Summary
We identified a choline, betaine and carnitine transporter, designated Cbc, from Pseudomonas syringae and Pseudomonas aeruginosa that is unusual among members of the ATP-binding cassette (ABC) transporter family in its use of multiple periplasmic substrate-binding proteins (SBPs) that are highly specific for their substrates. The SBP encoded by the cbcXWV operon, CbcX, binds choline with a high affinity (Km, 2.6 μM) and, although it also binds betaine (Km, 24.2 μM), CbcXWV-mediated betaine uptake did not occur in the presence of choline. The CbcX orthologue ChoX from Sinorhizobium meliloti was similar to CbcX in these binding properties. The core transporter CbcWV also interacts with the carnitine-specific SBP CaiX (Km, 24 μM) and the betaine-specific SBP BetX (Km, 0.6 μM). Unlike most ABC transporter loci, caiX, betX and cbcXWV are separated in the genome. CaiX-mediated carnitine uptake was reduced by CbcX and BetX only when they were bound by their individual ligands, providing the first in vivo evidence for a higher affinity for ligand-bound than ligand-free SBPs by an ABC transporter. These studies demonstrate not only that the Cbc transporter serves as a useful model for exploring ABC transporter component interactions, but also that the orphan SBP genes common to bacterial genomes can encode functional SBPs.
Introduction
Transporters in the ATP-binding cassette (ABC) family are vital among living organisms due to their role in translocating substrates ranging from ions, sugars and amino acids to high-molecular-weight proteins (Biemans-Oldehinkel et al., 2006; Davidson et al., 2008). The core transporter contains two transmembrane spanning domains (TMDs) that provide a substrate-specific pathway across the membrane, and two nucleotide-binding domains (NBDs, or ATPases) that power the translocation (Davidson et al., 2008). Prokaryotic ABC importers generally employ a third subunit, a substrate-binding protein (SBP), which is either free in the periplasm in Gram-negative bacteria or anchored in the external surface of the membrane in Gram-positive bacteria. SBPs are the determinants of substrate specificity and are indispensable to the import function of prokaryotic ABC transporters.
All characterized ABC transporter-associated SBPs involved in import consist of two lobes, or domains, that are linked by a flexible hinge (van der Heide and Poolman, 2002). The relative movement of these lobes creates open and closed conformations and allows SBPs to capture their ligands through a mechanism termed a Venus flytrap that involves very rapid ligand binding (Miller et al., 1983). The ligand-bound SBP is thought to transmit a signal via the TMD to the NBD on the other side of the membrane, probably through co-ordinated events including ATP binding and hydrolysis, to deliver the substrate into the cell (Orelle et al., 2008).
Structural and kinetic analyses of the ABC transporters crystallized to date have demonstrated that a single, monomeric SBP typically associates with the core transporter such that each SBP lobe interacts with one of the two subunits of the TMD (Davidson et al., 2008). These interaction sites, however, differ among transporters and range from simple salt bridges formed between amino acid residues of the SBP and the TMD (Braun and Herrmann, 2007; Rees et al., 2009) to extensive interactions such as a large periplasmic loop on the TMD that keeps the SBP closely associated even in the absence of substrate; this loop likely communicates substrate availability (Daus et al., 2009; Grote et al., 2009). Under optimal conditions in a Gram-negative bacterium, periplasmic SBPs can reach concentrations as high as 1 mM, or 50-fold more than the core transporter concentration (Bohl et al., 1995), thus potentially improving bacterial scavenging of substrates at low concentrations. Following rapid binding of the substrates by SBPs in the periplasm, the core transporter likely discriminates between the substrate-bound and substrate-free SBPs, preferentially allowing the bound SBPs to dock prior to substrate translocation.
Quaternary ammonium compounds (QACs), including glycine betaine, choline and carnitine, are found in many microbial environments where they can provide osmotic protection and nutritional benefit. Many transporters that take up QACs for osmoprotection are ABC transporters. For example, the Escherichia coli transporter ProU (ProVWX), which was characterized more than two decades ago, transports QACs to combat hyperosmotic stress (May et al., 1986). Since then many osmoregulatory QAC transporters have been characterized (Wood et al., 2001). In contrast, few QAC transporters are known that function primarily in uptake for nutrition. The partially characterized ChoXWV choline transporter from Sinorhizobium meliloti, and its orthologue GbcXWV from Rhizobium leguminosarum bv. viciae 3841, are the only known QAC transporters for catabolism (Dupont et al., 2004; Fox et al., 2008). While characterizing the osmoprotective QAC transporters in the phytopathogen Pseudomonas syringae, we identified the osmoregulatory QAC transporters OpuC (Chen and Beattie, 2007) and BetT (Chen and Beattie, 2008) but also found evidence for a transporter that functions in QAC uptake for catabolism.
The goal of this work was to characterize this catabolism-associated QAC transporter in P. syringae as well as in Pseudomonas aeruginosa. During our investigation, we identified the CbcXWV (choline/betaine/carnitine) transporter and found that the Cbc core transporter is atypical among ABC transporters in its ability to interact with at least three highly substrate-specific SBPs. A few ABC transporters are known to employ multiple SBPs with broader specificity; these include ArgT-HisJQMP of Salmonella typhimurium, which uses the two amino acid-binding SBPs HisJ and ArgT (Higgins and Ames, 1981; Wolf et al., 1996), and the oligopeptide transporter Opp of Borrelia burgdorferi, which uses as many as three chromosomally encoded SBPs (OppA1–3) and two plasmid-borne SBPs (OppA4 and OppA5) (Wang et al., 2004). Transport systems that involve multiple SBPs with distinct substrate specificity provide an opportunity to better understand some unanswered questions about the mechanism of ABC transport, including the mechanism of recognition between the core transporter and its cognate SBPs. Here, our in vivo studies involving simultaneous expression of the Cbc-associated SBPs and the Cbc core transporter provide the first in vivo demonstration of a core transporter preferentially interacting with ligand-bound versus ligand-free SBPs.
Results
The Cbc transporter of Pseudomonas spp. is required for the uptake of QACs under low-osmolarity conditions
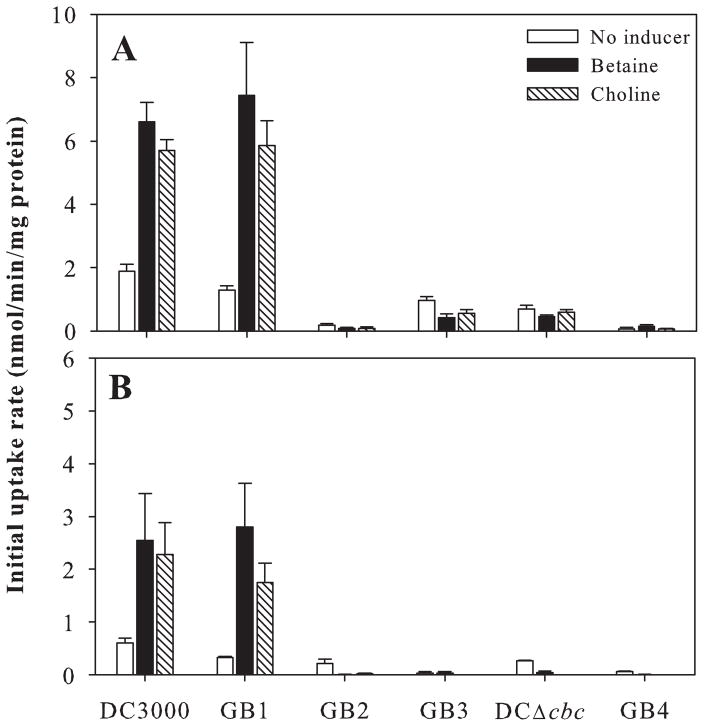
Previously, we described two QAC transporters in P. syringae: the ABC transporter OpuC, which transports glycine betaine, choline, carnitine and acetylcholine (Chen and Beattie, 2007), and the betaine/carnitine/choline family transporter BetT, which transports choline and acetylcholine (Chen and Beattie, 2008). Whereas OpuC and BetT are functional under hyperosmotic conditions, the loss of a third P. syringae locus, PSPTO_0464-PSPTO_0462, reduced choline uptake under low-osmolarity conditions (Chen and Beattie, 2007), suggesting a role in choline uptake for catabolism. We generated mutants of P. syringae pv. tomato strain DC3000 that lacked this locus, DCΔcbc, as well as the opuC and/or betT loci, strains GB1–GB4 (Table 1), and examined the uptake properties of these mutants. In the absence of choline and betaine as potential inducers during cell growth, DC3000 cells transported 1.8 and 0.6 nmol min−1 (mg protein)−1 of choline and betaine, respectively, when each was provided at a final concentration of 10 μM (Fig. 1). After growth with 10 mM choline or betaine as inducers, the initial uptake rate for these compounds increased three- to fourfold. GB1, a double mutant lacking opuC and betT, similarly exhibited a substrate-induced increase in uptake, whereas mutants GB2, GB3 and DCΔcbc, which all lack PSPTO_0464-PSPTO_0462, did not. This indicates that the transporter encoded by this locus was responsible for the inducible uptake. The triple transporter mutant GB4 did not transport detectable amounts of choline or betaine (Fig. 1), nor did it grow on these as sole C sources (Fig. S1), demonstrating that these three loci encode the full complement of transporters for choline and betaine uptake in DC3000. We designated the transporter encoded by PSPTO_0464-PSPTO_0462 as the Cbc transporter because of its ability to transport choline and betaine as well as carnitine, described below, and designated the genes cbcXWV for the SBP, TMD and NBD respectively.
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Description/relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| Acella | Host supporting pET15b-based expression | EdgeBio |
| MKH13 | ΔputPAΔproP2 ΔbetTIBA ΔproU::spc; Spr | Kempf and Bremer (1995) |
| Pseudomonas syringae pv. tomato | ||
| DC3000 | Wild type; RfR | Moore et al. (1989) |
| DCΔcbc | DC3000 ΔcbcXWV; Rfr | This work |
| GB1 | DC3000 ΔbetT ΔopuCA; Rfr | Chen and Beattie (2008) |
| GB2 | DC3000 ΔbetT ΔcbcXWV; Rfr | Chen and Beattie (2008) |
| GB3 | DC3000 ΔopuCA ΔcbcXWV; Rfr | Chen and Beattie (2008) |
| GB4 | DC3000 ΔbetT ΔopuCA ΔcbcXWV; Rfr | Chen and Beattie (2008) |
| Pseudomonas syringae pv. syringae | ||
| B728a | Wild type; RfR | Loper and Lindow (1987) |
| B0028Ko | B728a Psyr_0028::Tet; Rfr | This work |
| B2916Ko | B728a caiX::Tet; Rfr | This work |
| BT | B728a ΔbetT ΔopuC ΔcbcXWV; Rfr | This work |
| BT0028Ko | B728a ΔbetT ΔopuC ΔcbcXWV Psyr_0028::Tet | This work |
| BT2916Ko | B728a ΔbetT ΔopuC ΔcbcXWV caiX::Tet | This work |
| BT3758Ko | B728a ΔbetT ΔopuC ΔcbcXWV betX::Tet | This work |
| Pseudomonas aeruginosa | ||
| PA14 | Wild type | Rahme et al. (1995) |
| PA14ΔcaiX | PA14 ΔPA5388 | Wargo and Hogan (2009) |
| PA5376:tn | PA14 with transposon insertion in PA5376; Gmr | Liberati et al. (2006) |
| PA5377:tn | PA14 with transposon insertion in PA5377; Gmr | Liberati et al. (2006) |
| PA5378:tn | PA14 with transposon insertion in PA5378; Gmr | Liberati et al. (2006) |
| Plasmids | ||
| pTOK2 | Suicide vector; Tetr | Kitten and Willis (1996) |
| pTOK2T | pTOK2 with restored lacZ activity; Tetr | This work |
| pTsacB | pTOK2T with sacB; Tetr | This work |
| pT-PA5388 | pTOK2T with caiX from P. aeruginosa PAO1 | This work |
| pT-B2916 | pTOK2T with caiX from B728a | This work |
| pT-PA3236 | pTOK2T with betX from P. aeruginosa PAO1 | This work |
| pT-B3758 | pTOK2T with betX from B728a | This work |
| pT-D3758 | pTOK2T with betX from DC3000 | This work |
| pME6041 | Broad-host-range vector; Kmr | Heeb et al. (2000) |
| pCbcPAO1 | pME6041 with cbcXWV from P. aeruginosa PAO1 | This work |
| pCbcB728a | pME6041 with cbcXWV from B728a | This work |
| pCbcDC3000 | pME6041 with cbcXWV from DC3000 | This work |
| pME2916 | pME6041with caiX from B728a | This work |
| pME2915-16 | pME6041with caiX and cdhR from B728a | This work |
| pET15b | Expression vector | Novagen |
| pET-cbcB728a | pET15b with [His]6-cbcXWVB728a | This work |
| pET-cbcDC3000 | pET15b with [His]6-cbcXWVDC3000 | This work |
| pN | pME6041 with nptII promoter | This work |
| pNcbcWV | pN with cbcWV from P. aeruginosa PAO1 | This work |
| pNcbcXWV | pN with cbcXWV from P. aeruginosa PAO1 | This work |
| pNchoXWV | pN with choXWV from S. meliloti 1021 | This work |
Fig. 1.
Choline (A) and betaine (B) uptake by P. syringae pv. tomato DC3000 and mutants lacking one or more loci encoding the CbcXWV, OpuC and BetT transporters. The wild-type DC3000 and the strains GB1–GB4 and DCΔcbc (Table 1) were grown in the absence of choline or betaine, designated no inducer, or in the presence of betaine or choline (final concentration 10 mM), allowing these compounds to serve as inducers. As a catabolic product of choline, betaine may also serve as an inducer in the choline-amended cells. Uptake was measured using 10 μM [14C]choline (A) or [14C]betaine (B). Values are mean uptake rates + standard error of the mean (SE) (n = 3).
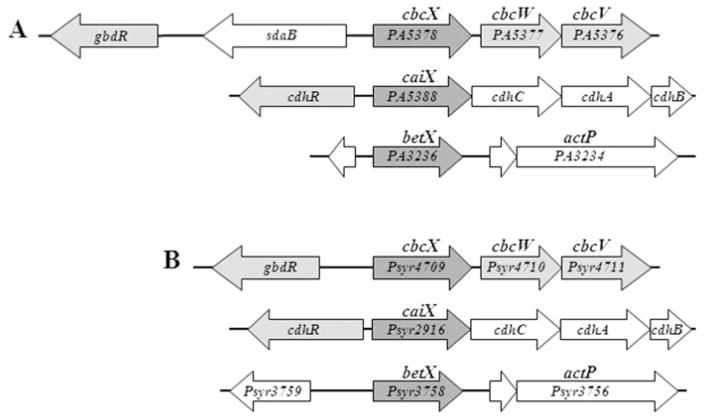
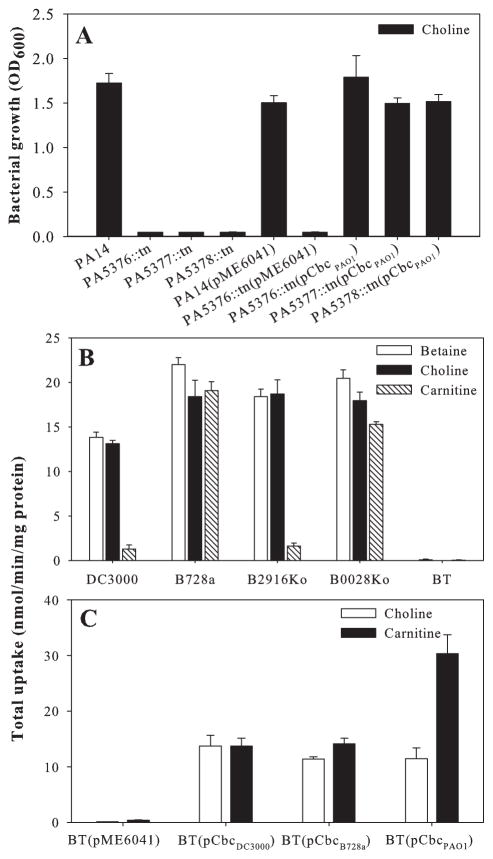
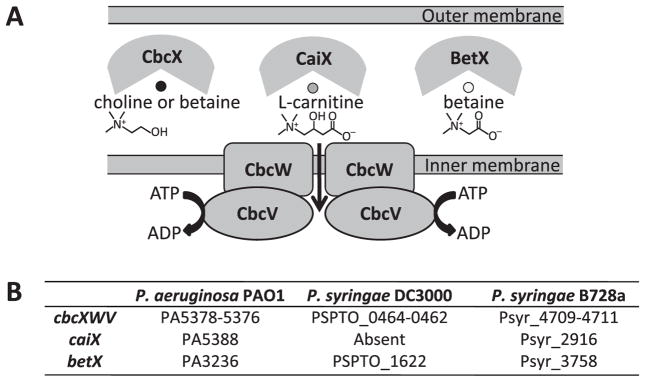
Orthologues of cbcXWV are present in all 14 strains of Pseudomonas spp. for which the complete genome sequences are available, as shown for three strains in Fig. 2. Although catabolism-associated QAC transporters have not been characterized in Pseudomonas species, Diab et al. (2006) found that the expression of the gene encoding the P. aeruginosa CbcX orthologue, PA5378, was highly induced in cells grown on betaine as a sole source of carbon. To test if the putative transporter encoded by PA5378-PA5376 in P. aeruginosa is functionally similar to the Cbc transporter, we evaluated growth on selected QACs of three mutants from the non-redundant transposon mutant library of P. aeruginosa strain PA14 (Liberati et al., 2006). The PA5376::tn, PA5377::tn and PA5378::tn mutants all failed to grow on choline (Fig. 3A) and betaine (Fig. S2) as sole C sources in the low-osmoticum medium ½-21C. These transposon mutants also failed to grow on L-carnitine (Fig. S2), suggesting that carnitine is an additional substrate for this transporter.
Fig. 2.
Genomic arrangement of the cbcXWV loci and related loci in (A) P. aeruginosa PA14 and PAO1 and (B) P. syringae strain B728a. The following genes are shown: gbdR [transcriptional regulator influencing betaine catabolism (Wargo et al., 2008)], sdaB (L-serine dehydratase), cbcXWV (ABC transporter), cdhR [transcriptional regulator influencing carnitine catabolism (Wargo and Hogan, 2009)], caiX (carnitine-specific binding protein), cdhCAB [carnitine catabolic proteins (Wargo and Hogan, 2009)], betX (betaine-specific binding protein) and actP (acetate permease).
Fig. 3.
Complementation of P. aeruginosa and P. syringae transporter-deficient mutants by plasmids expressing cbcXWV from various strains.
A. Growth of PA14, PA14 containing insertions in individual cbc genes, and the PA14 mutants containing pME6041 or pME6041 expressing cbcXWV from PAO1.
B. Uptake of radiolabelled betaine, choline or carnitine (1 mM) by DC3000, B728a, B728a with insertions in the putative SBP loci Psyr_2916 (B2916Ko) and Psyr_0028 (B0028Ko), and B728a lacking opuC, betT and cbc (BT).
C. Uptake of radiolabelled choline and carnitine (1 mM) by strain BT containing the vector pME6041 or pME6041 expressing cbcXWV from DC3000, B728a or PAO1.
Cells were grown with 20 mM choline (A) or with 10 mM choline and 10 mM betaine (B and C) as sole C sources and optical density at 600 nm (OD600) was recorded after 24 h (with an initial OD600 of 0.05). Values represent the mean + SE (n = 3).
DC3000 cannot grow on carnitine (Chen and Beattie, 2007) and therefore is not a good strain for characterizing transporters involved in carnitine uptake for catabolism. P. syringae pv. syringae strain B728a, however, grew well on carnitine and exhibited significant uptake activity (Fig. 3B). We deleted the B728a loci that were homologous to the DC3000 opuC, betT and cbc loci, generating the triple mutant strain BT. BT failed to transport choline, betaine and carnitine (Fig. 3B). The deficiencies in choline and carnitine uptake by BT were complemented by introduction of the vector pME6041 expressing cbcXWV from DC3000, B728a or PAO1 (Fig. 3C). Similarly, the deficiencies in choline uptake by PA14 mutants with insertions in individual cbc loci were complemented by cbcXWV from either PAO1 or B728a (Fig. S3), with the cbcXWV from DC3000 not evaluated, and deficiencies in the growth of the mutants in choline were complemented by cbcXWV from either PAO1 (Fig. 3A) or DC3000 (data not shown).
CbcXWV is sufficient to transport choline and betaine but not carnitine
The Cbc transporters in B728a and DC3000 share 98–100% identity at the amino acid level, yet B728a transports carnitine and DC3000 does not (Fig. 3B). To investigate the basis for this difference, we examined the function of the Cbc transporter when expressed in E. coli strain MKH13. MKH13 cells transformed with the vector pME6041 were deficient in choline and betaine uptake (Table 2), as expected based on the absence of transporters for these compounds (Kempf and Bremer, 1995). MKH13 has a carnitine transporter CaiT, which has no measurable activity under these conditions. This was expected based on the fact that caiT gene expression requires carnitine and anaerobic conditions (Buchet et al., 1998) and CaiT requires high internal concentrations of γ-butyrobetaine for uptake based on its L-carnitine/γ-butyrobetaine antiporter activity (Jung et al., 2002). Interestingly, cells of MKH13 transformed with pME6041 expressing cbcXWV from PAO1, pCbcPAO1, accumulated choline and betaine but little to no carnitine. Cells of MKH13 transformed with either pCbcDC3000 or pCbcB728a accumulated little to no choline, betaine or carnitine (data not shown); this low activity was probably due to poor expression of cbcXWV in MKH13, despite their clear expression in P. syringae (Fig. 3C). When these genes were expressed in E. coli strain Acella under the control of the T7lac promoter, they significantly increased choline and betaine uptake, indicating successful heterologous expression, but again did not affect carnitine uptake (Table 2). This finding that the Cbc transporter encoded by cbcXWV is sufficient for choline and betaine uptake but not for carnitine uptake suggests that Cbc-mediated transport of L-carnitine requires a component that is present in B728a and PA14 but is missing in DC3000 and the E. coli strains MKH13 and Acella.
Table 2.
Uptake activities of E. coli strains expressing cbcXWV from various Pseudomonas spp.
| Strainb | Plasmid | Total uptake [nmol (mg protein)−1]a
|
||
|---|---|---|---|---|
| Choline | Betaine | Carnitine | ||
| MKH13 | pME6041 | 0.2 (0.1) | 0.5 (0.5) | 0.5 (0.3) |
| MKH13 | pCbcPAO1 | 12.1 (1.1) | 5.8 (1.1) | 0.8 (0.4) |
| Acellac | pET15b | 0 | 8.2 | 0 |
| Acella | pET-cbcDC3000 | 15.0 (2.4) | 17.0 (1.8) | 0.59 (0.82) |
| Acellac | pET-cbcB728a | 19.2 | 16.2 | 0 |
Uptake was evaluated using radiolabelled choline, betaine or carnitine (1 mM). Unless otherwise indicated, average values (SE) are shown (n = 10).
The Acella cells were grown with 1 mM IPTG for promoter induction.
Uptake values for these constructs represent a single colony; these values were verified in a separate experiment.
CaiX, a carnitine-specific binding protein, is required for Cbc-mediated uptake of L-carnitine
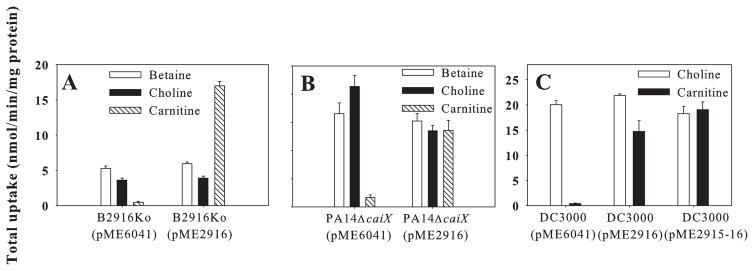
In previous studies with P. aeruginosa PA14, we demonstrated that a non-polar deletion of a putative SBP gene near a carnitine catabolic locus eliminated growth on L-carnitine (Wargo and Hogan, 2009). Among the four strains of P. syringae with sequenced genomes (B728a, DC3000, T1 and 1448A), an orthologue of this gene, PA5388, is present in B728a, which grows on carnitine, but is absent in the other three, which do not (data not shown). A knockout mutant of B728a lacking the SBP orthologue Psyr_2916, designated B2916Ko, was dramatically reduced in carnitine but not choline or betaine uptake (Fig. 3B), whereas a B728a mutant lacking an additional putative SBP, Psyr_0028, which exhibits 41% identity at the amino acid level to Psyr_2916, was not altered in QAC uptake (Fig. 3B). Introduction of pME6041 expressing Psyr_2916 complemented the carnitine-uptake deficiency of B2916Ko (Fig. 4A) as well as of a PA14 mutant lacking PA5388 (Fig. 4B). Introduction of Psyr_2916, as well as Psyr_2916 and an adjacent putative transcriptional regulator Psyr_2915 (Wargo and Hogan, 2009), into DC3000 conferred carnitine uptake (Fig. 4C) but not catabolism (data not shown). This inability to grow on carnitine was consistent with the absence of orthologues to carnitine oxidation genes, which were recently identified in PA14 (Wargo and Hogan, 2009). These findings provide compelling evidence that Psyr_2916 and PA5388, hereafter referred to as CaiX, enable Cbc-mediated carnitine uptake in P. syringae and P. aeruginosa respectively.
Fig. 4.
Complementation of uptake activity in P. syringae and P. aeruginosa strain derivatives by plasmids expressing Psyr_2916 (caiX) from P. syringae B728a. Uptake of radiolabelled betaine, choline and carnitine (1 mM) by derivatives of strain (A) B2916Ko, in which Psyr_2916 was insertionally inactivated, (B) PA14ΔcaiX, in which PA5388 was deleted, and (C) DC3000. Strain derivatives included those expressing only pME6041 or pME6041 containing caiXB728a (pME2916) or caiXB728a and the adjacent putative transcriptional regulator cdhR (pME2915-16) (Wargo and Hogan, 2009). Values are mean + SE (n = 3).
The lack of competition between choline and betaine for uptake by Cbc suggests the involvement of an additional choline- or betaine-binding SBP in P. syringae
To better understand the substrate specificity of the Cbc transporter, we performed competition uptake assays with strains expressing cbcXWV, using strain derivatives that differ in the origin of the cbcXWV and caiX components. Strain BT, which expressed an endogenous caiX, exhibited similar inhibition profiles regardless of whether cbcXWV was expressed via pCbcB728a (homologous expression) or pCbcDC3000 and pCbcPAO1 (heterologous expression) (Table 3). As expected, the uptake of radiolabelled betaine by all of the strains was strongly inhibited by the presence of a 100-fold excess of unlabelled betaine, with similar self-inhibition occurring for choline and carnitine. CbcXWV can transport both choline and betaine; therefore, we predicted that the uptake of one substrate would be inhibited by the other substrate, especially when the latter was supplied at a 100-fold excess. Surprisingly, the uptake of radiolabelled betaine was not inhibited, or was only marginally inhibited, by choline nor was the uptake of radiolabelled choline inhibited by betaine in any of the strains (Table 3). This lack of competition was also observed when cbcXWV was expressed in DC3000 mutant GB4 via pCbcB728a and pCbcPAO1, as well as in DC3000 mutant GB1 via the endogenous, single-copy cbcXWV (data not shown); caiX is absent in DC3000 and consequently had no influence. The mutual lack of competition between betaine and choline cannot be reconciled with CbcX being the sole SBP for these compounds. These results therefore suggest that the Cbc core transporter interacts with an additional choline- or betaine-binding SBP and that the substrate-bound SBPs do not significantly compete for Cbc-mediated transport; we discuss our investigation of these predictions below.
Table 3.
Abilities of various compounds to competitively inhibit the uptake of radiolabelled choline, betaine or carnitine by the QAC transporter-deficient BT strain expressing cbcXWV from B728a, DC3000 and PAO1.
| Competitor | [14C] substrate | % inhibition of uptake of [14C]substratea
|
||
|---|---|---|---|---|
| BT(pCbcB728a) | BT(pCbcDC3000) | BT(pCbcPAO1) | ||
| Water | Betaine | 0 | 0 | 0 |
| Betaine | Betaine | 93 | 89 | 83 |
| Choline | Betaine | 3 | −6 | −6 |
| Carnitine | Betaine | 5 | −2 | 2 |
| Water | Choline | 0 | 0 | 0 |
| Betaine | Choline | 19 | 12 | 9 |
| Choline | Choline | 93 | 89 | 84 |
| Carnitine | Choline | 5 | −6 | 1 |
| Water | Carnitine | 0 | 0 | 0 |
| Betaine | Carnitine | 83 | 88 | 84 |
| Choline | Carnitine | 75 | 81 | 77 |
| Carnitine | Carnitine | 93 | 94 | 94 |
Cells were grown with 10 mM choline and 10 mM betaine and uptake was evaluated using radiolabelled substrates (5 μM) in the presence of unlabelled competitors (0.5 mM) for 10 min. The steady-state total uptake in water, and thus in the absence of competitors, was: [14C]betaine uptake, 1.8, 1.8 and 1.6 nmol (mg protein)−1; [14C]choline, 5.8, 5.4 and 5.0 nmol (mg protein)−1; [14C]carnitine, 9.5, 5.0 and 15.2 nmol (mg protein)−1, for BT(pCbcDC3000), BT(pCbcB728a), BT(pCbcPAO1) respectively. Values are the mean % inhibition (n = 4), and the results are representative of two independent experiments. The standard deviation did not exceed 5%.
BetX is a betaine-specific binding protein that interacts with the Cbc core transporter CbcWV
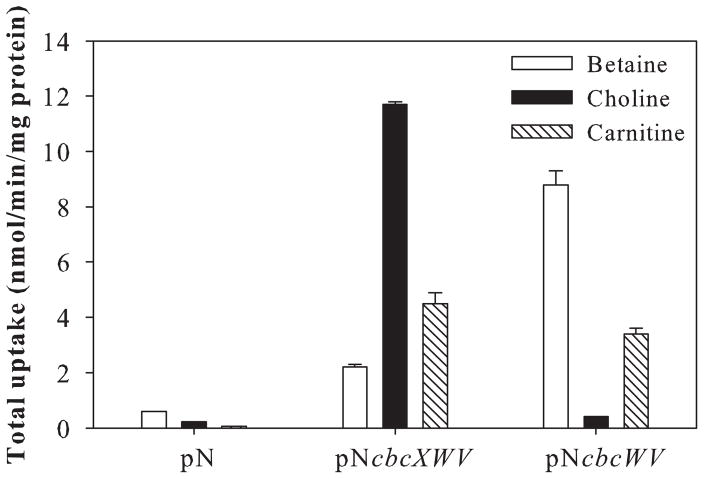
Plasmids that express a full-length cbcXWV locus were not useful for identifying additional choline- or betaine-binding SBPs because of overlap in the substrate specificities of these SBPs with CbcX (Table 2). To express cbcWV at a high level, but separately from the associated SBP-encoding cbcX, we first constructed the plasmid pNcbcXWV, which is a broad-host-range plasmid carrying cbcXWVPAO1 under the control of the nptII promoter. When expressed in the QAC transporter-deficient strain BT, pNcbcXWV conferred betaine, choline and carnitine uptake (Fig. 5). Moreover, when expressed in E. coli MKH13, pNcbcXWV conferred sufficiently high expression to increase the rate of choline and betaine uptake 10-fold more than was conferred by pCbcPAO1 (data not shown). A plasmid expressing only the core transporter, pNcbcWV, was constructed by deleting cbcX from pNcb-cXWV; it restored carnitine uptake to BT (Fig. 5), consistent with an association between the native CaiX SBP and the core transporter. The ability of pNcbcWV to restore betaine but not choline uptake to BT indicates that BT produces a Cbc-interacting SBP that specifically binds betaine.
Fig. 5.
Complementation of betaine, choline and carnitine uptake in the QAC transporter-deficient P. syringae strain BT by vectors expressing cbcWV and cbcXWV. Cells were grown in King’s B medium, washed with water, and re-suspended in ½-21C medium for the transport assay using radiolabelled substrates at a concentration of 10 μM. Values are mean + SE (n = 3).
We evaluated the genes Psyr_3758 and Psyr_0028 as candidates for a betaine-binding, Cbc-interacting SBP protein, and used caiX (Psyr_2916) as a control. Psyr_3758 and Psyr_0028 were annotated as putative SBPs in B728a (Feil et al., 2005) and were predicted to interact with CbcW based on their high scores (> 0.85) as predicted functional partners of CbcW using the STRING database of known and predicted protein–protein interactions (Jensen et al., 2009). Knockout mutants lacking these putative SBP genes, including mutants BT3758Ko, BT0028Ko and BT2916Ko, served as hosts for the SBP-expressing plasmids. As expected, the BT-derived mutants were all deficient in the uptake of betaine, carnitine and choline when expressing only the empty pME6041 vector with the nptII promoter, pN (Table 4). In the presence of the complete Cbc transporter encoded by pNcbcXWV, mutant BT2916Ko was deficient in carnitine uptake, consistent with the loss of CaiX; in contrast, the BT3758Ko and BT0028Ko mutants took up significant amounts of all of the substrates, consistent with the presence of chromosomally encoded CaiX and plasmid-borne CbcX (Table 4). In the presence of the Cbc core transporter encoded by pNcbcWV, mutant BT3758Ko, but not BT2916Ko and BT0028Ko, was deficient in betaine uptake, indicating that Psyr_3758 functions as a SBP for betaine; Psyr_3758 was thus designated BetX. The ability of BT3758Ko expressing pNcbcXWV to take up betaine is consistent with the presence of CbcX (PA5378) as a betaine-binding SBP; thus, the Cbc transporter appears to have redundant SBPs for betaine uptake. Uptake in these studies demonstrated expression of the endogenous caiX and betX genes in BT even in the absence of exogenous QACs as inducers. Although CbcX mediates approximately 99% of the choline uptake at low (10 μM) and high (1 mM) [14C]choline concentrations, residual choline uptake was detectable when pNcbcWV was expressed in P. syringae but not in E. coli MKH13 (data not shown), suggesting that one or more additional SBPs can contribute to choline uptake in P. syringae.
Table 4.
Uptake activities of strains expressing the core Cbc or Cho transporters in the presence and absence of the putative chromosomally encoded SBPs Psyr_2916, Psyr_3758 and Psyr_0028 and the plasmid-encoded SBPs CbcX and ChoX.
| Plasmid | Hostb | Total uptake [nmol (mg protein)−1]a
|
|||
|---|---|---|---|---|---|
| Betaine (10 μM) | Carnitine (10 μM) | Choline (10 μM) | Choline (1 mM) | ||
| pN | BT2916Ko | 0.0 | 0.1 | 0.0 | 0 |
| pN | BT3758Ko | 0.0 | 0.0 | 0.0 | 0 |
| pN | BT0028Ko | 0.0 | 0.0 | 0.0 | 0 |
| pNcbcXWV | BT2916Ko | 16.0 | 0.5 | 16.2 | 644 |
| pNcbcXWV | BT3758Ko | 14.3 (0.24) | 13.0 (0.3) | 16.4 (0.18) | 648 (27.5) |
| pNcbcXWV | BT0028Ko | 16.1 (0.41) | 7.0 (0.4) | 18.2 (0.36) | 562 (6.8) |
| pNcbcWV | BT2916Ko | 17.2 | 0.3 | 0.15 | 8.6 |
| pNcbcWV | BT3758Ko | 0.1 (0.01) | 15.1 (0.16) | 0.15 (0.05) | 7.1 (0.6) |
| pNcbcWV | BT0028Ko | 17.2 (0.2) | 16.3 (0.91) | 0.11 (0.05) | 8.9 (1.4) |
| pNchoXWVc | BT2916Ko | 3.0 | 0.1 | 6.5 | 6.1 |
| pNchoXW | BT3758Ko | 3.0 (0.64) | 1.8 (0.4) | 8.6 (1.28) | 7.4 (0.8) |
| pNchoXW | BT0028Ko | 2.8 (0.3) | 1.2 (0.1) | 7.8 (0.07) | 8.5 (1.4) |
Cells were re-suspended in ½-21C containing 0.3% glucose for the uptake measurements. Values represent a single replication for the BT2916Ko derivatives and the mean (SE) for the BT3758Ko (n = 3) and B0028Ko (n = 2) derivatives.
The host strains were derived from strain BT, and thus lacked OpuC, BetT and Cbc due to deletions, as well as lacked Psyr_2916/CaiX (BT2916Ko), Psyr_3758/BetX (BT3758Ko) and Psyr_0028 (B70028Ko) due to knockout insertions (Table 1).
The plasmid pNchoXWV expressed the S. meliloti ChoXWV transporter.
We compared the behaviour of the Cbc transporter and its interacting SBPs with the behaviour of the highly similar Cho transporter from S. meliloti, which mediates choline uptake (Dupont et al., 2004). The full-length choXWV locus has not yet been cloned and characterized. In P. syringae BT strain derivatives, which lacked all endogenous QAC transporters, ChoXWV was able to transport betaine and carnitine as well as choline (Table 4). Although betaine transport did not require BetX, carnitine transport required CaiX (Table 4), suggesting that the ChoWV core transporter interacted with CaiX from the phylogenetically distinct species P. syringae. CbcXWV and ChoXWV differed in their capacity for choline transport, with choline uptake by CbcXWV increasing from 16 to 18 nmol (mg protein)−1 to 562–648 nmol (mg protein)−1 when the choline concentration was increased from 10 μM to 1 mM, but not increasing by ChoXWV when the choline concentration was similarly increased (Table 4). Similar results were observed when both of them were expressed heterologously in E. coli MKH13 (data not shown). These data demonstrate an intrinsic difference between CbcXWV and ChoXWV in their uptake capacity.
The SBPs interacting with the Cbc core transporter are highly substrate specific and confer distinct uptake affinities and uptake capacities upon the Cbc transporter
To better understand the substrate specificities among the SBPs CbcX, BetX and CaiX, we expressed the Cbc core transporter and its associated SBPs in E. coli strain MKH13, which lacks orthologues of cbcX, betX and caiX, and evaluated the subsequent uptake activities. The absence of Cbc-interacting SBPs in MKH13 was confirmed using MKH13 (pNcbcWV) (Table 5). The caiX and betX genes from PAO1, B728a and DC3000 were introduced into the pBR322-based vector pTOK2T, which enables moderate expression of the cloned genes by an undescribed promoter. As expected, MKH13 cells coexpressing cbcWV (on pNcbcWV) and caiX (on a pTOK2T derivative) were able to take up carnitine but not choline and betaine, demonstrating that CaiX was sufficient for, and specific to, carnitine uptake (Table 5). Similarly, BetX was sufficient for, and specific to, betaine uptake. Consistent with earlier observations, CbcX conferred uptake for choline and betaine but not carnitine. ChoX was also sufficient for the transport of choline and betaine, in contrast to an earlier report that excluded betaine as a substrate (Dupont et al., 2004), but ChoWV, like CbcWV, depended on CaiX for carnitine uptake (Table 4). Cbc dependence on CaiX for carnitine uptake was also illustrated by the expansion of the substrate range when cbcX and caiX were coexpressed as compared with when cbcX was expressed (Table 5).
Table 5.
Uptake activities of the E. coli QAC transporter-deficient mutant MKH13 coexpressing components of the P. aeruginosa Cbc transporter or S. meliloti Cho transporter (Plasmid 1) and putative periplasmic substrate-binding proteins (SBPs) (Plasmid 2).
| Plasmid 1 | Plasmid 2 | SBP(s) present | Total uptake [nmol (mg protein)−1]a
|
||
|---|---|---|---|---|---|
| Choline | Betaine | Carnitine | |||
| pN | None | None | 0 | 0 | 0 |
| pNcbcWV | None | None | 0 | 0 | 0 |
| pNcbcWV | pTOK2T | None | 0 | 0 | 0 |
| pNcbcWV | pT-PA5388 | CaiXPAO1 | 0 | 0 | 3.8 |
| pNcbcWV | pT-B2916 | CaiXB728a | 0 | 0 | 3.7 |
| pNcbcWV | pT-PA3236 | BetXPAO1 | 0 | 21.6 | 0 |
| pNcbcWV | pT-B3758 | BetXB728a | 0 | 11.9 | 0 |
| pNcbcWV | pT-D3758 | BetXDC3000 | 0 | 6.1 | 0 |
| pNcbcXWV | None | CbcXPAO1 | 44 | 12 | 0 |
| pNchoXWV | None | ChoX | 4.7 | 2.7 | 0 |
| pNcbcXWV | pT-PA5388 | CbcX/CaiXPAO1 | 26.7 | 19.4 | 2.9 |
| pNcbcXWV | pT-B2916 | CbcX/CaiXB728a | 22.5 | 13.8 | 0.3 |
Uptake was evaluated using radiolabelled substrates (10 μM). Values represent a single replication, with multiple measurements for the SBPs cloned from distinct strains indicating variability among the measurements for each SBP tested.
We examined the uptake kinetics of the Cbc transporter system when it utilized each of the distinct SBPs. Using MKH13 cells and thus an in vivo assay, we found that CbcX and ChoX each conferred a similarly high affinity for choline, with Km values of 1.8–2.6 μM (Table 6). CbcX conferred an approximately 10-fold lower affinity for betaine than choline, and BetX exhibited a very high affinity for betaine, consistent with its role as a high-affinity betaine-binding protein. The CbcX-mediated choline and betaine transport activities were assayed using replicate samples of the same pool of cells to ensure similar concentrations of the transporter components, and thus the higher Vmax for choline than betaine [34.6 versus 11.6 nmol min−1 (mg protein)−1] suggested that CbcX-choline induced greater uptake activity in the core transporter than CbcX-betaine (Table 6). The E. coli MKH13 cells expressing BetX and the core transporter exhibited the lowest Vmax [4.8 nmol min−1 (mg protein)−1] among the choline and betaine transport activities of the Cbc transporter. Although the relative expression of BetX and CbcX was not determined, thus preventing a direct comparison of the Vmax values for BetX- and CbcX-mediated betaine uptake, we observed a lower betaine uptake capacity for BetX than CbcX even when comparing constructs with BetX expressed on a higher-copy plasmid than CbcX (data not shown). This suggests that BetX has a lower uptake capacity for betaine than CbcX, despite the fact that BetX bound betaine with a higher affinity than CbcX did. These relative kinetics of CbcX- versus BetX-mediated betaine uptake are consistent with a model predicting that a moderately lower binding affinity results in a faster release of substrate from the ligand-bound SBP into the core transporter, resulting in a higher uptake rate (Picon et al., 2000). The ChoXWV transporter exhibited the lowest Vmax among the transporter configurations shown in Table 6, suggesting that the interaction between ChoX-choline and ChoWV is less efficient than that between CbcX-choline or CbcX-betaine and CbcWV. A low capacity for choline uptake by ChoXWV was also indicated by the much lower level of choline uptake observed when choXWV was expressed on the expression vector pET15b in E. coli Acella cells (data not shown) as compared with the higher level of choline uptake by cbcXWVDC3000 and cbcXWVB728a when similarly expressed (Table 2). A low capacity for choline uptake by ChoXWV is also consistent with that observed when ChoXWV was expressed in P. syringae (Table 4) or P. aeruginosa (data not shown). We were unable to accurately assess the kinetics of CaiX-mediated carnitine uptake using MKH13; however, we did so using strain PA5378::tn(pNcbcWV) and observed a relatively low affinity and low uptake capacity of CbcWV-CaiX for carnitine. The apparent Km (1.8 ± 0.36 μM) for choline uptake by ChoXWV in MKH13 cells was in excellent agreement with the reported KD (2.3 μM) for choline binding to purified ChoX, as evaluated using a fluorescence-based equilibrium binding assay (Oswald et al., 2008), and to the reported KD (2.7 μM) from a binding assay with radiolabelled choline (Dupont et al., 2004). This similarity between KD and Km suggests that the rate-limiting step is the donation of the ligand by the ligand-bound SBP after docking onto the membrane complexes (or core transporters) (Lanfermeijer et al., 1999).
Table 6.
Summary of the uptake kinetics of the Cbc and Cho transporters.
| Host | Plasmid 1 | Plasmid 2 | SBP present | [14C] Substrate | Uptake kineticsa
|
|
|---|---|---|---|---|---|---|
| Km | Vmax | |||||
| MKH13 | pNcbcXWV | None | CbcX | Choline | 2.6 (0.66) | 34.6 (2.2) |
| MKH13 | pNcbcXWV | None | CbcX | Betaine | 24.2 (2.4) | 11.6 (0.6) |
| MKH13 | pNcbcWV | pTPA3236 | BetX | Betaine | 0.6 (0.16) | 4.8 (0.18) |
| MKH13 | pNchoXWV | None | ChoX | Choline | 1.8 (0.36) | 1.2 (0.05) |
| PA5378::tn | pNcbcWV | None | CaiXPAO1 | Carnitine | 24.2 (8.3) | 3.8 (0.33) |
The data were fit with the Michaelis–Menten equation (Fig. S4). The units for the apparent affinity constant (Km) are μM and for the maximal rate of uptake (Vmax) are nmol min−1 (mg protein)−1. Values shown are mean (SE) (n = 4).
Competition among the SBPs occurs both for binding to the substrates and for docking of ligand-bound SBPs to the core transporter
To better understand the substrate specificity of the individual SBPs as they interacted with the core CbcWV transporter, we examined the ability of selected substrates to inhibit the uptake of a radiolabelled substrate when supplied in a 10- or 100-fold excess over the radiolabelled substrate. As expected, BetX-mediated [14C]betaine uptake was inhibited by unlabelled betaine (94% reduction in uptake) (Table 7); it was also moderately inhibited (56%) by the betaine analogue dimethylglycine (DM) but not by choline, carnitine or the betaine analogue monomethylglycine (MM). CaiX-mediated [14C]carnitine uptake was inhibited by unlabelled carnitine (91%) but not by other compounds (Table 7). These results are consistent with BetX and CaiX being betaine- and carnitine-specific SBPs respectively. CbcX-mediated [14C]betaine uptake was strongly inhibited by unlabelled choline (99%), even more than by unlabelled betaine (87%), consistent with our kinetic data that CbcX binds choline with a much higher affinity than betaine. Interestingly, CbcX-mediated [14C]choline uptake was not inhibited by unlabelled betaine, even at a 100-fold excess. This finding was surprising in light of data indicating that Cbc can transport both betaine and choline (Tables 5 and 7). Together these data suggest that choline strongly competes against betaine for binding to CbcX; alternatively, CbcX-choline strongly competes against CbcX-betaine for docking to the core Cbc transporter. Unlike CbcX, ChoX-mediated choline uptake was inhibited by acetylcholine (Table 7), consistent with a previous report that ChoX binds acetylcholine (Dupont et al., 2004). ChoXWV- but not CbcXWV-mediated [14C]choline uptake was moderately inhibited (41%) by carnitine, reflecting another intrinsic difference between these transporters.
Table 7.
Competitive uptake of radiolabelled choline, betaine and carnitine in the E. coli transporter-deficient mutant MKH13 expressing selected transporter components.a
| Plasmid 1 | Plasmid 2 | SBP(s) | [14C] Substrate | % inhibition of uptake of substrate with the following unlabelled competitorsa
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| W | MM | DM | Bet | Cho | Car | AC | PC | ||||
| pNcbcWV | pT-PA3236 | BetX | Betaine | 0 | −13 | 56 | 94 | −8 | 3 | NT | NT |
| pNcbcWV | pT-PA5388 | CaiX | Car | 0 | 0 | 2 | 5 | 5 | 91 | NT | NT |
| pNcbcXWV | pT-PA5388 | CaiX/CbcX | Car | 0 | −1 | 7 | 57 | 66 | 97 | NT | NT |
| pNcbcXWV | None | CbcX | Betaine | 0 | −3 | −5 | 87 | 99 | 3 | NT | NT |
| pNcbcXWV | pT-PA5388 | CbcX/CaiX | Betaine | 0 | 3 | −4 | 88 | 99 | 10 | NT | NT |
| pNcbcXWV | None | CbcX | Choline | 0 | 2 | 4 | 2 | 92 | 6 | 9 | 1 |
| pNcbcXWV | pT-PA5388 | CbcX/CaiX | Choline | 0 | 2 | −8 | −1 | 96 | −2 | NT | NT |
| pNchoXWV | None | ChoX | Choline | 0 | −4 | −6 | 3 | 98 | 41 | 88 | 5 |
Uptake was evaluated using radiolabelled substrates (10 μM) in the presence of unlabelled competitors (1 mM). The steady-state total uptake in absence of competitors (Water) were, by row, 8.6, 14, 11, 9.8, 11.8, 17.7, 29.5 and 16.7 nmol (mg protein)−1. W, water; MM, monomethylglycine; DM, dimethylglycine; Bet, glycine betaine; Cho, choline; Car, carnitine; AC, acetylcholine; PC, phosphorylcholine; NT, not tested. Values are the mean percentage inhibition (n = 3–4), and the results are representative of two independent experiments.
To gain insight into the interactions among these SBPs and the core transporter, we coexpressed multiple SBPs and evaluated uptake of target substrates. When we coexpressed caiX and cbcX in the presence of cbcWV, CaiX-mediated [14C]carnitine uptake was moderately inhibited by choline (66%) and betaine (57%) (Table 7), which are the ligands for CbcX. Choline and betaine do not compete with carnitine for binding to CaiX, as indicated by the lack of competition by choline or betaine in the absence of CbcX (Table 7) and by the lack of choline or betaine uptake when CaiX was the sole SBP (Table 5); therefore, the reduction in [14C]carnitine uptake likely resulted from reduced CaiX-carnitine docking to the core transporter. This suggests that CbcX-choline and CbcX-betaine have a higher affinity for docking, as supported by the fact that carnitine, even at a 100-fold excess, did not reduce CbcX-mediated choline or betaine uptake in the presence of CaiX (Table 7). Furthermore, the lack of inhibition of CaiX-mediated carnitine uptake by ligand-free CbcX, as was observed in a control with only water rather than choline or betaine (Table 7), indicates that ligand-free CbcX did not compete with ligand-bound CaiX for docking. This result is consistent with the Cbc core transporter having a higher affinity for a ligand-bound than a ligand-free SBP.
Although we did not evaluate whether BetX-betaine similarly reduces CaiX-mediated carnitine uptake in MKH13, we observed that CaiX-mediated carnitine uptake was reduced more by betaine than by choline in each of the three BT strain derivatives examined (Table 3). Given that betaine uptake involves BetX and CbcX whereas choline uptake involves only CbcX, this suggests that BetX-betaine reduced CaiX-mediated carnitine uptake. We also observed higher inhibition (75–88%) of carnitine uptake when betX, caiX and cbcX were simultaneously expressed, with betX and caiX chromosomally encoded (Table 3), than when only caiX and cbcX were expressed (57–66%) (Table 7), despite similar self-inhibition by unlabelled carnitine (94% versus 97%). This result also suggests that BetX-betaine reduces CaiX-mediated carnitine uptake. Together, this result suggests that BetX-betaine competes for docking with CaiX-carnitine.
Discussion
The Cbc transporter described here represents the first catabolism-associated QAC transporter to be characterized in Pseudomonas spp., despite the fact that catabolic uptake of QACs was reported in P. aeruginosa more than four decades ago (Kleber and Aurich, 1967). In addition to the core transporter comprised of a transmembrane protein (CbcW) and an ATPase (CbcV), the Cbc transporter system interacts with at least three SBPs: CbcX, which binds choline with a high affinity and betaine with a much lower affinity; BetX, which binds betaine with a very high affinity; and CaiX, which binds carnitine with a moderate affinity and represents the first carnitine-specific SBP to be reported. These three SBPs are encoded by genes located in separate regions of the genome. The complexity of this Cbc transporter, compounded by the redundancy of osmoregulatory QAC transporters [e.g. OpuC, BetT and BetT-like transporters such as PA5291 in P. aeruginosa (Chen and Beattie, 2007; 2008)], may have hindered previous attempts to identify such transporters (Aurich and Kleber, 1970; Salvano et al., 1989; Diab et al., 2006; Galvao et al., 2006). To minimize any compounding effects, we expressed Cbc transporter configurations in E. coli mutant MKH13, in which all QAC transporters and SBPs have been deleted or remained inactive under the test conditions (Kempf and Bremer, 1995). In addition, enzymes that are responsible for the conversion of choline into betaine were also deleted in MKH13. We characterized the components of the Cbc transporter in three different Pseudomonas spp. strains, as shown in Fig. 6.
Fig. 6.
Model of the interacting components comprising the Cbc transporter of P. aeruginosa and P. syringae. (A) Diagrammatic model and (B) list of Cbc transporter components. This study has provided functional evidence for all of the components shown.
Among characterized transporters, the S. meliloti ChoXWV transporter (Dupont et al., 2004) shares the greatest sequence similarity with CbcXWV as well as shares several functional similarities. ChoXWV was initially reported to transport only choline (Dupont et al., 2004); however, a subsequent report showed that the SBP ChoX binds betaine, albeit with a lower affinity than choline (Oswald et al., 2008). Here, we provide direct evidence that ChoXWV transports betaine as well as choline and, moreover, that the core transporter ChoWV is similar to CbcWV in interacting with CaiX to transport carnitine. The complete genomes of strains of Rhizobiaceae do not contain genes encoding CaiX orthologues. The SBPs CbcX and ChoX share a high structural similarity, including the amino acid residues identified in ChoX as required for substrate binding (Fig. S5), and they bind choline and betaine with comparable affinity (Oswald et al., 2008). Although CbcX mediates betaine uptake, betaine surprisingly did not inhibit CbcX-mediated choline uptake even when it was present at a 100-fold excess over choline. A similar lack of inhibition was reported for the transporter ChoXWV (Dupont et al., 2004) and its orthologue GbcXWV (Fox et al., 2008). This lack of competition by one substrate against another substrate has not been observed in osmoregulatory QAC transporters, such as E. coli ProU (Cosquer et al., 1999a) and P. syringae OpuC (Chen and Beattie, 2007), and probably contributed to the initial conclusion that betaine was not a substrate for the Cho transporter. Such a lack of competition in uptake can be explained by better adaptation of ChoX, and potentially CbcX, to bind choline than betaine (Dupont et al., 2004); for example, perhaps choline binding induces a conformational change that excludes betaine binding. Interestingly, among the CbcXWVDC3000 and ChoXWV transporter components, the SBPs exhibited the highest sequence similarity, contrary to those of other ABC transporters in which the NBDs exhibit the highest and the SBP the lowest similarity (Davidson et al., 2008). Based on the fact that P. syringae and S. meliloti have evolved to live in close association with plants, this unusual conservation profile may reflect a selection pressure for choline utilization in plants, as suggested by previous studies (de Rudder et al., 2000; Chen and Beattie, 2008). Differences between the Cbc and Cho transporter include that the capacity for choline uptake is much higher by Cbc than Cho, that acetylcholine and carnitine inhibit choline uptake by Cho but not by Cbc, and that the encoding loci differ in their genomic context. The cbcXWV operon is located proximal to the AraC family transcriptional regulator GbdR (Fig. 2), which is required for betaine catabolism (Wargo et al., 2008), whereas the choXWV operon is not located near an obvious transcriptional regulator.
Sequence alignments among the QAC-specific SBPs characterized here reveal high conservation in many amino acid residues that are involved in substrate binding by ChoX (Horn et al., 2006; Oswald et al., 2008). This includes the three Trp residues Trp-43, Trp-90 and Trp-205 of ChoX (Fig. S5) that are critical to the ‘cation–π’ interaction characteristic of the binding of QAC ligands to their cognate SBPs. This cation–π interaction results from the trimethylammonium head moiety, which promotes delocalization of the positive charge over the three methyl groups and forms a bulky cation that interacts with three to five aromatic residues in the ligand-binding box of a SBP. This type of interaction contrasts with the H bonding that typically mediates ligand binding to SBPs. The differential recognition by the Cbc-associated SBPs of QACs that share a strong structural similarity demonstrates that these SBPs have exceptional specificity. Although unusual among the characterized SBPs of ABC transporters, perhaps the best example of such exquisite specificity is that of oxyanion-binding proteins, which have evolved to discriminate among highly similar ions such as phosphate and sulphate; they do this based on the protonation of phosphate at physiological pH (Quiocho, 1996). The specificity of the Cbc-associated SBPs for QAC binding must be based on recognition of the carboxyl versus hydroxyl group distinguishing betaine from choline (Smits et al., 2008), and in the shorter versus longer acyl chain distinguishing betaine from carnitine. Future crystallization and binding affinity studies may provide insight into the molecular basis for SBP recognition of distinct QACs.
The Cbc transporter system offers many advantages as a model system for investigating the mechanisms underlying uptake by prokaryotic ABC transporters. The distinct substrate specificity of certain pairs of SBPs, such as CaiX and CbcX, provides an opportunity to use in vivo studies to examine SBP competition for docking to the core transporter. In vivo studies avoid some of the limitations inherent to purified-component studies, including altered behaviour due to purification and reconstitution processes (Rees et al., 2009) and due to the absence of a full transporter when individual components are examined (Orelle et al., 2008). For example, our studies demonstrated that the Cbc core transporter preferentially supported docking of a ligand-bound SBP, CbcX-choline or CbcX-betaine, over the ligand-free SBP CbcX based on the greater inhibition of CaiX-carnitine docking and transport by the ligand-bound SBPs. Although we predict that a core transporter would preferentially permit docking of a ligand-bound SBP over a ligand-free SBP, surprisingly, such preference has not been unambiguously demonstrated. On the contrary, studies using detergent-solubilized proteins or permeabilized proteoliposomes have commonly found that ligand-bound and ligand-free SBPs compete equally well for docking (Bohl et al., 1995; Ames et al., 1996). Another advantage of the Cbc transporter as a model system is that the QAC substrates are generally available or easily produced in radiolabelled form. They also have low molecular masses, which allow them to enter and rapidly diffuse in the periplasm; this avoids entry and diffusion as rate-limiting steps during transport. Moreover, bacterial strains that do not synthesize the QAC substrates, such as MKH13, can be easily obtained; this allows in vivo studies to be performed with the Cbc transporter without interference from endogenous substrates. This contrasts with most or all of the other ABC transporters that use multiple SBPs, most of which transport amino acids and oligopeptides.
The finding that BetX-betaine did not detectably compete with CbcX-choline for docking is intriguing and suggests the need for a better understanding of docking. It is possible that docking and substrate delivery are exceedingly fast and thus significant competition for docking is avoided. It is also possible that the Cbc core transporter was present in a similar or greater concentration than the SBPs and thus the number of docking sites was not limited. This latter possibility, however, seems unlikely for several reasons. First, uptake was not inhibited by the alternate substrate when CbcX and BetX were expressed in a variety of constructs that probably varied in the relative production of the core transporter and the SBPs; this included expression from the DC3000 genome as well as expression of homologous and heterologous plasmid-borne cbcWV in DC3000 and B728a. Second, the location of cbcX as the first gene in the cbcXWV operon suggests equal or greater transcript levels of cbcX versus the downstream core transporter genes, although the stability of the transcripts may vary. And third, SBP synthesis is generally subject to less feedback inhibition than the core transporter components because of SBP localization in the periplasm, which generally contributes to the much greater abundance of SBPs than their core transporter components (Bohl et al., 1995). If we assume that the docking sites were in fact limited, then another possible explanation for the two-way lack of competition of ligand-bound CbcX and BetX is the presence of cooperative interactions, such as heterodimerization of the two SBPs. Although such heterodimerization has not been reported, the possibility of homodimerization has been elegantly demonstrated in another QAC transporter, the osmoregulatory OpuA transporter of Lactococcus lactis (Biemans-Oldehinkel and Poolman, 2003). This dimerization resulted from the association of two TMDs, each of which was fused to a SBP. Kinetic studies performed with purified transporters in proteoliposomes have suggested cooperative interactions between SBPs that stimulated either the docking of the SBPs on the core transporter or the translocation of the substrate.
Betaine and choline are known components of plant and animal tissues, with the former most often associated with osmoprotection and the latter present as a precursor for betaine as well as the major membrane lipid phosphatidylcholine. In contrast, carnitine is prevalent in animal tissues but rarely reported in plant tissues; thus, carnitine transport and metabolism has been characterized in the animal pathogen P. aeruginosa (Kleber and Aurich, 1966; 1967; Wargo and Hogan, 2009) but not in P. syringae or other phytopathogens. In fact, of the four P. syringae strains for which the genome is fully sequenced, only B728a can grow on carnitine and has the predicted carnitine catabolism genes. Among 30 P. syringae strains collected from at least 13 distinct host plants, however, 50% were capable of growth on carnitine (C. Chen and G.A. Beattie, unpubl. data). Almost all of the strains grew on betaine, choline and acetylcholine, although, surprisingly, carnitine typically supported growth that was the fastest and to the highest cell densities. Thus, for carnitine-catabolizing strains, adaptation of the Cbc transporter to interact with a carnitine-binding SBP could have provided a significant ecological fitness benefit. For phytopathogens, this benefit may be realized on germinating seedlings (Hirano et al., 1997), where carnitine may be present due to the active oxidation of fatty acids (Bourdin et al., 2007).
We can only speculate on the ecological advantages that led to a transporter that recruits multiple substrate-specific SBPs rather than a single SBP with broad substrate specificity. These substrate-specific SBPs appear to be conserved across Pseudomonas spp. based on that all of the Pseudomonas spp. strains sequenced to date have orthologues of CbcX, BetX and CaiX, with the exception that the P. syringae strains DC3000, T1 and 1448A lack CaiX orthologues. Burkholderia spp. such as B. amifaria and B. vietnamiensis have up to seven SBPs, and Rhizobiaceae members have up to three SBPs, that are predicted to interact with Cbc-like transporters. These bacterial species are commonly found as living free in the soil or in association with plants or animals. The chemical complexity of these habitats suggests the possibility of a diversity of QAC compounds, some of which could be toxic to the bacteria (Anthoni et al., 1991). Such toxicity of QAC analogues has been demonstrated in studies with S. meliloti (Cosquer et al., 1999b). Thus, one possible ecological advantage for evolving SBPs that are highly discriminating among QACs is to allow them to avoid binding toxic QAC derivatives. Moreover, the finding that toxic betaine analogues inhibited the growth of bacteria that were capable of catabolizing QACs but not of those using QACs solely as osmoprotectants (Cosquer et al., 1999b) suggests that it would be more advantageous to evolve highly substrate-specific transporters for catabolic uptake than for osmoprotective uptake. Thus, assuming that these proteobacterial genera encounter multiple QACs, including toxic QAC analogues, the presence of QAC transporters such as OpuC with broad specificity may provide osmoprotection, but the presence of a transporter such as Cbc with multiple, highly substrate-specific SBPs may maximize catabolic potential while minimizing damage due to the import of toxic compounds. Interestingly, the predicted result of increased tolerance to toxic QAC compounds may be reflected in the observation that, among microorganisms, Pseudomonas spp. are particularly tolerant to biocidal QACs that are widely used as sanitizing agents (Langsrud et al., 2003).
Experimental procedures
Bacterial strains and growth conditions
The bacterial strains and plasmids used are listed in Table 1. P. syringae strains were maintained in King’s B medium (King et al., 1954) at 28°C, whereas E. coli strains and P. aeruginosa strains were maintained in LB at 37°C. To evaluate growth on selected QACs, cells were grown in the low-osmoticum medium ½-21C (Halverson and Firestone, 2000; Chen and Beattie, 2007). Unless otherwise indicated, Pseudomonas spp. strains were grown in ½-21C containing 20 mM pyruvate, a non-repressive carbon source (Sage and Vasil, 1997), for the transport assays. E. coli strains were grown in M63 medium (Silhavy et al., 1984) containing glucose (0.2%) and vitamin B1 (0.0005%) for the transport assays. When necessary, antibiotics were used at the following concentrations (μg ml−1): ampicillin (Amp), 100; gentamicin (Gm), 60; kanamycin (Km), 500 for P. aeruginosa, 50 for P. syringae and 20 for E. coli; rifampin (Rf), 100; and tetracycline (Tet), 20.
Construction of plasmids
The integration vectors pKnockout-Ω (Windgassen et al., 2000), pTOK2T and pTsacB were used for the generation of P. syringae mutants. pTOK2T was constructed by modifying the vector pTOK2, which is a pBR322 derivative containing the highly effective mob region from RSF1010 (Kitten and Willis, 1996), to allow blue-white screening. To repair the dysfunctional lacZα gene, a 60 bp DNA fragment from pKnockout-Ω was inserted into pTOK2 and a blue colony was identified; the sequence of the modified region is shown in Fig. S6. PCR products were introduced into the SmaI site of pTOK2T to generate the pT-based plasmids (Table 1). The counter-selectable vector pTsacB was constructed by inserting a 3.5 kb XmnI–EcoRV fragment containing the sacB gene from pFlp2 (Hoang et al., 1998) into the XmnI site of pTOK2T.
The Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA) was used to amplify all DNA fragments. The expression plasmids pET-cbcB728a and pET-cbcDC3000 were constructed by introducing NdeI- and BamHI-restricted PCR products into pET15b (Novagen). The plasmids pCbcB728a, pCbcDC3000 and pCbcPAO1 were constructed by introducing the cbc loci from B728a, DC3000 and PAO1, respectively, which were amplified by PCR using the primers shown in Table S1, into the EcoRV site of pME6041 (Heeb et al., 2000). The plasmids pME2916 and pME2915-16 were similarly constructed (Table 1). The high-level expression vector pN was constructed by introducing a 733 bp DNA fragment containing the nptII promoter (Axtell and Beattie, 2002) (Fig. S7) into the EcoRV site of pME6041 such that it regenerated the EcoRV site. Plasmid pNcbcXWV was constructed by introducing a DNA fragment containing cbcXWVPAO1 into the EcoRV site of pN. Primers facing outward from cbcX (PA5378) in pNcbcXWV, PAproXF and PAproXR (Table S1), were treated with T4 polynucleotide kinase (Promega, Madison, WI) and were used with pNcbcXWV to create pNcbcWV; the PCR product was digested with DpnI to remove residual plasmid DNA before incubation with T4 DNA ligase (Promega, Madison, WI). Successful construction was verified by PCR and partial sequencing.
Generation of mutants
The generation of DC3000 strains GB1, GB2, GB3 and GB4 has been described previously (Chen and Beattie, 2008). The cbcXWV locus was deleted in DC3000 by using splice-overlap-extension (SOE) PCR to amplify 1–2 kb of the regions immediately flanking the locus and replacing the locus with a kan cassette surrounded by FRT sites, as we have done previously (Chen and Beattie, 2008). The amplified fragment was cloned into pKnockout-Ω, which was transformed into DC3000. A screen for double recombinants followed by eviction of the kan cassette by introducing pFlp2 (Hoang et al., 1998) resulted in the generation of DCΔcbc (Table 1).
Knockout mutants of B728a were generated by amplifying the internal region of the target genes and cloning the fragment into the SmaI site of pTOK2T (Table 1). The resulting plasmids were conjugated into B728a and mutants were selected as single recombinants. Attempts to generate deletion mutants of B728a using pKnockout-Ω derivatives were not successful. The betT gene was deleted in B728a by using SOE-PCR and a kan cassette surrounded by FRT sites, similar to the strategy used for the DC3000 mutants, but with the integration vector pTOK2T and plasmid mobilization via tri-parental mating, as described previously (Chen and Beattie, 2007). The opuC and cbcXWV loci were deleted by performing SOE-PCR to generate an unmarked deletion of the target loci between the flanking regions, cloning it into pTsacB, mobilizing it into B728a, and selecting initially for single recombinants, with a subsequent screen for a second recombination event following growth in King’s B broth containing 10% sucrose for 1 day and plating on King’s B agar plates containing 10% sucrose. The triple deletion mutant BT was obtained by generating an ΔopuC ΔcbcXWV double mutant, followed by introduction of the marked betT deletion and subsequent eviction of the kan cassette. The quadruple mutants BT0028Ko, BT2916Ko and BT3758Ko were obtained by conjugating the integration vectors pT0028Ko, pT2916Ko and pT3758Ko, respectively, into the BT strain.
The PA14 transposon mutants were obtained from the non-redundant PA14 transposon mutant library. The transposon insertion sites were verified using the two-round arbitrary PCR protocol described by Liberati et al. (2006).
Transport assays
[Methyl-14C]choline and L-carnitine (specific activity of 55 mCi mmol−1) were obtained from American Radiolabelled Chemicals (St. Louis, MO). [Methyl-14C]glycine betaine was prepared by the oxidation of [methyl-14C]choline (Chen and Beattie, 2007). Unless indicated otherwise, P. syringae and P. aeruginosa cells were grown in ½-21C medium to an optical density at 600 nm (OD600) of 0.3–0.5 (mid-log phase) and E. coli cells were grown in M63 medium to an OD600 of 0.6–0.7 (mid-log phase). For bacterial cells grown in the presence of QACs, cells were washed 3× with ½-21C medium to remove residual QACs. Bacterial cells grown in the absence of QACs were generally washed twice with ½-21C medium (Pseudomonas spp.) or M63 medium (E. coli), and re-suspended with the same medium to an OD600 of 1 for evaluation of QAC uptake. Total uptake, initial uptake rates, inhibition of uptake and uptake kinetics were obtained as previously described (Chen and Beattie, 2007; Chen and Beattie, 2008).
Supplementary Material
Acknowledgments
We thank Tim Denny for the pTOK2 plasmid, Erhard Bremer and Bert Poolman for helpful discussions, and Yun Li and Kelly Peterson for technical assistance. This work was supported by National Science Foundation award No. MCB-0524300 (G.A.B.), by National Institutes of Health Grants P20-RR018787 (D.A.H.) and P20-RR021905 (M.J.W.) from the IDeA Program of the National Center for Research Resources, and by a Cystic Fibrosis Foundation Postdoctoral Research Fellowship (M.J.W.).
Footnotes
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ames GF, Liu CE, Joshi AK, Nikaido K. Liganded and unliganded receptors interact with equal affinity with the membrane complex of periplasmic permeases, a subfamily of traffic ATPases. J Biol Chem. 1996;271:14264–14270. doi: 10.1074/jbc.271.24.14264. [DOI] [PubMed] [Google Scholar]
- Anthoni U, Christophersen C, Hougaard L, Nielsen PH. Quaternary ammonium compounds in the biosphere – an example of versatile adaptive strategy. Comp Biochem Physiol. 1991;99B:1–18. [Google Scholar]
- Aurich H, Kleber HP. Kinetics of active carnitine transport in Pseudomonas aeruginosa. Acta Biol Med Ger. 1970;24:559–568. [PubMed] [Google Scholar]
- Axtell CA, Beattie GA. Construction and characterization of a proU–gfp transcriptional fusion that measures water availability in a microbial habitat. Appl Environ Microbiol. 2002;68:4604–4612. doi: 10.1128/AEM.68.9.4604-4612.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemans-Oldehinkel E, Poolman B. On the role of the two extracytoplasmic substrate-binding domains in the ABC transporter OpuA. EMBO J. 2003;22:5983–5993. doi: 10.1093/emboj/cdg581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemans-Oldehinkel E, Doeven MK, Poolman B. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 2006;580:1023–1035. doi: 10.1016/j.febslet.2005.11.079. [DOI] [PubMed] [Google Scholar]
- Bohl E, Shuman HA, Boos W. Mathematical treatment of the kinetics of binding protein dependent transport systems reveals that both the substrate loaded and unloaded binding proteins interact with the membrane components. J Theor Biol. 1995;172:83–94. doi: 10.1006/jtbi.1995.0006. [DOI] [PubMed] [Google Scholar]
- Bourdin B, Adenier H, Perrin Y. Carnitine is associated with fatty acid metabolism in plants. Plant Physiol Biochem. 2007;45:926–931. doi: 10.1016/j.plaphy.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Braun V, Herrmann C. Docking of the periplasmic FecB binding protein to the FecCD transmembrane proteins in the ferric citrate transport system of Escherichia coli. J Bacteriol. 2007;189:6913–6918. doi: 10.1128/JB.00884-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchet A, Eichler K, Mandrand-Berthelot MA. Regulation of the carnitine pathway in Escherichia coli: investigation of the cai-fix divergent promoter region. J Bacteriol. 1998;180:2599–2608. doi: 10.1128/jb.180.10.2599-2608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Beattie GA. Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-β-synthase domains are required for its osmoregulatory function. J Bacteriol. 2007;189:6901–6912. doi: 10.1128/JB.00763-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Beattie GA. Pseudomonas syringae BetT is a low-affinity choline transporter that is responsible for superior osmoprotection by choline over glycine betaine. J Bacteriol. 2008;190:2717–2725. doi: 10.1128/JB.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosquer A, Pichereau V, Pocard JA, Minet J, Cormier M, Bernard T. Nanomolar levels of dimethylsulfoniopropionate, dimethylsulfonioacetate, and glycine betaine are sufficient to confer osmoprotection to Escherichia coli. Appl Environ Microbiol. 1999a;65:3304–3311. doi: 10.1128/aem.65.8.3304-3311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosquer A, Pichereau V, Le Mee D, Le Roch M, Renault J, Carboni B, et al. Toxicity and osmoprotective activities of analogues of glycine betaine obtained by solid phase organic synthesis towards Sinorhizobium meliloti. Bioorg Med Chem Lett. 1999b;9:49–54. doi: 10.1016/s0960-894x(98)00679-9. [DOI] [PubMed] [Google Scholar]
- Daus ML, Grote M, Schneider E. The MalF P2 loop of the ATP-binding cassette transporter MalFGK2 from Escherichia coli and Salmonella enterica serovar typhimurium interacts with maltose binding protein (MalE) throughout the catalytic cycle. J Bacteriol. 2009;191:754–761. doi: 10.1128/JB.01439-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diab F, Bernard T, Bazire A, Haras D, Blanco C, Jebbar M. Succinate-mediated catabolite repression control on the production of glycine betaine catabolic enzymes in Pseudomonas aeruginosa PAO1 under low and elevated salinities. Microbiology. 2006;152:1395–1406. doi: 10.1099/mic.0.28652-0. [DOI] [PubMed] [Google Scholar]
- Dupont L, Garcia I, Poggi MC, Alloing G, Mandon K, Le Rudulier D. The Sinorhizobium meliloti ABC transporter Cho is highly specific for choline and expressed in bacteroids from Medicago sativa nodules. J Bacteriol. 2004;186:5988–5996. doi: 10.1128/JB.186.18.5988-5996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A, et al. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc Natl Acad Sci USA. 2005;102:11064–11069. doi: 10.1073/pnas.0504930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MA, Karunakaran R, Leonard ME, Mouhsine B, Williams A, East AK, et al. Characterization of the quaternary amine transporters of Rhizobium leguminosarum bv. viciae 3841. FEMS Microbiol Lett. 2008;287:212–220. doi: 10.1111/j.1574-6968.2008.01307.x. [DOI] [PubMed] [Google Scholar]
- Galvao TC, de Lorenzo V, Canovas D. Uncoupling of choline-O-sulphate utilization from osmoprotection in Pseudomonas putida. Mol Microbiol. 2006;62:1643–1654. doi: 10.1111/j.1365-2958.2006.05488.x. [DOI] [PubMed] [Google Scholar]
- Grote M, Polyhach Y, Jeschke G, Steinhoff HJ, Schneider E, Bordignon E. Transmembrane signaling in the maltose ABC transporter MalFGK2-E: peri-plasmic MalF-P2 loop communicates substrate availability to the ATP-bound MalK dimer. J Biol Chem. 2009;284:17521–17526. doi: 10.1074/jbc.M109.006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson LJ, Firestone MK. Differential effects of permeating and nonpermeating solutes on the fatty acid composition of Pseudomonas putida. Appl Environ Microbiol. 2000;66:2414–2421. doi: 10.1128/aem.66.6.2414-2421.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, et al. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- van der Heide T, Poolman B. ABC transporters: one, two or four extracytoplasmic substrate-binding sites? EMBO Rep. 2002;3:938–943. doi: 10.1093/embo-reports/kvf201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF, Ames GF. Two periplasmic transport proteins which interact with a common membrane receptor show extensive homology: complete nucleotide sequences. Proc Natl Acad Sci USA. 1981;78:6038–6042. doi: 10.1073/pnas.78.10.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano SS, Ostertag EM, Savage SA, Baker LS, Willis DK, Upper CD. Contribution of the regulatory gene lemA to field fitness of Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1997;63:4304–4312. doi: 10.1128/aem.63.11.4304-4312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Horn C, Sohn-Bosser L, Breed J, Welte W, Schmitt L, Bremer E. Molecular determinants for substrate specificity of the ligand-binding protein OpuAC from Bacillus subtilis for the compatible solutes glycine betaine and proline betaine. J Mol Biol. 2006;357:592–606. doi: 10.1016/j.jmb.2005.12.085. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8 – a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Buchholz M, Clausen J, Nietschke M, Revermann A, Schmid R, Jung K. CaiT of Escherichia coli, a new transporter catalyzing L-carnitine/gamma-butyrobetaine exchange. J Biol Chem. 2002;277:39251–39258. doi: 10.1074/jbc.M206319200. [DOI] [PubMed] [Google Scholar]
- Kempf B, Bremer E. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J Biol Chem. 1995;270:16701–16713. doi: 10.1074/jbc.270.28.16701. [DOI] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- Kitten T, Willis DK. Suppression of a sensor kinase-dependent phenotype in Pseudomonas syringae by ribosomal proteins L35 and L20. J Bacteriol. 1996;178:1548–1555. doi: 10.1128/jb.178.6.1548-1555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleber HP, Aurich H. Inhibition of enzyme induction for oxidation of quaternary compounds by chloramphenicol in Pseudomonas aeruginosa. Naturwissenschaften. 1966;53:234. doi: 10.1007/BF00633912. [DOI] [PubMed] [Google Scholar]
- Kleber HP, Aurich H. Evidence for an inducible active transport of carnitine in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1967;26:255–260. doi: 10.1016/0006-291x(67)90114-3. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Picon A, Konings WN, Poolman B. Kinetics and consequences of binding of nona-and dodecapeptides to the oligopeptide binding protein (OppA) of Lactococcus lactis. Biochemistry. 1999;38:14440–14450. doi: 10.1021/bi9914715. [DOI] [PubMed] [Google Scholar]
- Langsrud S, Sundheim G, Borgmann-Strahsen R. Intrinsic and acquired resistance to quaternary ammonium compounds in food-related Pseudomonas spp. J Appl Microbiol. 2003;95:874–882. doi: 10.1046/j.1365-2672.2003.02064.x. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper JE, Lindow SE. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology. 1987;77:1449–1454. [Google Scholar]
- May G, Faatz E, Villarejo M, Bremer E. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli K12. Mol Gen Genet. 1986;205:225–233. doi: 10.1007/BF00430432. [DOI] [PubMed] [Google Scholar]
- Miller DM, 3rd, Olson JS, Pflugrath JW, Quiocho FA. Rates of ligand binding to periplasmic proteins involved in bacterial transport and chemotaxis. J Biol Chem. 1983;258:13665–13672. [PubMed] [Google Scholar]
- Moore RA, Starratt AN, Ma SW, Morris VL, Cuppels DA. Identification of a chromosomal region required for biosynthesis of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Can J Microbiol. 1989;35:910–917. [Google Scholar]
- Orelle C, Ayvaz T, Everly RM, Klug CS, Davidson AL. Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proc Natl Acad Sci USA. 2008;105:12837–12842. doi: 10.1073/pnas.0803799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald C, Smits SH, Hoing M, Sohn-Bosser L, Dupont L, Le Rudulier D, et al. Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states. J Biol Chem. 2008;283:32848–32859. doi: 10.1074/jbc.M806021200. [DOI] [PubMed] [Google Scholar]
- Picon A, Kunji ER, Lanfermeijer FC, Konings WN, Poolman B. Specificity mutants of the binding protein of the oligopeptide transport system of Lactococcus lactis. J Bacteriol. 2000;182:1600–1608. doi: 10.1128/jb.182.6.1600-1608.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiocho FA. Atomic basis of the exquisite specificity of phosphate and sulfate transport receptors. Kidney Int. 1996;49:943–946. doi: 10.1038/ki.1996.132. [DOI] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat Rev. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rudder KE, Lopez-Lara IM, Geiger O. Inactivation of the gene for phospholipid N-methyltransferase in Sinorhizobium meliloti: phosphatidylcholine is required for normal growth. Mol Microbiol. 2000;37:763–772. doi: 10.1046/j.1365-2958.2000.02032.x. [DOI] [PubMed] [Google Scholar]
- Sage AE, Vasil ML. Osmoprotectant-dependent expression of plcH, encoding the hemolytic phospholipase C, is subject to novel catabolite repression control in Pseudomonas aeruginosa PAO1. J Bacteriol. 1997;179:4874–4881. doi: 10.1128/jb.179.15.4874-4881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvano MA, Lisa TA, Domenech CE. Choline transport in Pseudomonas aeruginosa. Mol Cell Biochem. 1989;85:81–89. doi: 10.1007/BF00223517. [DOI] [PubMed] [Google Scholar]
- Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- Smits SH, Hoing M, Lecher J, Jebbar M, Schmitt L, Bremer E. The compatible-solute-binding protein OpuAC from Bacillus subtilis: ligand binding, site-directed mutagenesis, and crystallographic studies. J Bacteriol. 2008;190:5663–5671. doi: 10.1128/JB.00346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XG, Kidder JM, Scagliotti JP, Klempner MS, Noring R, Hu LT. Analysis of differences in the functional properties of the substrate binding proteins of the Borrelia burgdorferi oligopeptide permease (Opp) operon. J Bacteriol. 2004;186:51–60. doi: 10.1128/JB.186.1.51-60.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo MJ, Hogan DA. Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology. 2009;155:2411–2419. doi: 10.1099/mic.0.028787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo MJ, Szwergold BS, Hogan DA. Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J Bacteriol. 2008;190:2690–2699. doi: 10.1128/JB.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen M, Urban A, Jaeger KE. Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2000;193:201–205. doi: 10.1111/j.1574-6968.2000.tb09424.x. [DOI] [PubMed] [Google Scholar]
- Wolf A, Lee KC, Kirsch JF, Ames GF. Ligand-dependent conformational plasticity of the periplasmic histidine-binding protein HisJ. Involvement in transport specificity. J Biol Chem. 1996;271:21243–21250. doi: 10.1074/jbc.271.35.21243. [DOI] [PubMed] [Google Scholar]
- Wood JM, Bremer E, Csonka LN, Kraemer R, Poolman B, van der Heide T, Smith LT. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp Biochem Physiol A Mol Integr Physiol. 2001;130:437–460. doi: 10.1016/s1095-6433(01)00442-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.