed sequence alignments. A simple phasing algorithm was developed to identify phased SNPs (see Supplementary Note and Supplementary Fig. 15). Along each contig, the algorithm assigns phasing-blocks where chained phased SNPs can be identified. Within each block, if a raw read contains a sufficient number of het-SNPs, it assigns a haplotype phase for the read unambiguously. Combined with the block and the haplotype phase information, it assigns a “block-phase” tag for each phased read in each phasing block. Some reads might not have enough phasing information. For example, if there are not enough het-SNP sites covered by a read, it assigns a special “un-phased tag” for each un-phased read.
Overview of the algorithm constructing haplotype specific contigs
The algorithm to construct the haplotype specific contigs (haplotigs) is summarized in Fig. 1 and Supplementary Fig. 13. Briefly, for each contig c, it constructs a haplotype-specific assembly graph from all reads that mapped to it, denoted as Hc, by ignoring the overlaps between any two reads from the same block but different phases. It then combines this graph Hc to the fused assembly sub-graph that contains the paths of contig c to construct a complete contig sub-graph . Unlike the initial subgraph , where some reads are masked out by reads from different phases, the complete contig sub-graph rescues such masked-out reads and have complete read representation from both haplotypes.
In the fused assembly graph , there is a path that is corresponding to the original contig c starting from node s to node t. It is desirable to generate a new locally phased contig that also starts from the same node s and ends at the same node t as new primary contig pc. While such primary contig pc may not be fully phased end-to-end, the collection of pc of all contig c can serve as a haploid assembly representation with annotated locally phased regions. And, the variations between the two haplotypes can be identified by aligning other haplotigs to the primary contigs. Once pc is identified, the corresponding edges of pc in are removed. It also removes all other edges connecting different phases of the same block. Namely, it constructs a subgraph of by removing edges which are already in pc or connect distinctly phased nodes. We identify all linear paths within as the haplotigs hc,i=1..n, where n is the total number of haplotigs associated with the primary contig. Some of the haplotigs might be caused by missing overlaps or sequence errors. The haplotig sequences are aligned to the primary contig. If the alignment identity is high and no phased-reads are associated with the haplotig, the haplotig will be marked as duplicated and removed. Note that a haplotig may contain multiple haplotype-phased blocks. For example, haplotype-specific SVs may affect the initial mapping such that the phasing algorithm cannot connect two neighboring blocks. However, reads from different phasing blocks might be uniquely overlapped if the SVs between the haplotypes are distinguishable. Such haplotype-specific overlaps can connect broken haplotype-phased blocks into to larger haplotigs.
Polishing partially phased primary contigs and their associated haplotigs
Conceptually, FALCON-Unzip generates one new primary contig pc and n haplotigs hc,i=1..n from the original assembly graph of the contig c. It uses the phasing information to decide whether a phased read belongs to the primary contig pc or one of the haplotigs hc,i=1..n. Each un-phased read may also contain structural level variations that are the same as in a particular haplotig. In such case, by examining the overlaps between the read to those in the haplotigs, it can find the best hit from the un-phased read to one haplotig. In the end, each raw-read will be augmented with the information which haplotig or primary contig it belongs to and will be mapped accordingly. This ensures that the haplotig consensus is generated from the appropriate reads belonging to the correct haplotype. Finally, it uses the Quiver algorithm23 to remove residual errors in the haplotig consensus from the haplotype specific alignments.
FALCON-Unzip outputs a set of partially phased primary contigs (p-contigs) and the associated haplotigs (h-contigs) for each primary contig. The phased regions in the primary contig can be identified by simply aligning the associated haplotigs to the primary contig or directly examine the assembly graph identifying the anchoring nodes from the haplotigs to the primary contig.
Software Availability
FALCON and FALCON-unzip are written in C and Python. FALCON and its dependences are hosted open-source on GitHub® (https://github.com/PacificBiosciences/falcon). FALCON-Unzip is also hosted open-source on GitHub®(https://github.com/PacificBiosciences/FALCON_unzip). The specific git repositories of the various modules used for generating the assemblies presented in this paper are listed in the supplementary material. We have also prepared an Amazon Web Services EBS volume that contains all of the preconfigured software and example C. pyxidata dataset (See Supplementary Note for a walkthrough).
Supplementary Material
Acknowledgments
The sequencing of the Cabernet Sauvignon genome was supported in part by a gift of the J. Lohr Vineyards and Wines to DC. We also like to thank Felipe Simao Neto providing early release BUSCO plant data set. Clavicorona pyxidata DNA was provided by L. Nagy (Institute of Biochemistry Biological Research Centre of the Hungarian Academy of Sciences). We thank Joseph D. Puglisi, Florian Jupe, Alex Copeland and Aaron Wenger for reading and critique of the manuscript. The project was supported in part by National Institutes of Health award (R01-HG006677) and by National Science Foundation awards (DBI-1350041 and IOS-1237880 to MCS; MCB 0929402 and MCB 1122246 to J.R.E)/J.R.E is an investigator of the Howard Hughes Medical Institute and Gordon and Betty Moore Foundation (GBMF 3034).
Footnotes
Data Accession
Arabidopsis data: PRJNA314706
V. vinifera cv. Cabernet Sauvignon: PRJNA316730
Clavicorona pyxidata: PRJNA336540
The assemblies can be downloaded from https://downloads.pacbcloud.com/public/dataset/PhasedDiploidAsmPaperData/FUNZIP-PhasedDiploidAssemblies.tgz
Author Contributions
C-S. C., P.P., A.C., D.R.R., and M.C.S. conceived the idea of FALCON/FALCON-Unzip assembler. C-S. C, P.P., F.J.S. M.N., G.T.C., D.R.R., D.C, and M.C.S. designed the experiments and perform the analysis. P.P., D.C, D.R.R, M.C.S collected the sequencing data. R.O., C.L. and J.R.E constructed the Col-0 x Cvi-1. A.C., R.O., R. F-B., A. M-C., G.R.C., M.D., C.L., J.R.E., D.C. collected the samples and prepared DNA for sequencing. C-S. C., P.P., F.S., M.N. G.T.C., D.C., D.R.R., M.C.S. wrote the manuscript. C-S. C. and C. D. implemented the computer code.
Competing Financial Interests
C.-S. C., P.P., G. C., C. D., and D. R. are employees and shareholder of Pacific Biosciences, a company commercializing DNA sequencing technologies.
References
- 1.Goffeau A, et al. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 2.Myers EW, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 3.Bonfield JK, Smith KF, Staden R. A new DNA sequence assembly program. Nucleic acids research. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stamatoyannopoulos JA, Guigó Serra R, Djebali S, Lagarde J, Adams LB. An encyclopedia of mouse DNA elements (Mouse ENCODE) 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consortium GP. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earl D, et al. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome research. 2011;21:2224–2241. doi: 10.1101/gr.126599.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Church DM, et al. Extending reference assembly models. Genome biology. 2015;16:13. doi: 10.1186/s13059-015-0587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tewhey R, Bansal V, Torkamani A, Topol EJ, Schork NJ. The importance of phase information for human genomics. Nat Rev Genet. 2011;12:215–223. doi: 10.1038/nrg2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henson J, Tischler G, Ning Z. Next-generation sequencing and large genome assemblies. Pharmacogenomics. 2012;13:901–915. doi: 10.2217/pgs.12.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nature methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinson JP, et al. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 2005;15:1127–1135. doi: 10.1101/gr.3722605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nature genetics. 2012;44:226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kajitani R, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome research. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roach J, et al. Chromosomal haplotypes by genetic phasing of human families. Am J Hum Genet. 2011;89:382–397. doi: 10.1016/j.ajhg.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkness E, et al. Sequencing of isolated sperm cells for direct haplotyping of a human genome. Genome Res. 2013;23:826–832. doi: 10.1101/gr.144600.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitzman J, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol. 2011;29:59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCoy RC, et al. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly-repetitive transposable elements. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mostovoy Y, et al. A hybrid approach for de novo human genome sequence assembly and phasing. Nature Methods. 2016 doi: 10.1038/nmeth.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berlin K, et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nature biotechnology. 2015 doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- 22.Gordon D, et al. Long-read sequence assembly of the gorilla genome. Science. 2016;352:aae0344. doi: 10.1126/science.aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 24.Fasulo D, Halpern A, Dew I, Mobarry C. Efficiently detecting polymorphisms during the fragment assembly process. Bioinformatics. 2002;18:S294–S302. doi: 10.1093/bioinformatics/18.suppl_1.s294. [DOI] [PubMed] [Google Scholar]
- 25.Initiative AG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. nature. 2000;408:796. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 26.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 28.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome research. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajitani R, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 31.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 32.Patel S, Swaminathan P, Fennell A, Zeng E. Bioinformatics and Biomedicine (BIBM), 2015 IEEE International Conference on; IEEE; 2015. pp. 1771–1773. [Google Scholar]
- 33.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012 arXiv preprint arXiv:1207.3907. [Google Scholar]
- 34.Bansal V, Bafna V. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics. 2008;24:i153–159. doi: 10.1093/bioinformatics/btn298. [DOI] [PubMed] [Google Scholar]
- 35.Degner JF, et al. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu YG, Whittier RF. Rapid preparation of megabase plant DNA from nuclei in agarose plugs and microbeads. Nucleic acids research. 1994;22:2168. doi: 10.1093/nar/22.11.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayward GS. Unique double-stranded fragments of bacteriophage T5 DNA resulting from preferential shear-induced breakage at nicks. Proceedings of the National Academy of Sciences. 1974;71:2108–2112. doi: 10.1073/pnas.71.5.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers G. Algorithms in Bioinformatics. Springer; 2014. pp. 52–67. [Google Scholar]
- 39.Myers EW. The fragment assembly string graph. Bioinformatics. 2005;21:ii79–ii85. doi: 10.1093/bioinformatics/bti1114. [DOI] [PubMed] [Google Scholar]
- 40.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.