Abstract
Enterocytes, the absorptive cells of the small intestine, mediate efficient absorption of dietary fat (triacylglycerol, TAG). The digestive products of dietary fat are taken up by enterocytes, re-esterified into TAG, and packaged on chylomicrons (CMs) for secretion into blood or temporarily stored within cytoplasmic lipid droplets (CLDs). Altered enterocyte TAG distribution impacts susceptibility to high fat diet associated diseases, but molecular mechanisms directing TAG toward these fates are unclear. Two enzymes, acyl CoA: diacylglycerol acyltransferase 1 (Dgat1) and Dgat2, catalyze the final, committed step of TAG synthesis within enterocytes. Mice with intestine-specific overexpression of Dgat1 (Dgat1Int) or Dgat2 (Dgat2Int), or lack of Dgat1 (Dgat1−/−), were previously found to have altered intestinal TAG secretion and storage. We hypothesized that varying intestinal Dgat1 and Dgat2 levels alters TAG distribution in subcellular pools for CM synthesis as well as the morphology and proteome of CLDs. To test this we used ultrastructural and proteomic methods to investigate intracellular TAG distribution and CLD-associated proteins in enterocytes from Dgat1Int, Dgat2Int, and Dgat1−/− mice 2 hours after a 200 μl oral olive oil gavage. We found that varying levels of intestinal Dgat1 and Dgat2 altered TAG pools involved in CM assembly and secretion, the number or size of CLDs present in enterocytes, and the enterocyte CLD proteome. Overall, these results support a model where Dgat1 and Dgat2 function coordinately to regulate the process of dietary fat absorption by preferentially synthesizing TAG for incorporation into distinct subcellular TAG pools in enterocytes.
Keywords: DGAT, enterocyte, triacylglycerol, chylomicron, cytoplasmic lipid droplet
1. Introduction
The highly efficient absorption of dietary fat (triacylglycerol, TAG) is mediated by enterocytes, the absorptive cells of the small intestine. This process is important in the promotion of health as well as in contributing to disease when dysregulated [1]. Within this process, TAG is first hydrolyzed to fatty acids and monoacylglycerol in the intestinal lumen. These digestive products are then taken up by enterocytes and rapidly re-synthesized into TAG at the ER membrane. The resulting TAG is packaged in chylomicrons (CMs) for secretion into blood or in cytoplasmic lipid droplets (CLDs) for temporary storage within enterocytes [2]. The number and size of CLDs increase and then decrease over time in response to dietary fat consumption [3], suggesting that the stored TAG is hydrolyzed at later time points and either re-esterified into TAG at the ER membrane and used for CM synthesis or used for other cellular functions. The mechanism(s) through which TAG is partitioned for CMs and CLDs in enterocytes is unclear.
The partitioning of TAG for CM and CLD synthesis may be mediated by specific TAG synthesis enzymes. Acyl CoA: diacylglycerol acyltransferase 1 (DGAT1) and DGAT2 both catalyze the final, committed step of TAG synthesis (the acylation of diacylglycerol with a fatty acyl-CoA); however, they share no sequence homology, are members of different gene families and exhibit different biochemical, cellular, and physiological functions [4]. Although both DGAT1 and DGAT2 are expressed ubiquitously, they have different tissue expression patterns [5, 6]. Within the cell, both DGAT1 and DGAT2 localize to the ER, but DGAT2 has also been identified on CLDs and mitochondria-associated membranes [7, 8]. DGAT activity occurs on the lumenal and cytosolic sides of the ER membrane[9]. Topological studies demonstrate that the active site of DGAT1 may be present on either side of the ER membrane as the protein has dual topology [10], while the active site of DGAT2 is oriented toward the cytoplasm [11]. However, additional studies investigating the roles of DGAT1 and DGAT2 in hepatocytes suggest that this partitioning may be more complex [12–15]. Furthermore, the distinct physiological roles of Dgat1 and Dgat2 have been demonstrated in knockout mouse models. Dgat1−/− mice are viable and resistant to diet-induced obesity [16]. Dgat2−/− mice die within a few hours, likely due to extremely low whole body TAG content and an impaired skin barrier [17]. Together, these differences suggest that Dgat1 and Dgat2 have non-redundant roles, but their specific cellular functions remain unclear.
Targeted studies investigating the role of Dgat1 and Dgat2 in intestinal lipid metabolism in mice indicated that these enzymes synthesize TAG for specific cellular fates within enterocytes and contribute to whole body physiology. Dgat1−/− mice secrete TAG in smaller-sized CMs at a reduced rate and exhibit increased TAG storage in enterocyte CLDs compared to wild-type (WT) mice [18]. This intestine phenotype was shown to be critical for their resistance to diet-induced obesity and hepatic steatosis by using a mouse model where Dgat1 was only expressed in the intestine [19]. Mice with intestine-specific overexpression of Dgat1 (Dgat1Int) secrete TAG from the intestine at a similar rate within similar sized CMs compared to WT mice [20]. Interestingly, we were barely able to detect TAG accumulation in enterocytes of Dgat1Int mice by coherent anti-Stokes Raman scattering microscopy, but by biochemical analysis TAG levels were similar to WT mice [19, 20]. This result suggested that using a more sensitive technique to detect intracellular distribution of TAG, such as transmission electron microscopy (TEM) would be necessary to understand the role of Dgat1 and Dgat2 in synthesizing TAG for specific intracellular fates. Mice with intestine-specific overexpression of Dgat2 (Dgat2Int) have higher intestinal TAG secretion rates but unchanged CM size and similar TAG accumulation in enterocytes compared to WT mice [20]. The higher TAG secretion rate seen in Dgat2Int mice contributes to postprandial hypertriglyceridemia and an increased susceptibility to hepatic steatosis [20]. Together, these differences suggest that Dgat1 and Dgat2 have non-redundant roles in dietary fat absorption, but their specific cellular functions in this process requires further investigation.
A major difference within enterocytes of mice with varying levels of Dgat1 and Dgat2 is the storage of TAG within CLDs. Emerging evidence suggests that proteins present on CLDs play important roles in regulating CLD morphology and potentially the balance between TAG storage and secretion [2]. Both targeted and global proteomic approaches have been used to identify proteins associated with enterocyte CLDs that are important in their metabolism [21–25]. These proteins include enzymes involved in lipid synthesis, lipolysis and CM metabolism, as well as other diverse cellular pathways. This localization of these proteins may exert regulatory effects on CM and CLD metabolism directly or indirectly. Determining the differences in proteins that associate with enterocyte CLDs in models with varying levels of Dgat1 and Dgat2 (Dgat1Int, Dgat2Int, and Dgat1−/− mice) will provide further insight into the mechanism through which Dgat1 and Dgat2 synthesize TAG for secretion and storage within enterocytes.
Based on our previous observations in the intestinal phenotypes of mouse models with varying Dgat1 and Dgat2 levels, we hypothesize that varying intestinal Dgat1 and Dgat2 levels alters TAG distribution in subcellular pools for CM synthesis as well as the morphology and proteome of CLDs. To test this hypothesis, enterocytes from the proximal region of the small intestine of mouse models with varying levels of Dgat1 and Dgat2 (Dgat1Int, Dgat2Int, and Dgat1−/−) were analyzed 2 hours after a 200 μl oral olive oil gavage. An ultrastructural examination of enterocytes by TEM and a proteomic analysis of isolated CLDs using LC-MS/MS were performed. Identifying differences in subcellular TAG distribution and the proteins associated with CLDs in enterocytes from mouse models with varying Dgat levels will illustrate unique cellular contributions of Dgat1 and Dgat2 to the process of dietary fat absorption.
2. Methods
2.1 Diet and Animals
All procedures were approved by the Purdue Animal Care and Use Committee. Mice with intestine-specific overexpression of Dgat1 (Dgat1Int), intestine-specific overexpression of Dgat2 (Dgat2Int) and whole body Dgat1 deficient (Dgat1−/−) mice were generated as previous described [16, 19, 20]. Male Dgat1Int, Dgat2Int and Dgat1−/− mice, 4–6 months of age were used in this study. The mice were housed in a specific pathogen-free barrier facility with a 12 hour light/dark cycle (6AM/6PM) and fed a low-fat, rodent chow diet (PicoLab 5053, Lab Diets, Richmond, IN). On the day of the experiment the mice were fasted for 4 hours at the beginning of the light cycle and administered 200 μl of olive oil by oral gavage. No food was available after the gavage.
2.2 Intestinal TAG Secretion
Intestinal TAG secretion in these models of altered intestinal Dgat expression was assessed previously in our lab [19, 20]. Mice were fasted for 4 h starting at the beginning of the light cycle and administered 500 mg/kg Tyloxapol (T0307, Sigma-Aldrich, St. Louis, MO) via IP injection to block lipase activity in circulation. At 30 min post-Tyloxapol injection, mice were given a 200 μl oral olive oil gavage and blood was collected via the submandibular vein at 2 and 4 h post gavage. Plasma TAG concentration was determined using the Wako L-Type TG M determination kit (Wako Chemicals USA).
2.3 Transmission Electron Microscopy (TEM)
Mice were anesthetized using inhaled isoflurane 2 h after a 200 μl oral olive oil gavage. We performed whole mouse perfusion fixation by cardiac infusion of 2% glutaraldehyde and 2% paraformaldehyde in 0.1M sodium cacodylate (pH 7.4). The small intestine was harvested and divided into five equal length segments and labeled S1–S5 (proximal to distal) in relation to the stomach, with S2–3 representing the jejunum. Small pieces of the jejunum were stored in freshly prepared 2% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.4 for at least 2 h at 4°C. Tissues were diced into 1 x 5 mm pieces then fixed with 1% osmium tetroxide in 0.1 M sodium cacodylate, pH 7.4, for 1 h at room temperature. Tissues were then washed repeatedly in distilled deionized water. Dehydration was completed with a graded series of ethanol and specimens were embedded in Embed 812 resin. Thick sections (0.5 μm) were stained with 1% toluidine blue and examined by light microscopy to confirm tissue orientation. Thin sections (80 nm) were cut on a Leica UC6 ultramicrotome and stained in 2% uranyl acetate and lead citrate. Images were acquired on a Tecnai T20 transmission electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 source and operating at 100 kV. The quantitative measurements with acquired micrographs were made using ImageJ software (NIH, USA). Sixty intact enterocytes from the middle area of villi in each group were examined for quantitative measurements (20 cells/mouse; 3 mice/group). All the enterocytes selected for quantitative measurements had TAG present in the subcellular pools for both secretion (ER lumen and Golgi/secretory vesicles (SVs)) and storage (CLDs). The identification of these subcellular TAG pools is based on previously published electron microscopy work [26–30]. The TAG for secretion was assessed by measuring areas of expanded, TAG-containing ER vesicles and Golgi/SVs within an individual cell. The TAG for storage was assessed by counting the number of CLDs and measuring the diameter of CLDs within an individual cell. The amount of TAG stored in CLDs within an individual cell was indicated by the sum of the areas of identified CLDs. The area of a CLD was estimated from its diameter by applying the equation of (mean number of CLDs) x π x (mean radius)2.
2.4 Enterocyte Isolation
The proximal third of the small intestine was excised 2 h after a 200 μl oral olive oil gavage and enterocytes were isolated as previously described [21, 24, 31]. Briefly, intestinal tissue was washed in cold tissue buffer (Hank’s Balanced Salt Solution containing 25 mM HEPES and 1% fetal bovine serum), then incubated in isolation buffer (Calcium and magnesium free Hanks’s Balanced Salt Solution with 1.5 mM EDTA) for 15 minutes at 37°C with rotation. The sample was vortexed briefly and the supernatant, which contained isolated enterocytes, was obtained. This isolation process was repeated, the supernatants were combined, and isolated enterocytes were pelleted.
2.5 CLD Isolation
CLDs were isolated from enterocytes using a previously validated sucrose gradient ultracentrifugation protocol that was modified for mouse enterocytes [24, 32, 33]. Enterocytes were lysed using ice cold lysis buffer containing sucrose (175 mM sucrose, 10 mM HEPES and 1 mM EDTA) and disrupted by passing through a 27 gauge 1 inch needle eight times. The 2 mls of cell lysate was transferred to a centrifuge tube and overlaid with 6 mls of sucrose free lysis buffer. The sample was centrifuged at 20,000 x g at 4°C for 2 h. Following the centrifugation, the sample was frozen at −80°C and then sliced into seven fractions of approximately 1 cm in length, with the top fraction containing isolated CLDs. A sample from the CLD enriched fraction was negatively stained with 2% w/v aqueous uranyl acetate and imaged using a Technai T20 transmission electron microscope (FEI, Hillsboro, OR). Protein concentrations of these fractions were determined using a BCA assay.
2.6 Protein Digestion and LC-MS/MS Analysis
CLD fractions were delipidated using 2:1 chloroform:methanol, followed by protein precipitation using ice cold acetone. In preparation for the digest, samples were denatured using 8M urea and 10 mM DTT for 1 h at 37°C, followed by alkylation (using a 2% 2–iodoethanol, 97.5% acetonitrile, 0.5% TEP solution). Peptides were digested using Trypsin/Lys-C Mix (Promega, Madison, WI, USA) using the Barocycler NEP2320 (Pressure Biosciences, Inc., South Easton, MA, USA) at 50°C under 20,000 psi for 120 cycles (2 h). One cycle consisted of 50 seconds at 20,000 psi followed by 10 seconds at atmospheric pressure. The digestion was quenched using trifluoroacetic acid. Injection volumes were adjusted to load 1 μg of sample onto the column for nanoLC-MS/MS analysis.
Peptides were separated on a nanoLC system (1100 Series LC, Agilent Technologies, Santa Clara, CA). The peptides were loaded on the Agilent 300SB-C18 enrichment column for concentration and the enrichment column was switched in-line with the analytical column after 5 min. Peptides were separated with the C18 reversed phase ZORBAX 300SB-C18 analytical column (0.75 μm × 150 mm, 3.5um) from Agilent. The column was connected to an emission tip from New Objective and coupled to the nano electrospray ionization (ESI) source of the high resolution hybrid ion trap mass spectrometer LTQ Orbitrap XL (Thermo Scientific). The peptides were eluted from the column using acetonitrile (ACN)/0.1% formic acid (mobile phase B). For the first 5 minutes, the column was equilibrated with 95% purified H2O /0.1% formic acid (mobile phase A) followed by the linear gradient of 5% B to 40% B in 65 minutes at 0.3ul/min and from 40% B to 95% B in an additional 10 minutes. The column was washed with 95% of ACN/0.1% formic acid and equilibrated with 95% purified H2O/0.1% formic acid before the next sample was injected. A blank injection was run between samples to avoid carryover and keep the system clean.
The LTQ-Orbitrap mass spectrometer was operated in the data dependent positive acquisition mode at a resolution of 30,000. Each full MS scan was followed by eight MS/MS scans where these abundant molecular ions were selected and fragmented by collision induced dissociation using a normalized collision energy of 35%.
2.7 Proteomic data analysis
The MS and MS/MS peak list files were analyzed using MaxQuant Version 1.5.2.8 [34]. To identify proteins, the MS/MS spectra were searched against the UniProt protein database, which combines the SwissProt (manually annotated and reviewed) and TrEMBL (automatically annotated and not reviewed) databases [35]. The following settings were used for conducting the search: trypsin and Lys-C digestion enzymes with a maximum of two missed cleavages, fixed modification of ethanolyl addition to cysteine, and variable modifications of N-terminal acetylation and oxidation of methionine. The MS tolerance was set at 4.5 ppm with a maximum of five modifications. A false discovery rate of 0.01 was used for protein and peptide identifications, and the spectra were searched against a reverse decoy database. The minimum peptide length was set at seven amino acids, and a minimum score of 40 was required for modified peptides. The MS/MS match tolerance was set at 40 ppm for protein identification, and at least one unique/razor peptide was required for identification.
The MaxQuant output was analyzed using the bioinformatics statistical analysis program Perseus 1.5.1.6. Potential contaminants, such as keratin, were removed, and then LFQ intensities transformed log2(x). Protein intensities falling below the level of detection were assigned a value of 12.0941. Only proteins identified in at least 4 of 6 biological replicates were considered present in a particular genotype and used for the analysis of relative protein levels across genotypes. The identified proteins were grouped into broad clusters based on Gene Ontology (GO) terms for biological process or molecular function using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v 6.7 and the analysis tool in STRING version 10.0. Protein-protein interactions were visualized using STRING version 10.0 using the confidence view (medium confidence, score 0.400).
2.8 qPCR
Total RNA was extracted from jejunal mucosa with RNA STAT60 (Tel-Test, Friendswood, TX) and then DNase treated with a Turbo DNA-free Kit (Ambion, Austin, TX). cDNA was synthesized from 1μg DNase-treated RNA using the AffinityScript QPCR cDNA Synthesis Kit (Stratagene, La Jolla, CA). Total DNA was extracted from tissues using DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). SYBR green qPCR was performed using the Mx3000P QPCR System (Stratagene) and Brilliant III SYBR Green Master Mix (Stratagene). Primers used for determination of relative mRNA levels were produced by Integrated DNA technologies (Coralville, IA) and validated for efficiency and correct product size in cDNA from mouse intestinal mucosa (see Table 1). Mitochondrial content was assessed by determining the ratio of mitochondrial DNA to nuclear genomic DNA. Primers for mitochondrial specific gene are forward 5’-CCCATTCGCGTTATTCTT-3’ and reverse 5’-AAGTTGATCGTAACGGAAGC-3’. Primers for nuclear specific gene are forward 5’-TGGTAAAGCAAAGAGGCCTAA-3’ and reverse 5’-AGAAGTAGCCACAGGGTTGG-3’. The level of each gene was calculated with the comparative Ct method using WT mice as the reference group.
Table 1.
Primers used for QPCR
| Gene | Primer Sequences |
|---|---|
| Cpt1α | F 5’-TGTGGTGTCCAAGTATCTGGCAGT-3’ |
| R 5’-AACACCATAGCCGTCATCAGAAC-3’ | |
| Acox1 | F 5’-ATATTTACGTCACGTTTACCCCGG-3’ |
| R 5’-GGCAGGTCATTCAAGTACGACAC-3’ | |
| Plin2 | F 5’-AAGAGGCCAAACAAAAGAGCCAGGAGACCA-3’ |
| R 5’-ACCCTGAATTTTCTGGTTGGCACTGTGCAT-3’ | |
| Plin3 | F 5’-ATGGAATCCGTGAAACAGGGTGTG-3’ |
| R 5’-TGAGAGGTCCTGGAAGGAGTGAAT-3’ | |
| Seipin | F 5’-TTTGGCTTCGCTGAACAGAAGCAG-3’ |
| R 5’-ATAGCTGAAGAGCACGATGACGCT-3’ | |
| β-actin | F 5’-AGGCCCAGAGCAAGAGAGGTA-3’ |
| R 5’-GGGGTGTTGAAGGTCTCAAACA-3’ |
2.9 Statistical analysis
Values of quantitative data were reported as means ± standard error of the mean (SEM). Statistical analyses of data were performed in SAS (SAS Institute, Inc., Cary, NC). One-way analysis of variance (ANOVA) with Tukey’s HSD post hoc test was used for multiple comparisons. Type 3 tests of fixed effects and Kolmogorov-Smirnov tests were used for characterization of CLD size distribution. A p value < 0.05 denotes a statistically significant difference.
3. Results
3.1 Varying levels of intestinal Dgat1 and Dgat2 alters intracellular TAG distribution in enterocytes
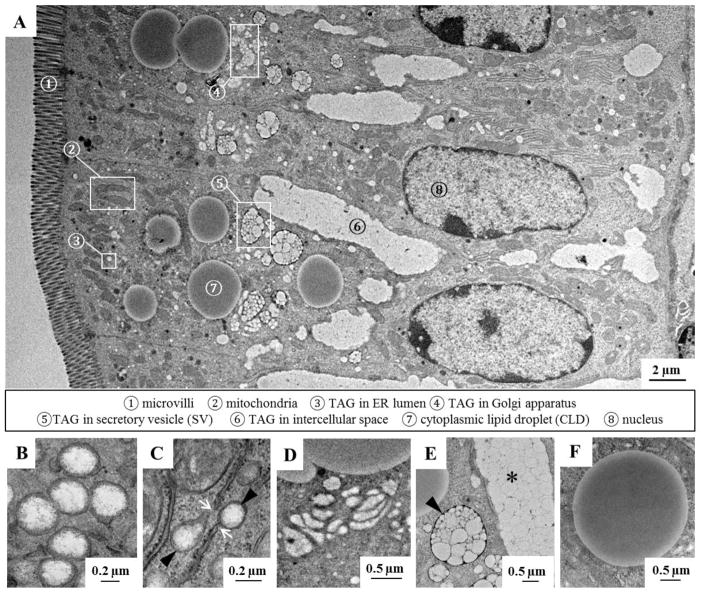
Multiple subcellular TAG pools were identified in enterocytes of WT mice 2 hours after a 200 μl oral olive oil gavage using TEM (Figure 1A), and the ultrastructural morphology of these TAG pools were consistent with previously published work [26–30]. The subcellular TAG pools present in the lumen of the ER, Golgi and Golgi-derived SVs) are involved in CM synthesis and secretion, highlighting the complexity of this process. CMs are first assembled and expanded in the lumen of the ER (Figure 1A–➂, B and C), then transported to the Golgi (Figure 1A–➃ and D) and carried by SVs (Figure 1A–➄ and E) to the basolateral side of enterocytes for secretion. CM-sized lipid particles were also present in the intercellular space between adjacent enterocytes (Figure1A–➅ and E), showing that CMs were being secreted into the lymph. In addition, CLDs (Figure1A–➆ and F), which serve as a temporary TAG storage pool, were present in enterocytes. The subcellular location of TAG pools for secretion and storage were also distinct. TAG in the lumen of the ER was usually found in the apical region of enterocytes beneath the microvilli, whereas TAG in the Golgi and SVs was usually found in the central region of enterocytes just above the nucleus. CLDs of various sizes were primarily found in the cytoplasmic area above the nucleus and occasionally on the basolateral side of enterocytes. In addition, enterocyte CLDs were observed to have physical interactions with multiple organelles including the ER, nucleus, degradative vacuoles (possibly lysosomes) and mitochondria (Supplementary Figure 1).
Figure 1. Identification of subcellular TAG pools within enterocytes by TEM.
(A) An ultrastructural overview of enterocytes from the jejunum of WT mice 2 hours after a 200 μl oral olive oil gavage. At this time point during dietary fat absorption, TAG is found in several subcellular pools involved in either CM or CLD synthesis. CMs are assembled and further expanded in the ER lumen ➂, then transported to the Golgi ➃ and Golgi-derived SVs ➄, and finally secreted from enterocytes into the lymph ➅. CLDs serve as temporary TAG storage pools in enterocytes ➆. (B) TAG present in the lumen of the ER is surrounded by smooth ER membrane. (C) The smooth ER containing TAG (black arrows) is continuous with the rough ER membrane, which can be distinguished by the presence of ribosomes (white arrows). (D) TAG is present within stacks of the Golgi apparatus. (E) CMs are carried within SVs (black arrow) and secreted into the intercellular space (*). (F) CLDs are surrounded by a phospholipid monolayer.
To investigate the role of individual Dgats in directing synthesized TAG into specific subcellular pools, an ultrastructural examination of enterocytes from jejunum segments of WT, Dgat1Int, Dgat2Int and Dgat1−/− mice was performed 2 hours after a 200 μl oral olive oil gavage. No abnormal villus morphology was observed in any of the mouse models, as assessed using light microscopy (Supplementary Figure 2). The pattern of TAG distribution along the villi was similar among the mouse models, with the greatest TAG content in the tip and the least in the crypt (Supplementary Figure 3). The abundance of intracellular TAG in the tip area made it difficult to clearly observe membrane structures in this region. Thus, the enterocytes in the middle area of villi, which were structurally intact and contained moderate amounts of intracellular lipids, were further examined.
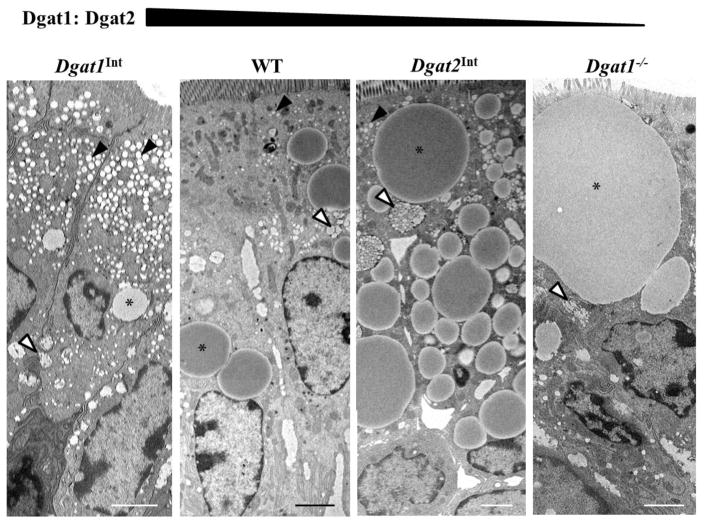
Varying levels of intestinal Dgat1 and Dgat2 altered TAG distribution within enterocytes (Figure 2). Among these models, almost all enterocytes presented with TAG in at least one of the pools involved in the secretion process (ER lumen, Golgi, and/or SVs); however, not every enterocyte contained TAG stored in CLDs. Thus, more than fifty enterocytes representing the middle area of villi from each mouse were viewed to determine CLD presence. We found that when Dgat1 levels were increased (Dgat1Int mice), the percentage of enterocytes containing CLDs was significantly lower than in WT, Dgat2Int and Dgat1−/− mice (Supplementary Figure 4).
Figure 2. Varying levels of intestinal Dgat1 and Dgat2 alters intracellular TAG distribution in enterocytes.
Representative TEM images of enterocytes from the jejunum of Dgat1Int, WT, Dgat2Int and Dgat1−/− mice 2 hours after a 200 μl oral olive oil gavage. Representative CLD (*), TAG in ER lumen (black arrows), and TAG in Golgi/SVs (white arrows) are highlighted. Scale bar = 2 μm.
3.2 Varying levels of intestinal Dgat1 and Dgat2 alters the distribution of TAG in subcellular pools for CM synthesis and secretion
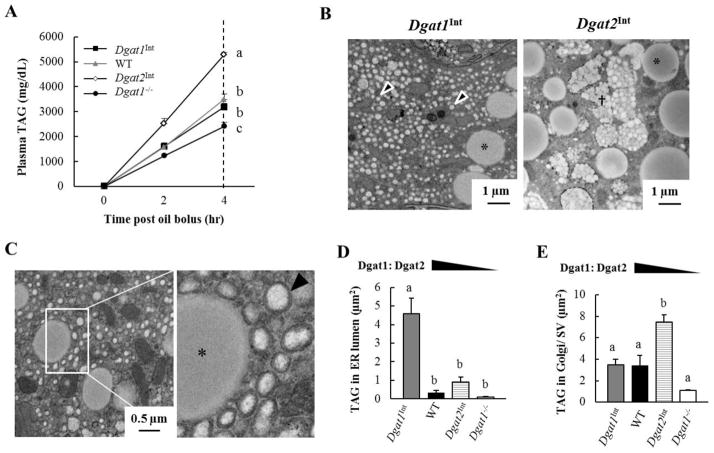
In our previous studies, the intestinal TAG secretion rates of WT, Dgat1Int, Dgat2Int and Dgat1−/− mice were assessed by administering an IP injection of Tyloxapol (to block TAG clearance from circulation) and then measuring plasma TAG levels in response to an oral olive oil gavage. We found that Dgat1Int, Dgat2Int and Dgat1−/− mice had unchanged, increased and decreased rates of intestinal TAG secretion compared to WT mice, respectively (Figure 3A) [19, 20].
Figure 3. Varying levels of intestinal Dgat1 and Dgat2 alters the distribution of TAG in subcellular pools for CM synthesis and secretion.
(A) TAG secretion was assessed by measuring plasma TAG levels in Dgat1Int, WT, Dgat2Int and Dgat1−/− mice given an IP injection of Tyloxapol at 0, 2 and 4 hours after a 200 μl oral olive oil gavage (n = 6–16 mice/group). (B) Representative TEM images of the area above the nucleus in enterocytes from Dgat1Int and Dgat2Int mice 2 hours after a 200 μl oral olive oil gavage. A greater amount of TAG was found in the ER lumen of Dgat1Int enterocytes, whereas a greater amount of TAG was found in Golgi/SVs of Dgat2Int enterocytes. Representative CLD (*), TAG in ER lumen (black arrows), and TAG in Golgi/SVs (†) are highlighted. (C) TEM images highlighting distinct, small CLDs (*) and TAG within the ER lumen (black arrow) in enterocytes of Dgat1Int mice 2 hours after a 200 μl olive oil oral gavage. (D) Mean area of TAG in the ER lumen and (E) mean area of TAG in Golgi/ SVs per cell were quantified using ImageJ. The value of each bar is the average of three biological replicates (20 cells/mouse, n=3 mice/group). Data are represented as mean ± SEM. Different letters denote significant differences, p < 0.05 (one-way ANOVA, Tukey HSD test), N.S = not significant.
To determine whether subcellular TAG pools involved in CM synthesis and secretion are altered by varying the levels of intestinal Dgat1 and Dgat2, we imaged enterocytes 2 hours after a 200 μl oral olive oil gavage using TEM and quantified the area of TAG in the lumen of the ER, as well as in the Golgi/SVs within individual cells from these models. In Dgat1Int mice, the mean area of TAG in the ER is significantly greater than in WT mice (Figure 3B–D), while the mean area of TAG in Golgi/SVs is similar (Figure 3E). In Dgat2Int mice, the mean area of TAG in the Golgi/SVs is significantly greater than in WT mice (Figure 3B and E), while the mean area of TAG in the ER is similar (Figure 3D). In Dgat1−/− mice the mean area of TAG in the ER and Golgi/SVs trended lower than in WT mice (Figure 3D and E). Taken together, varying intestinal levels of Dgat1 and Dgat2 alters subcellular pools of TAG in the ER and Golgi/SVs as well as TAG secretion rate.
3.3 Varying levels of intestinal Dgat1 and Dgat2 alters enterocyte CLD morphology
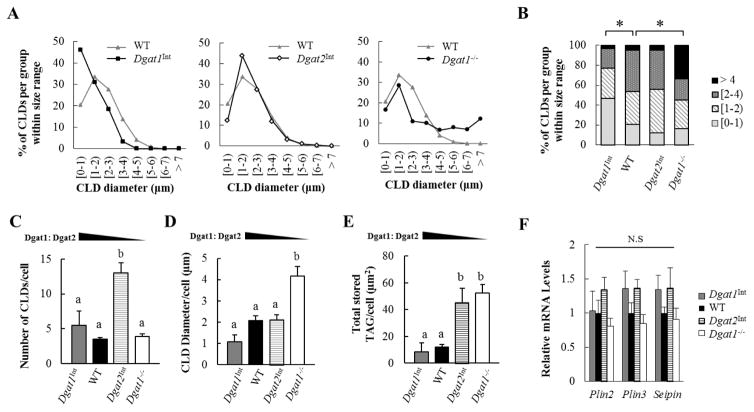
We also identified alterations in CLD morphology in models with varying levels of intestinal Dgat1 and Dgat2. We found that enterocytes of WT, Dgat1Int, Dgat2Int, and Dgat1−/− mice all contained CLDs of various sizes. However, Dgat1Int and Dgat1−/−, but not Dgat2Int mice, had significantly altered CLD size distributions compared to WT mice (Figure 4A and B). In WT and Dgat2Int mice the majority of CLDs were 1–3 μm in diameter. Dgat1Int mice had a right skewed distribution in CLD size, with CLDs smaller than 1 μm in diameter observed most frequently. On the other hand, Dgat1−/− mice exhibited the widest range of CLD sizes and were the only model that could synthesize CLDs greater than 7 μm in diameter (maximum ~10μm in diameter).
Figure 4. Varying levels of intestinal Dgat1 and Dgat2 alters enterocyte CLD morphology.
(A, B) Size distribution of CLDs within enterocytes of mice 2 hours after a 200 μl oral olive oil gavage. All of the CLDs identified from 60 enterocytes (20 enterocytes/mouse, n=3 mice/group) were examined by TEM and the diameter of CLDs were determined using ImageJ. Data is reported as % of all the CLDs measured per group. * denotes a statistically significant difference, p < 0.05 (Kolmogorov-Smirnov tests). (C) Mean number of CLDs and (D) mean size of CLDs per cell (diameter) were determined. (E) The amount of TAG stored in CLDs is estimated by the equation of (mean number of CLD) x π x (mean radius)2. The value of each bar is the average of three biological replicates (20 cells/mouse, n=3 mice/group). (F) qPCR analysis of mRNA levels of genes involved in CLD metabolism. WT mice were the reference group, with their mRNA levels set as 1 (n=3–5 mice/group). Different letters denote significant differences, p < 0.05 (one-way ANOVA, Tukey HSD test), N.S = not significant.
We further analyzed changes in CLD morphology within individual cells containing CLDs. In Dgat1Int mice, the mean number, diameter and total area of CLDs is similar to WT mice (Figure 4C, D, and E). In Dgat2Int mice, the mean diameter of individual CLDs is similar to WT mice (Figure 4D); however, the mean number and total area of CLDs per cell are significantly greater than in WT mice (Figure 4C and E). In Dgat1−/− mice, the mean number of CLDs is similar to WT mice (Figure 4C); however, the mean diameter and total area of CLDs per cell are significantly greater than in WT mice (Figure 4D and E). Even though varying levels of intestinal Dgat1 and Dgat2 alters the size or the number of CLDs per cell, intestinal mRNA levels of proteins involved in CLD metabolism or morphology (Plin2, Plin3, and Seipin) [21, 36] were similar in Dgat1Int, Dgat2Int and Dgat1−/− compared to WT mice 2 hours after a 200 μl oral olive oil gavage (Figure 4F).
3.4 Varying levels of intestinal Dgat1 and Dgat2 alters the enterocyte CLD proteome
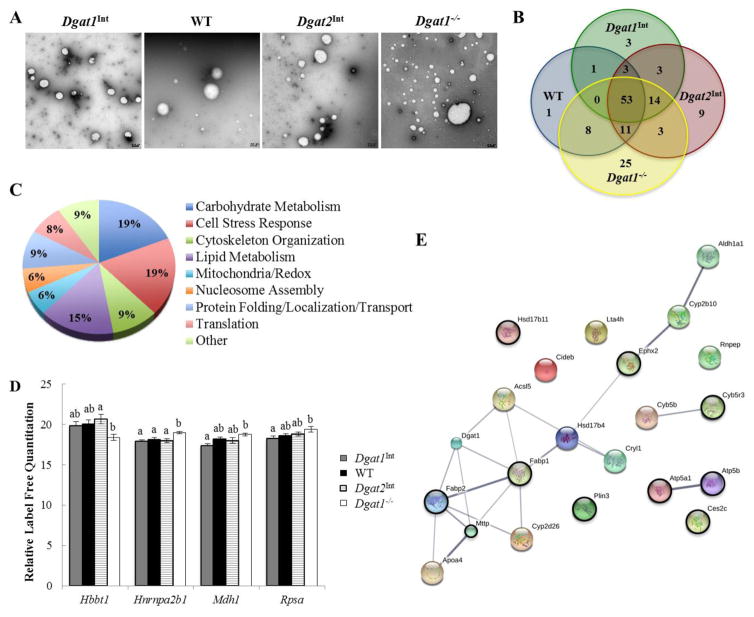
Since proteins associated with CLDs have been shown to regulate their synthesis and catabolism, we hypothesized that varying intestinal levels of Dgat1 and Dgat2 alters the proteins that associate with enterocyte CLDs. These alterations may contribute to the altered subcellular TAG distribution in enterocytes of these models. To determine this, CLDs were isolated from enterocytes of WT, Dgat1Int, Dgat2Int, and Dgat1−/− mice 2 hours after a 200 μl oral olive oil gavage using density gradient ultracentrifugation [24, 32, 33]. This method enriches for CLDs, as demonstrated by immunoblotting of the obtained fractions for representative CLD, cytosolic, and membrane proteins [24]. A challenge in isolating CLDs from cells that make both CLDs and lipoproteins is the potential for ER lumenal lipids and lipoproteins to contaminate the CLD fraction. To minimize this, CLDs were isolated at a much lower speed than those used to isolate ER lumenal lipids or CMs [37–39]. In addition, using negative stain electron microscopy we observed that the isolated CLDs in WT, Dgat1Int, and Dgat2Int mice were similar in size, while the isolated CLDs from Dgat1−/− mice spanned a greater size distribution, with the presence of small as well as larger CLDs compared to the other models (Figure 5A). This result is consistent with the ultrastructural observation in Figure 4A and the images also suggest a lack of broader contamination in the CLD enriched fraction.
Figure 5. Varying levels of intestinal Dgat1 and Dgat2 alters the enterocyte CLD proteome.
CLDs were isolated from enterocytes of Dgat1Int, WT, Dgat2Int, and Dgat1−/− mice 2 hours after a 200μl oral olive oil gavage. (A) Negative stain EM of the intestinal CLD-enriched fraction from a representative sample for each mouse model. Scale bar = 0.5 μm. (B) Overlap of CLD-associated proteins for each mouse model, identified by proteomics. (C) Biological functions of CLD-associated proteins common to all four mouse models, classified based on GO terms (for biological process & molecular function). (D) Differentially expressed CLD-associated proteins common among all mouse models. Different letters denote significantly different relative protein levels (p < 0.05). (E) String map of all proteins with a GO term/functional classification related to lipid/lipoprotein metabolism. Black circles indicate proteins identified in all four genotypes.
Proteins present in the CLD fraction were identified using high resolution tandem mass spectrometry. A total of 158 proteins were identified in at least 4 of 6 biological replicates of at least one of the four mouse models. Of these 158 proteins, 53 of them were present in all four models (Figure 5B, Table 2), and the remainder were unique to one, two, or three genotypes (Figure 5B, Table 3). For further characterization, the gene functional classification tool DAVID and the protein analysis tool STRING were used to classify proteins into broad categories based on gene ontology (GO) terms for biological process or molecular function. Since several proteins had multiple functions that fell under more than one of the broad classifications, their functions were further investigated by performing individual protein searches using the UniProt database. The available information about the function of the protein was used to determine the most appropriate broad classification (Table 2 & 3, Function column).
Table 2.
CLD-associated proteins common among all genotypes.
| Protein | Gene | UniProt ID | Function |
|---|---|---|---|
| Alpha-enolase | Eno1 | P17182 | Carbohydrate Metabolism |
| ATP synthase subunit alpha, mitochondrial | Atp5a1 | Q03265 | Carbohydrate Metabolism |
| ATP synthase subunit beta, mitochondrial | Atp5b | P56480 | Carbohydrate Metabolism |
| Bifunctional ATP-dependent dihydroxyacetone kinase/FAD-AMP lyase (cyclizing) | Dak | Q8VC30 | Carbohydrate Metabolism |
| Fructose-bisphosphate aldolase B | Aldob | Q91Y97 | Carbohydrate Metabolism |
| Glyceraldehyde-3-phosphate dehydrogenase | Gm3839 | P16858 | Carbohydrate Metabolism |
| Malate dehydrogenase, cytoplasmic | Mdh1 | P14152 | Carbohydrate Metabolism |
| Malate dehydrogenase, mitochondrial | Mdh2 | P08249 | Carbohydrate Metabolism |
| Pyrethroid hydrolase Ces2e | Ces2e | Q8BK48 | Carbohydrate Metabolism |
| Pyruvate kinase PKM | Pkm | P52480 | Carbohydrate Metabolism |
| 78 kDa glucose-regulated protein | Hspa5 | P20029 | Cell Stress Response |
| Apoptosis-associated speck-like protein containing a CARD | Pycard | Q9EPB4 | Cell Stress Response |
| Bifunctional 3-phosphoadenosine 5- phosphosulfate synthase 2 | Papss2 | O88428 | Cell Stress Response |
| Bifunctional epoxide hydrolase 2 | Ephx2 | P34914 | Cell Stress Response |
| Glutathione S-transferase Mu 3 | Gstm3 | P19639 | Cell Stress Response |
| Glutathione S-transferase P 1 | Gstp1 | P19157 | Cell Stress Response |
| Hemoglobin subunit beta-1 | Hbbt1 | A8DUK4 | Cell Stress Response |
| Protein disulfide-isomerase | P4hb | P09103 | Cell Stress Response |
| Superoxide dismutase [Cu-Zn] | Sod1 | P08228 | Cell Stress Response |
| Tpm1 | G5E8R0 | Cell Stress Response | |
| Actin, cytoplasmic 2 | Actg1 | P63260 | Cytoskeleton Organization |
| Annexin | Anxa2 | B0V2N7 | Cytoskeleton Organization |
| Tubulin alpha-1C chain | Tuba1c | P68373 | Cytoskeleton Organization |
| Tubulin beta-4B chain | Tubb4b | P68372 | Cytoskeleton Organization |
| Villin-1 | Vil1 | Q62468 | Cytoskeleton Organization |
| Acylcarnitine hydrolase | Ces2c | Q91WG0 | Lipid Metabolism |
| Apolipoprotein A-I | Apoa1 | Q00623 | Lipid Metabolism |
| Estradiol 17-beta-dehydrogenase 11 | Hsd17b11 | Q9EQ06 | Lipid Metabolism |
| Fatty acid-binding protein, intestinal | Fabp2 | P55050 | Lipid Metabolism |
| Fatty acid-binding protein, liver | Fabp1 | P12710 | Lipid Metabolism |
| Microsomal triglyceride transfer protein large subunit | Mttp | O08601 | Lipid Metabolism |
| NADH-cytochrome b5 reductase 3 | Cyb5r3 | Q9DCN2 | Lipid Metabolism |
| Perilipin-3 | Plin3 | Q9DBG5 | Lipid Metabolism |
| Creatine kinase U-type, mitochondrial | Ckmt1 | P30275 | Mitochondria/Redox |
| Cytochrome b5 | Cyb5a | G5E850 | Mitochondria/Redox |
| Dehydrogenase/reductase SDR family member 1 | Dhrs1 | Q99L04 | Mitochondria/Redox |
| Histone H2A type 1-H | Hist1h2ah | Q8CGP6 | Nucleosome Assembly |
| Histone H2B type 1-P | Hist1h2bp | Q8CGP2 | Nucleosome Assembly |
| Histone H4 | Hist1h4a | P62806 | Nucleosome Assembly |
| Heat shock protein HSP 90-beta | Hsp90ab1 | P11499 | Protein Folding |
| 14-3-3 protein zeta/delta | Ywhaz | P63101 | Protein Localization/Transport |
| Alpha-actinin-4 | Actn4 | P57780 | Protein Localization/Transport |
| Annexin A4 | Anxa4 | P97429 | Protein Localization/Transport |
| Clathrin heavy chain | Cltc | Q5SXR6 | Protein Localization/Transport |
| 40S ribosomal protein SA | Rpsa | P14206 | Translation |
| Elongation factor 1-alpha 1 | Eef1a1 | P10126 | Translation |
| Elongation factor 1-delta | Eef1d | D3Z7N2 | Translation |
| Heterogeneous nuclear ribonucleoproteins A2/B1 | Hnrnpa2b1 O88569 | Translation | |
| Calmodulin | Calm1 | P62204 | Other (Calcium Mediated Signaling) |
| Galectin-2 | Lgals2 | Q9CQW5 | Other (Carbohydrate Binding, Unknown Function) |
| Galectin-4 | Lgals4 | Q8K419 | Other (Carbohydrate Binding, Unknown Function) |
| Heat shock cognate 71 kDa protein | Hspa8 | P63017 | Other (Protein Metabolism) |
| Sodium/potassium-transporting ATPase subunit alpha-1 | Atp1a1 | Q8VDN2 | Other (Ion Transport) |
Table 3.
CLD-associated proteins unique to one, two, or three genotypes.
| Protein | Gene | UniProt ID | Function |
|---|---|---|---|
| Dgat1Int | |||
| Cytochrome b-c1 complex subunit 1, mitochondrial | Uqcrc1 | Q9CZ13 | Carbohydrate Metabolism, Mitochondria/Redox |
| Guanine deaminase | Gda | Q9R111 | Carbohydrate Metabolism |
| Diacylglycerol O-acyltransferase 1 | Dgat1 | Q9Z2A7 | Lipid Metabolism |
| WT | |||
| Carbonyl reductase [NADPH] 1 | Cbr1 | P48758 | Carbohydrate Metabolism |
| Dgat2Int | |||
| Aldehyde dehydrogenase X, mitochondrial | Aldh1b1 | Q9CZS1 | Carbohydrate Metabolism |
| Cytochrome c oxidase subunit 2 | Mtco2 | P00405 | Carbohydrate Metabolism |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit | Ddost | O54734 | Carbohydrate Metabolism |
| Ornithine aminotransferase, mitochondrial | Oat | P29758 | Carbohydrate Metabolism |
| Ces2g | E9PV38 | Hydrolase | |
| Peroxisomal multifunctional enzyme type 2 | Hsd17b4 | P51660 | Lipid Metabolism |
| V-type proton ATPase catalytic subunit A | Atp6v1a | P50516 | Nucleotide Biosynthesis |
| Zymogen granule membrane protein 16 | Zg16 | Q8K0C5 | Protein Localization/Transport |
| T-complex protein 1 subunit delta | Cct4 | G5E839 | Protein Metabolism |
| Dgat1−/− | |||
| 4-trimethylaminobutyraldehyde dehydrogenase | Aldh9a1 | Q9JLJ2 | Carbohydrate Metabolism |
| 6-phosphogluconate dehydrogenase, decarboxylating | Pgd | Q9DCD0 | Carbohydrate Metabolism |
| Aldehyde dehydrogenase family 16 member A1 | Aldh16a1 | D3Z0B9 | Carbohydrate Metabolism |
| Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | Gfpt1 | P47856 | Carbohydrate Metabolism |
| Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | Gpd1 | E0CXN5 | Carbohydrate Metabolism |
| Isocitrate dehydrogenase [NADP] cytoplasmic | Idh1 | O88844 | Carbohydrate Metabolism, Cell Stress Response |
| Cell death activator CIDE-B | Cideb | O70303 | Cell Stress Response |
| Nucleophosmin | Npm1 | Q5SQB0 | Cell Stress Response, Cytoskeletal Organization |
| Peroxiredoxin-1 | Prdx1 | B1AXW5 | Cell Stress Response |
| Peroxiredoxin-2 | Prdx2 | Q61171 | Cell Stress Response |
| Peroxiredoxin-5, mitochondrial | Prdx5 | G3UZJ4 | Cell Stress Response |
| Adenylyl cyclase-associated protein 1 | Cap1 | P40124 | Cytoskeleton Organization |
| Destrin | Dstn | Q9R0P5 | Cytoskeletal Organization |
| Cytochrome P450 2D26 | Cyp2d26 | Q8CIM7 | Mitochondria/Redox |
| Bifunctional purine biosynthesis protein PURH | Atic | Q9CWJ9 | Nucleotide Biosynthesis, Mitochondria/Redox |
| Eukaryotic translation initiation factor 5A-1 | Eif5a | J3QPS8 | Protein Localization/Transport |
| Poly(rC)-binding protein 1 | Pcbp1 | P60335 | RNA Processing |
| Small nuclear ribonucleoprotein Sm D3 | Snrpd3 | P62320 | RNA Processing |
| Dgat1Int, WT | |||
| 40S ribosomal protein S8 | Rps8 | P62242 | Translation |
| Dgat1Int, Dgat2Int | |||
| Beta-1,4 N-acetylgalactosaminyltransferase 2 | B4galnt2 | Q09199 | Carbohydrate Metabolism |
| Catalase | Cat | P24270 | Cell Stress Response |
| Transmembrane 9 superfamily member 2 | Tm9sf2 | P58021 | Unknown Function |
| WT, Dgat2Int | |||
| Heat shock protein HSP 90-alpha | Hsp90aa1 | P07901 | Protein Localization/Transport |
| WT, Dgat1−/− | |||
| L-lactate dehydrogenase A chain | Ldha | P06151 | Carbohydrate Metabolism |
| NSFL1 cofactor p47 | Nsfl1c | Q9CZ44 | Cell Stress Response |
| Cofilin-1 | Cfl1 | F8WGL3 | Cytoskeleton Organization |
| Creatine kinase B-type | Ckb | Q04447 | Kinase, Transferase |
| Glutathione S-transferase | Gsta2 | D3Z6A6 | Transferase |
| 40S ribosomal protein S14 | Rps14 | P62264 | Translation |
| 40S ribosomal protein S3 | Rps3 | D3YV43 | Translation |
| Fcgbp | E9Q9C6 | Unknown Function | |
| Dgat2Int, Dgat1−/− | |||
| 14-3-3 protein epsilon | Ywhae | P62259 | Protein Localization/ Transport |
| 40S ribosomal protein S15 | Rps15 | P62843 | Translation |
| 40S ribosomal protein S18 | Rps18 | F6YVP7 | Translation |
| Dgat1Int, WT, Dgat2Int | |||
| Nucleobindin-1 | Nucb1 | H3BK79 | Calcium Ion Binding, Unknown Function |
| Apolipoprotein A-IV | Apoa4 | P06728 | Lipid Metabolism |
| Retinal dehydrogenase 1;Aldehyde dehydrogenase, cytosolic 1 | Aldh1a1 | O35945 | Lipid Metabolism |
| Dgat1Int, Dgat2Int, Dgat1−/− | |||
| Protein disulfide-isomerase A3 | Pdia3 | P27773 | Cell Stress Response |
| Protein disulfide-isomerase A6 | Pdia6 | Q3TML0 | Cell Stress Response |
| Myosin-14 | Myh14 | K3W4R2 | Cytoskeletal Organization |
| Myosin-9 | Myh9 | Q8VDD5 | Cytoskeletal Organization |
| Cytochrome P450 2B10 | Cyp2b10 | Q9WUD0 | Lipid Metabolism |
| Long-chain-fatty-acid--CoA ligase 5 | Acsl5 | Q8JZR0 | Lipid Metabolism |
| Aconitate hydratase, mitochondrial | Aco2 | Q99KI0 | Mitochondria/Redox |
| Cytochrome b5 type B | Cyb5b | Q9CQX2 | Mitochondria/Redox |
| Synaptic vesicle membrane protein VAT-1 homolog | Vat1 | Q62465 | Mitochondria/Redox |
| Ras-related protein Rab-2A | Rab2a | P53994 | Protein Localization/Transport |
| Peptidyl-prolyl cis-trans isomerase A | Ppia | P17742 | Protein Metabolism |
| 40S ribosomal protein S3a | Rps3a | P97351 | Translation |
| 60S ribosomal protein L13 | Rpl13 | P47963 | Translation |
| 60S ribosomal protein L7 | Rpl7 | P14148 | Translation |
| WT, Dgat2Int, Dgat1−/− | |||
| Fructose-1,6-bisphosphatase isozyme 2 | Fbp2 | P70695 | Carbohydrate Metabolism |
| Transketolase | Tkt | P40142 | Carbohydrate Metabolism |
| Heterogeneous nuclear ribonucleoprotein K | Hnrnpk | B2M1R6 | Cell Stress Response |
| Purine nucleoside phosphorylase | Pnp | Q543K9 | Cell Stress Response |
| Ubiquitin-like modifier-activating enzyme 1 | Uba1 | Q02053 | Cell Stress Response |
| Plastin-1 | Pls1 | Q3V0K9 | Cytoskeletal Organization |
| Leukotriene A-4 hydrolase | Lta4h | P24527 | Lipid Metabolism |
| Guanine nucleotide-binding protein subunit beta- 2-like 1 | Gnb2l1 | P68040 | Protein Localization/Transport |
| Transitional endoplasmic reticulum ATPase | Vcp | Q01853 | Protein Localization/Transport |
| Leukocyte elastase inhibitor A | Serpinb1a | Q9D154 | Protein Metabolism |
| Elongation factor 2 | Eef2 | P58252 | Translation |
The 53 proteins common among the four mouse models are involved in multiple biological processes (Figure 5C). The majority of these common proteins are involved in carbohydrate metabolism (19%), cell stress response (19%), or lipid metabolism (15%). The relative abundances of these common proteins were also compared among the genotypes, and four proteins were found to be differentially expressed among the models (Figure 5D). Hemoglobin subunit beta-1 (Hbbt), which was classified as a cell stress response-related protein [40], had significantly higher relative levels in Dgat2Int enterocyte CLD fractions compared to Dgat1−/− fractions. In addition, heterogeneous nuclear ribonucleoproteins A2/B1 (Hnrnpa2b1), which is involved regulating transcription/translation , was expressed at higher levels in Dgat1−/− mice compared to all the other genotypes. Furthermore, 40S ribosomal protein SA (Rpsa), which also plays a role in translation [41], was expressed at higher levels in Dgat1−/− compared to Dgat1Int mice. Additionally, the carbohydrate metabolism-related protein malate dehydrogenase (Mdh1) [42] was expressed at higher levels in Dgat1−/− compared to Dgat1Int mice.
To better understand the role of CLDs in intestinal lipid metabolism in these models with varying levels of intestinal Dgat1 and Dgat2, further analysis was performed on all of the identified proteins with biological process or molecular function GO terms related to lipid metabolism. Overall, 13% of all of the proteins identified in this study were associated with lipid/lipoprotein metabolism. The majority of these proteins have been previously identified as CLD-associated proteins in various cell types, and those identified in enterocyte models are highlighted (Table 4, Previously Identified column). In addition, just under half of these lipid metabolism related proteins were common to WT, Dgat1Int, Dgat2Int, and Dgat1−/− mice, while the remainder were unique to one, two, or three of these models (Table 4). A few of these differentially expressed proteins, including Dgat1, Lambda-crystallin homolog (Cryl1), and Aminopeptidase B (Rnpep), were identified for the first time as being associated with CLDs in these models. Predicted interactions between the identified lipid metabolism proteins were visualized by performing a STRING analysis. The STRING network shows several protein clusters, including one large cluster containing proteins involved in fatty acid modification as well as TAG and CM synthesis (Figure 5E).
Table 4.
CLD-associated proteins involved in lipid/lipoprotein metabolism.
| Protein | Gene | Previously Identified | Dgat1Int | WT | Dgat2Int | Dgat1−/− |
|---|---|---|---|---|---|---|
| Acylcarnitine hydrolase | Ces2c | [24, 55] | ✓ | ✓ | ✓ | ✓ |
| ATP synthase subunit alpha, mitochondrial | Atp5a1 | [24, 52, 54–56, 72, 73] | ✓ | ✓ | ✓ | ✓ |
| ATP synthase subunit beta, mitochondrial | Atp5b | [22, 24, 52, 54–56, 72, 73] | ✓ | ✓ | ✓ | ✓ |
| NADH-cytochrome b5 reductase 3 | Cyb5r3 | [22–24, 52, 54, 55, 72–74] | ✓ | ✓ | ✓ | ✓ |
| Bifunctional epoxide hydrolase 2 | Ephx2 | [24, 54, 55] | ✓ | ✓ | ✓ | ✓ |
| Fatty acid-binding protein, liver Fabp1 | [24, 54, 55] | ✓ | ✓ | ✓ | ||
| Fatty acid-binding protein, intestinal | Fabp2 | [24, 55] | ✓ | ✓ | ✓ | ✓ |
| Estradiol 17-beta- dehydrogenase 11* | Hsd17b11 | [22, 24, 53–56, 72–77] | ✓ | ✓ | ✓ | ✓ |
| Microsomal triglyceride transfer protein large subunit* | Mttp | [22–24, 55] | ✓ | ✓ | ✓ | ✓ |
| Perilipin-3* | Plin3 | [22–24, 53, 55, 56, 72–75, 78–81] | ✓ | ✓ | ✓ | ✓ |
| Retinal dehydrogenase 1 | Aldh1a1 | [24, 55] | ✓ | ✓ | ✓ | |
| Apolipoprotein A-IV* | Apoa4 | [22, 24, 55, 73] | ✓ | |||
| Long-chain-fatty-acid--CoA ligase 5* | Acsl5 | [23, 24, 55] | ✓ | ✓ | ✓ | |
| Cytochrome b5 type B | Cyb5b | [52, 54, 55, 72–75, 77, 79, 81] | ✓ | ✓ | ✓ | |
| Cytochrome P450 2B10 | Cyp2b10 | [24] | ✓ | ✓ | ✓ | |
| Leukotriene A-4 hydrolase | Lta4h | ✓ | ✓ | ✓ | ||
| Aminopeptidase B | Rnpep | ✓ | ✓ | |||
| Lambda-crystallin homolog | Cryl1 | ✓ | ✓ | |||
| Diacylglycerol O-acyltransferase 1 | Dgat1 | ✓ | ||||
| Peroxisomal multifunctional enzyme type 2 | Hsd17b4 | [55, 72, 73] | ✓ | |||
| Cell death activator CIDE-B* | Cideb | ✓ | ||||
| Cytochrome P450 2D26 | Cyp2d26 | [24, 55] | ✓ |
3.5 Varying levels of intestinal Dgat1 and Dgat2 impacts enterocyte mitochondrial biology
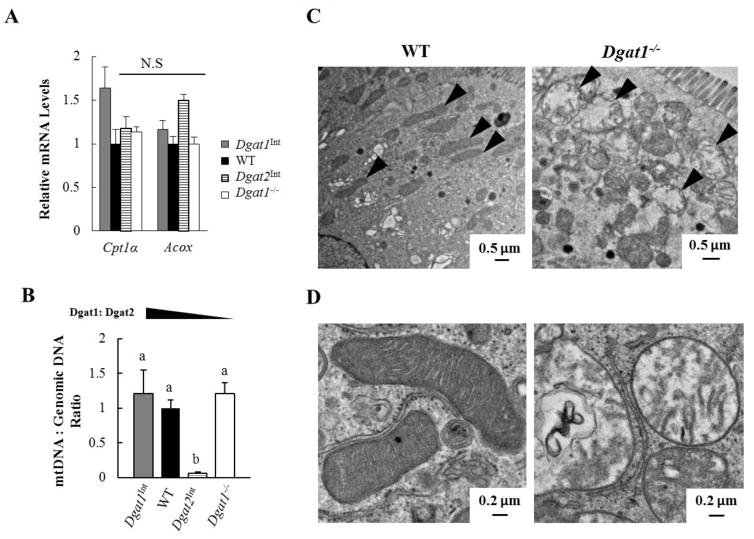
Fatty acid oxidation (FAO) takes place mainly within mitochondria and has the potential to reduce the availability of fatty acids for TAG synthesis and incorporation into CMs and CLDs [43–45]. Therefore, we determined mRNA levels of genes involved in FAO and the ratio of mitochondrial DNA to genomic DNA (indicator of mitochondrial content) in enterocytes of WT, Dgat1Int, Dgat2Int, and Dgat1−/− mice 2 hours after a 200 μl oral olive oil gavage. We found similar mRNA levels of FAO genes (Cpt1 and Acox) in mice with varying intestinal Dgat1 and Dgat2 levels (Figure 6A). In Dgat2Int mice, however, mitochondrial content was significantly lower than that observed in all the other mouse models (Figure 6B). In addition, we frequently observed round, swollen-looking mitochondria in Dgat1−/− mice by TEM (Figure 6C and D). Taken together, varying levels of intestinal Dgat1 and Dgat2 affects enterocyte mitochondrial morphology and content without affecting mRNA levels of FAO genes in response to acute fat challenges.
Figure 6. Varying levels of intestinal Dgat1 and Dgat2 impacts enterocyte mitochondrial biology.
(A) qPCR analysis of mRNA levels of genes involved in FAO. (B) qPCR analysis of the ratio of mitochondrial DNA to nuclear DNA for the assessment of mitochondrial content. WT mice was the reference group, with its DNA level set as 1 (n=3–4 mice/group). Different letters denote significant differences, p < 0.05 (one-way ANOVA, Tukey HSD test), N.S = not significant. (C) Representative TEM images of mitochondria (black arrow) within enterocytes of WT and Dgat1− /− mice 2 hours after a 200 μl oral olive oil gavage. (D) Representative TEM images of normal mitochondria (left) and round, swollen mitochondria (right) found in Dgat1−/− mice 2 hours after a 200 μl oral olive oil gavage.
4. Discussion
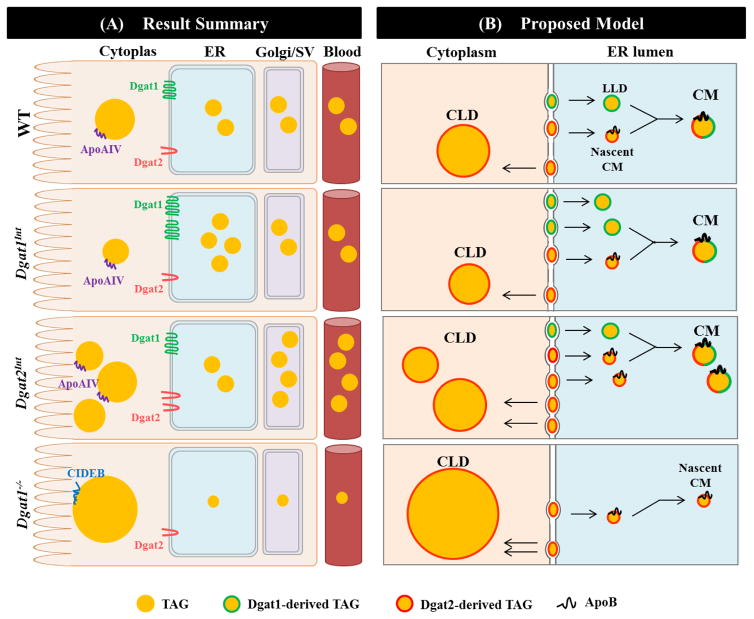
In the present study, we investigated whether varying intestinal Dgat1 and Dgat2 levels alters TAG distribution in subcellular pools for CM synthesis as well as the morphology and proteome of CLDs. The multi-step and dynamic nature of this process makes it challenging to determine distinct roles of Dgat enzymes in CM and CLD synthesis; however the results presented here highlight that Dgat1 and Dgat2 play non-redundant, but coordinated roles in dietary fat absorption. A proposed model for the roles of Dgat1 and Dgat2 in enterocytes during dietary fat absorption was generated based on these as well as previous results (Figure 7). Overall, we found that Dgat1 preferentially synthesizes TAG that supports the synthesis of TAG-rich CMs, whereas Dgat2 preferentially synthesizes TAG that is incorporated into nascent CMs and CLDs.
Figure 7. Summary of results and proposed model of cellular roles of Dgat1 and Dgat2 in synthesizing TAG for CM and CLDs within enterocytes.
(A) Results summary: In Dgat1Int mice, greater amounts of TAG in the ER lumen, similar amounts of TAG in Golgi/SVs, a trend toward smaller CLDs, and a similar intestinal TAG secretion rate were found compared to WT mice. In Dgat2Int mice, similar amounts of TAG in the ER lumen, greater amounts of TAG in Golgi/SVs, an increased number of CLDs, and a higher intestinal TAG secretion rate were observed compared to WT mice. In Dgat1−/− mice, a trend towards less TAG in the ER lumen and Golgi/SVs and an increase in CLD size were found in enterocytes compared to WT mice. Dgat1−/− mice also secrete TAG from the intestine at a slower rate within smaller sized CMs compared to WT mice. Some of the CLD-associated proteins are present in all genotypes, whereas others, such as ApoAIV and Cideb, are unique to certain genotype(s) and may be contributing to the altered subcellular distribution of TAG observed in these models. (B) Proposed model: A proposed model of coordinated, non-redundant roles of Dgat1 and Dgat2 in CM synthesis was generated by combining the results of the current study with previous knowledge of the CM synthesis pathway. CM assembly in enterocytes requires two distinct pools of TAG within the lumen of the ER: ApoB-containing nascent CMs and ApoB-free LLDs. In this model, Dgat1 is proposed to preferentially synthesize TAG for incorporation into ApoB-free LLDs, which determines the availability of TAG for nascent CM expansion and thus the size of CMs. Dgat2 is proposed to preferentially synthesize TAG for incorporation into ApoB-containing nascent CMs, which likely regulates the rate of intestinal TAG secretion. In addition, TAG storage in CLDs and its subsequent catabolism provides substrates for re-synthesis of TAG at the ER membrane that may contribute to CM synthesis and secretion at later times. Based on the results from the current study, Dgat1 is proposed to restrict CLD growth in enterocytes by preferentially synthesizing TAG that is delivered into the ER lumen, thus limiting TAG storage in CLDs. Dgat2, on the other hand, is proposed to preferentially increase TAG storage in CLDs, which may provide fatty acids for re-synthesis of TAG at the ER membrane for incorporation into CMs at later time points. Differential associations of proteins with enterocyte CLDs in these models may serve to regulate the localization and activity of proteins involved in dietary fat absorption.
4.1 Varying levels of intestinal Dgat1 and Dgat2 alters the distribution of TAG in subcellular pools for CM synthesis and secretion
Consistent with earlier ultrastructural studies of enterocytes after consumption of dietary fat [26–29], we observed TAG present along the secretory pathway (in the ER, Golgi and SVs) in enterocytes from WT mice as well as mice with altered intestinal Dgat levels. CMs are first synthesized in ER lumen by a two-step process that involves two distinct pools of TAG: apolipoprotein B (ApoB)-containing nascent CMs and ApoB-free lumenal lipid droplets (LLDs) [46] (Figure 7B). ApoB-containing nascent CMs are generated by incorporation of a small amount of TAG onto ApoB, the main structural protein of CMs. ApoB-free LLDs are used in the second step of CM synthesis for expansion of nascent CMs, yielding TAG-rich pre-CM particles. ApoB-free LLDs were initially identified in mice deficient in ApoB by TEM [30], and have a distinct appearance similar to what was observed predominantly in the Dgat1Int mice in this study (Figure 3B and C). ApoB-containing nascent CMs are challenging to visualize due to their poor lipidation, however only particles containing ApoB are able to be transported from the ER to the Golgi for secretion [47]. Pre-CM particles are transported to the Golgi by pre-CM transport vesicles and ultimately to SVs for exocytosis from the basolateral side of enterocytes into lymph [48]. By quantitatively assessing TEM images we observed alterations in TAG distribution within these subcellular locations in enterocytes of mouse models with varying intestinal Dgat1 and Dgat2 levels (Figure 3). The proposed roles of Dgat1 and Dgat2 in synthesizing TAG for CM synthesis and secretion are discussed below.
We propose that Dgat1 drives the synthesis of TAG for incorporation into ApoB-free LLDs based on the results from Dgat1Int and Dgat1−/− mice (Figure 7). First, the accumulation of TAG in the ER in Dgat1Int enterocytes suggests more ApoB-free LLDs than ApoB-containing particles are produced when Dgat1 is overexpressed (Figure 2 and 3). Although the presence/absence of ApoB on these particles was not assessed, the result mimics the accumulation of LLDs seen in enterocytes of mice with intestine-specific deficiency of ApoB [30] and in response to pharmaceutical inhibition of protein synthesis [26], where only LLDs can be synthesized. However, the greater amount of TAG present in the ER lumen in Dgat1Int enterocytes has little effect on the amount of TAG reaching the Golgi/SVs within enterocytes, the size of CMs in the blood, or intestinal TAG secretion rate, since these are all similar to WT mice [20]. This suggests the presence of other mechanisms that determine which or how many ApoB-free LLDs are able to fuse with ApoB-containing nascent CMs within the ER. Second, significant reductions in the size of CMs secreted into the blood are seen in Dgat1−/− compared to WT mice [20]. This was supported by a trend, but not significant reduction in TAG content in the ER and Golgi/SVs within enterocytes of Dgat1−/− compared to WT mice in this study (Figure 3D and E). This suggests that the absence of Dgat1 leads to a defect in LLD synthesis that prevents the synthesis of TAG-rich CMs. Taken together, our results support a role of Dgat1 in synthesizing TAG that is incorporated into LLDs, which determines the availability of TAG for nascent CM expansion and contributes to the synthesis of TAG-rich CMs.
We propose that Dgat2 drives the synthesis of TAG for incorporation into ApoB-containing nascent CMs, as supported by the results from Dgat2Int and Dgat1−/− mice (Figure 7). First, since lipid particles must acquire ApoB before being transported from the ER to Golgi, the increased TAG content in the Golgi/SVs in Dgat2Int enterocytes suggests that Dgat2 is involved in synthesizing TAG for incorporation into ApoB-containing nascent CMs. In addition, TAG present in the Golgi/SVs within enterocytes positively correlates with CM secretion from enterocytes [26, 29, 30], so the increased TAG content in the Golgi/SVs of Dgat2Int enterocytes is also consistent with the increased intestinal TAG secretion rate in this model [20]. This increased TAG secretion in Dgat2Int mice is likely due to an increased number of CMs being secreted since there are no differences in CM size compared to WT mice [20]. Second, mice lacking Dgat2 die within a few hours of birth [17, 49]. This supports an essential role of Dgat2 in the synthesis and secretion of ApoB-containing lipoproteins from enterocytes for dietary fat absorption and survival. Furthermore, Dgat1−/− mice, which only express Dgat2, have nascent CMs present in the Golgi of enterocytes and are capable of secreting CMs into blood [18]. Taken together, these results suggest that Dgat2 plays an important role in driving the synthesis of ApoB-containing nascent CMs that fuse with LLDs synthesized by Dgat1, and together these enzymes regulate the rate of intestinal TAG secretion.
4.2 Varying levels of intestinal Dgat1 and Dgat2 alters enterocyte CLD morphology
Varying levels of intestinal Dgat1 and Dgat2 affects the number and size of CLDs in enterocytes after dietary fat is consumed, which may in part be due to their contribution to CM synthesis and secretion. The CLD morphology observed in enterocytes of Dgat1Int and Dgat1−/− mice (Figure 4 and 7) support the hypothesis that Dgat1 restricts and Dgat2 promotes CLD expansion reported in other models [12, 50]. In primary hepatocytes, which express both Dgat1 and Dgat2, inhibition of Dgat2 reduced CLD size whereas inhibition of Dgat1 increased CLD size [12]. In addition, the overexpression of Dgat2 in HEK293T cells resulted in increased CLD size [50]. This observation is thought to be due to the localization of Dgat2 to CLDs and its ability to synthesize TAG directly at the CLD surface [50, 51]. Although we did see larger CLDs in Dgat1−/− mice, we did not see an increase in CLD size when Dgat2 was overexpressed in the intestine. We hypothesize that this is due to the combination of high endogenous levels of Dgat1 and high transgenic levels of Dgat2, which is different than in the liver or other cell types that have been used to assess the roles of these enzymes. In this situation the presence of high amounts of Dgat1 can direct synthesized TAG to LLDs and thus restrict CLD expansion. In addition, high amounts of Dgat2 can synthesize TAG for incorporation into both nascent CMs and CLDs, which may also limit CLD expansion.
4.3 Varying levels of intestinal Dgat1 and Dgat2 alters the enterocyte CLD proteome
Consistent with other CLD proteomic studies, proteins involved in diverse cellular functions were found to be associated with enterocyte CLDs among the mouse models. Interestingly, the greatest percentage of the proteins identified in all four mouse models were classified as playing roles in carbohydrate metabolism or cell stress response (Figure 5C). It is unknown whether or not these proteins play functional roles at the CLD surface, are being sequestered at this location to regulate their function/activity in the cell, or are contaminants resulting from the lipid droplet isolation procedure. However, proteins involved in these processes have been identified in CLD proteomics studies from multiple cell types [22–24, 52–56].
Several proteins associated with lipid and lipoprotein metabolism were found to associate with CLDs in the current study, and some were differentially expressed among mice with altered intestinal Dgat levels. We have selected a few of these proteins to highlight below. They include: Cideb, ApoAIV, Dgat1, Cryl1, and Rnpep. Among them, only the localization of Cideb and ApoAIV to CLDs has been validated using additional methods (Western blot or immunofluorescence) [22, 24, 57, 58]. Although validation of the localization of the other proteins to enterocyte CLDs is still needed, the findings suggest that these lipid related proteins may be uniquely contributing to the altered susceptibilities to obesity and related metabolic diseases in these mouse models.
Interestingly, we found that Cideb uniquely associates with enterocyte CLDs in Dgat1−/− mice, while ApoAIV was identified in all of the models except Dgat1−/− mice. Cideb is a member of the CIDE protein family, which has been shown to regulate CLD size in multiple cell types [59, 60]. Cideb has been show to localize to CLDs in mouse liver and when exogenously expressed in Caco-2 cells [58]. Cideb-deficient mice exhibit a decreased TAG secretion rate, decreased CM size, and increased TAG storage within enterocyte CLDs, suggesting Cideb is important for CM lipidation. Since Dgat1−/− and Cideb-deficient mice exhibit a similar phenotype, this suggests that Cideb localization to CLDs may prevent it from promoting CM lipidation at the ER membrane. Consistent with previous studies that validated the localization of ApoAIV to enterocyte CLDs [22, 24], ApoAIV was identified on enterocyte CLDs of WT, Dgat1Int, and Dgat2Int mice in the current study. However ApoAIV was not identified on enterocyte CLDs of Dgat1−/− mice, which exhibit decreased TAG secretion. Since ApoAIV is known to play a role in regulating CM metabolism, it is possible that ApoAIV plays a functional role at the CLD surface that promotes CM secretion. Taken together, the differential association of these proteins that regulate CM metabolism with enterocyte CLDs in Dgat1−/− mice compared to the other models suggests that they may contribute to the altered TAG storage and secretion in this model.
Dgat1 was only found to associate with CLDs in enterocytes of Dgat1Int mice. Dgat1 is thought to localize exclusively to the ER membrane and has not previously been identified in other CLD proteomic studies. However, other TAG synthesis enzymes that were initially thought to be ER resident proteins (Mgat2 [18], Dgat2 [7, 50], and several glycerol-3-phosphate pathway enzymes [7]) have also been found to localize to CLDs and play a role in local CLD growth. Since Dgat1 is a multi-spanning integral membrane protein [61], it is unlikely that it directly associates with the CLD monolayer, and thus may only be identified in Dgat1Int mice due to its overexpression. However we cannot rule out the possibility that Dgat1associates with CLDs through an alternative domain or mechanism, as is the case for Dgat2 [50]. Alternatively, it could be misfolded or non-functional at the CLD surface and targeted to CLDs for degradation (as is the case for apoB [62]) as a way to regulate Dgat1 activity at the ER.
Two lipid related proteins, Cryl1 and Rnpep, were only found to associate with enterocyte CLDs in models with a higher than normal ratio of Dgat2 to Dgat1 (Dgat2Int and Dgat1−/− mice). Cryl1 shares sequence homology with 3-hydroxyacyl-CoA dehydrogenase, an enzyme that catalyzes the third reaction of fatty acid beta oxidation [18, 63]. Human CRYL1 mRNA is expressed at low levels in the small intestine and Cryl1 has been detected at moderate levels in mouse large intestine [64], but very little is known about its role in intestinal lipid metabolism. Therefore, further investigation of this protein’s function in enterocytes is needed. In addition to exhibiting aminopeptidase activity, Rnpep also hydrolyzes the lipid mediator leukotriene A4 (LTA4) [65], resulting in the formation of the pro-inflammatory LTB4 [66]. Interestingly, we also identified Leukotriene A-4 hydrolase (Lta4h) associated with enterocyte CLDs in all except the Dgat1Int model, and this protein has also been identified in other CLD proteomics studies (see Table 4). The presence of these enzymes at the CLD surface suggests that an acute high dietary fat load has the potential to induce an inflammatory response, consistent with previous studies [67].
4.4 Varying levels of intestinal Dgat1 and Dgat2 impacts enterocyte mitochondrial biology
The changes in mitochondrial biology observed in enterocytes of Dgat2Int and Dgat1−/− compared to WT mice have the potential to contribute to the alterations in intestinal TAG metabolism observed in these models. We found that Dgat2Int mice have significantly reduced mitochondrial content, but similar mRNA levels of genes involved in FAO in response to an acute fat challenge. Interestingly, we previously found that Dgat2Int mice have reduced mRNA levels of FAO genes in a fed state after chronic high fat diet feeding [20]. We are uncertain why we observed differences in the chronic high fat diet fed state but not after an acute high fat challenge. However, these results highlight the potential for FAO to alter the availability of fatty acids for TAG synthesis and incorporation into CMs and CLDs. In addition, we found that Dgat1−/− mice have altered mitochondrial morphology but no change in mitochondrial content in enterocytes. The poor membrane integrity of mitochondria in Dgat1−/− enterocytes indicates possible impaired mitochondrial function, which may contribute to or result from the abnormal TAG storage in CLDs observed in this model. Although FAO was not directly measured in these studies, these observations suggest that regulation of FAO is important in these models. Future studies are needed to further assess the role of FAO in intestinal TAG metabolism.
4.5 Intestinal DGAT1 and DGAT2 in mice versus humans
Although it is known that DGAT1 is the major intestinal DGAT enzyme in both mice and humans, it is still unclear whether or not DGAT2 plays an important physiological role within this tissue in humans. Dgat2 is present at significantly lower levels compared to Dgat1 in the intestine of mice, and whether DGAT2 is expressed in human intestine is questionable [6]. One of the reasons for this is that a DGAT1 loss of function mutation has been identified in humans and observed to cause congenital diarrheal disorder resulting in severe outcomes [68, 69]. Dgat1−/− mice, on the other hand, have no obvious increase in fecal fat or diarrhea, and their intestine phenotype contributes to their beneficial resistance to diet induced obesity [16, 18, 19]. In addition, gastrointestinal side effects observed in humans with DGAT1 mutations are also observed in humans treated with DGAT1 inhibitors, leading to the conclusion that these effects may be target specific. More recently, however, the DGAT1 inhibitor pradigastat has been well tolerated in overweight/obese individuals and patients with familial chylomicronemia syndrome [70, 71], suggesting these inhibitors may still be beneficial for some patients. Although mice and humans may differ in terms of the relative contributions of DGAT1 and DGAT2 to dietary fat absorption, knowledge gained from investigating the altered intestinal lipid metabolism in models with varying intestinal levels of Dgat1 and Dgat2 in mice will contribute to novel knowledge of additional players in the process of dietary fat absorption that may be significant in human health and disease.
5. Conclusion
Overall, the results from this study show that Dgat1 and Dgat2 have non-redundant roles in intestinal lipid metabolism that are required for proper regulation of dietary fat absorption. The ultrastructural analyses of enterocytes suggest that Dgat1 preferentially synthesizes TAG for incorporation into ApoB-free LLDs, promoting CM expansion. The results also support that Dgat2 preferentially synthesizes TAG for incorporation into ApoB-containing nascent CMs and CLDs. The defect in CM synthesis and secretion due to the deficiency of Dgat1 highlights the importance of the coordinated functions of Dgat1 and Dgat2 in the process of dietary fat absorption. Furthermore, varying levels of intestinal Dgat1 and Dgat2 impacts the proteins that associate with CLDs, which may play a role in regulating both TAG storage and secretion. Further investigation of these identified proteins may lead to the identification of novel regulators of dietary fat absorption, which have the potential to serve as therapeutic targets for the treatment of obesity, diabetes, and cardiovascular disease.
Supplementary Material
Highlights.
Dgat1 and Dgat2 preferentially synthesize TAG for distinct subcellular pools within enterocytes.
Dgat1 and Dgat2 regulate CM assembly and secretion in enterocytes.
Dgat1 and Dgat2 alter the morphology and proteome of enterocyte CLDs.
Dgat1 and Dgat2 function coordinately to regulate dietary fat absorption.
Acknowledgments
We acknowledge the Multi-Scale Imaging Center, Purdue University, particularly Christopher Gilpin and Laurie Mueller for helping with the TEM imaging. We thank Vicki Hedrick, Mercedes Laland, Tiago Paschoal Sobreira, and the Purdue Proteomics facility at the Bindley Bioscience Center for helping with mass spectrometry and proteomic analysis.
Funding: This work was supported by the National Institutes of Health [5T32DK076540-06, 5T32DK076540-07] and the American Diabetes Association [7-13-IN-05].
Abbreviations
- ApoB
apolipoprotein B
- CLD
cytoplasmic lipid droplet
- CM
chylomicron
- Dgat
diacylglycerol acyltransferase
- FAO
fatty acid oxidation
- LLD
lumenal lipid droplet
- SV
secretory vesicle
- TAG
triacylglycerol
- TEM
transmission electron microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nikolopoulou A, Kadoglou NP. Obesity and metabolic syndrome as related to cardiovascular disease. Expert Rev Cardiovasc Ther. 2012;10:933–939. doi: 10.1586/erc.12.74. [DOI] [PubMed] [Google Scholar]
- 2.D'Aquila T, Hung YH, Carreiro A, Buhman KK. Recent discoveries on absorption of dietary fat: Presence, synthesis, and metabolism of cytoplasmic lipid droplets within enterocytes. Biochimica et biophysica acta. 2016;1861:730–747. doi: 10.1016/j.bbalip.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uchida A, Whitsitt MC, Eustaquio T, Slipchenko MN, Leary JF, Cheng JX, Buhman KK. Reduced triglyceride secretion in response to an acute dietary fat challenge in obese compared to lean mice. Front Physiol. 2012;3:26. doi: 10.3389/fphys.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. Journal of lipid research. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, Erickson SK, Farese RV., Jr Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV., Jr Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- 7.Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV, Jr, Walther TC. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Developmental cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone SJ, Levin MC, Zhou P, Han J, Walther TC, Farese RV., Jr The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J Biol Chem. 2009;284:5352–5361. doi: 10.1074/jbc.M805768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen MR, Corstorphine CC, Zammit VA. Overt and latent activities of diacylglycerol acytransferase in rat liver microsomes: possible roles in very-low-density lipoprotein triacylglycerol secretion. Biochem J. 1997;323(Pt 1):17–21. doi: 10.1042/bj3230017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurie HR, Buckett L, Zammit VA. Evidence that diacylglycerol acyltransferase 1 (DGAT1) has dual membrane topology in the endoplasmic reticulum of HepG2 cells. J Biol Chem. 2011;286:36238–36247. doi: 10.1074/jbc.M111.251900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone SJ, Levin MC, Farese RV., Jr Membrane topology and identification of key functional amino acid residues of murine acyl-CoA:diacylglycerol acyltransferase-2. J Biol Chem. 2006;281:40273–40282. doi: 10.1074/jbc.M607986200. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Li L, Lian J, Watts R, Nelson R, Goodwin B, Lehner R. Roles of Acyl-CoA:Diacylglycerol Acyltransferases 1 and 2 in Triacylglycerol Synthesis and Secretion in Primary Hepatocytes. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:1080–1091. doi: 10.1161/ATVBAHA.114.304584. [DOI] [PubMed] [Google Scholar]
- 13.Yu XX, Murray SF, Pandey SK, Booten SL, Bao D, Song XZ, Kelly S, Chen S, McKay R, Monia BP, Bhanot S. Antisense oligonucleotide reduction of DGAT2 expression improves hepatic steatosis and hyperlipidemia in obese mice. Hepatology. 2005;42:362–371. doi: 10.1002/hep.20783. [DOI] [PubMed] [Google Scholar]
- 14.Choi CS, Savage DB, Kulkarni A, Yu XX, Liu ZX, Morino K, Kim S, Distefano A, Samuel VT, Neschen S, Zhang D, Wang A, Zhang XM, Kahn M, Cline GW, Pandey SK, Geisler JG, Bhanot S, Monia BP, Shulman GI. Suppression of diacylglycerol acyltransferase-2 (DGAT2), but not DGAT1, with antisense oligonucleotides reverses diet-induced hepatic steatosis and insulin resistance. The Journal of biological chemistry. 2007;282:22678–22688. doi: 10.1074/jbc.M704213200. [DOI] [PubMed] [Google Scholar]
- 15.Villanueva CJ, Monetti M, Shih M, Zhou P, Watkins SM, Bhanot S, Farese RV., Jr Specific role for acyl CoA:Diacylglycerol acyltransferase 1 (Dgat1) in hepatic steatosis due to exogenous fatty acids. Hepatology. 2009;50:434–442. doi: 10.1002/hep.22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith SJ, Cases S, Jensen DR, Chen HC, Sande E, Tow B, Sanan DA, Raber J, Eckel RH, Farese RV., Jr Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nature genetics. 2000;25:87–90. doi: 10.1038/75651. [DOI] [PubMed] [Google Scholar]
- 17.Stone SJ, Myers HM, Watkins SM, Brown BE, Feingold KR, Elias PM, Farese RV., Jr Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. The Journal of biological chemistry. 2004;279:11767–11776. doi: 10.1074/jbc.M311000200. [DOI] [PubMed] [Google Scholar]
- 18.Buhman KK, Smith SJ, Stone SJ, Repa JJ, Wong JS, Knapp FF, Jr, Burri BJ, Hamilton RL, Abumrad NA, Farese RV., Jr DGAT1 is not essential for intestinal triacylglycerol absorption or chylomicron synthesis. The Journal of biological chemistry. 2002;277:25474–25479. doi: 10.1074/jbc.M202013200. [DOI] [PubMed] [Google Scholar]
- 19.Lee B, Fast AM, Zhu J, Cheng JX, Buhman KK. Intestine-specific expression of acyl CoA:diacylglycerol acyltransferase 1 reverses resistance to diet-induced hepatic steatosis and obesity in Dgat1-/- mice. Journal of lipid research. 2010;51:1770–1780. doi: 10.1194/jlr.M002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchida A, Slipchenko MN, Eustaquio T, Leary JF, Cheng JX, Buhman KK. Intestinal acyl-CoA:diacylglycerol acyltransferase 2 overexpression enhances postprandial triglyceridemic response and exacerbates high fat diet-induced hepatic triacylglycerol storage. Biochim Biophys Acta. 2013;1831:1377–1385. doi: 10.1016/j.bbalip.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee B, Zhu J, Wolins NE, Cheng JX, Buhman KK. Differential association of adipophilin and TIP47 proteins with cytoplasmic lipid droplets in mouse enterocytes during dietary fat absorption. Biochim Biophys Acta. 2009;1791:1173–1180. doi: 10.1016/j.bbalip.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Bouchoux J, Beilstein F, Pauquai T, Guerrera IC, Chateau D, Ly N, Alqub M, Klein C, Chambaz J, Rousset M, Lacorte JM, Morel E, Demignot S. The proteome of cytosolic lipid droplets isolated from differentiated Caco-2/TC7 enterocytes reveals cell-specific characteristics. Biology of the cell / under the auspices of the European Cell Biology Organization. 2011;103:499–517. doi: 10.1042/BC20110024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beilstein F, Bouchoux J, Rousset M, Demignot S. Proteomic analysis of lipid droplets from Caco-2/TC7 enterocytes identifies novel modulators of lipid secretion. PloS one. 2013;8:e53017. doi: 10.1371/journal.pone.0053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Aquila T, Sirohi D, Grabowski JM, Hedrick VE, Paul LN, Greenberg AS, Kuhn RJ, Buhman KK. Characterization of the proteome of cytoplasmic lipid droplets in mouse enterocytes after a dietary fat challenge. PLoS One. 2015;10:e0126823. doi: 10.1371/journal.pone.0126823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seyer A, Cantiello M, Bertrand-Michel J, Roques V, Nauze M, Bezirard V, Collet X, Touboul D, Brunelle A, Comera C. Lipidomic and spatio-temporal imaging of fat by mass spectrometry in mice duodenum during lipid digestion. PloS one. 2013;8:e58224. doi: 10.1371/journal.pone.0058224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman HI, Cardell RR., Jr Effects of puromycin on the structure of rat intestinal epithelial cells during fat absorption. The Journal of cell biology. 1972;52:15–40. doi: 10.1083/jcb.52.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabesin SM, Frase S. Electron microscopic studies of the assembly, intracellular transport, and secretion of chylomicrons by rat intestine. Journal of lipid research. 1977;18:496–511. [PubMed] [Google Scholar]
- 28.Jersild RA., Jr A time sequence study of fat absorption in the rat jejunum. Am J Anat. 1966;118:135–162. doi: 10.1002/aja.1001180108. [DOI] [PubMed] [Google Scholar]
- 29.Cardell RR, Jr, Badenhausen S, Porter KR. Intestinal triglyceride absorption in the rat. An electron microscopical study. The Journal of cell biology. 1967;34:123–155. doi: 10.1083/jcb.34.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young SG, Cham CM, Pitas RE, Burri BJ, Connolly A, Flynn L, Pappu AS, Wong JS, Hamilton RL, Farese RV., Jr A genetic model for absent chylomicron formation: mice producing apolipoprotein B in the liver, but not in the intestine. The Journal of clinical investigation. 1995;96:2932–2946. doi: 10.1172/JCI118365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie Y, Nassir F, Luo J, Buhman K, Davidson NO. Intestinal lipoprotein assembly in apobec-1−/− mice reveals subtle alterations in triglyceride secretion coupled with a shift to larger lipoproteins. American journal of physiology Gastrointestinal and liver physiology. 2003;285:G735–746. doi: 10.1152/ajpgi.00202.2003. [DOI] [PubMed] [Google Scholar]
- 32.Brasaemle DL, Wolins NE. Isolation of lipid droplets from cells by density gradient centrifugation, Current protocols in cell biology / editorial board. Juan S Bonifacino .. [et al.] 2006;Chapter 3(Unit 3.15) doi: 10.1002/0471143030.cb0315s29. [DOI] [PubMed] [Google Scholar]
- 33.Harris LA, Shew TM, Skinner JR, Wolins NE. A single centrifugation method for isolating fat droplets from cells and tissues. Journal of lipid research. 2012;53:1021–1025. doi: 10.1194/jlr.D023598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 35.UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:27. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK, Garg A, Olarte MJ, Lin Q, Frohlich F, Hannibal-Bach HK, Upadhyayula S, Perrimon N, Kirchhausen T, Ejsing CS, Walther TC, Farese RV. Seipin is required for converting nascent to mature lipid droplets. Elife. 2016;5 doi: 10.7554/eLife.16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Gilham D, Lehner R. Proteomic and lipid characterization of apolipoprotein B-free luminal lipid droplets from mouse liver microsomes: implications for very low density lipoprotein assembly. J Biol Chem. 2007;282:33218–33226. doi: 10.1074/jbc.M706841200. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Quiroga AD, Lehner R. Analysis of lipid droplets in hepatocytes. Methods in cell biology. 2013;116:107–127. doi: 10.1016/B978-0-12-408051-5.00007-3. [DOI] [PubMed] [Google Scholar]
- 39.Potts JL, Fisher RM, Humphreys SM, Gibbons GF, Frayn KN. Separation of lipoprotein fractions by ultracentrifugation: investigation of analytical recovery with sequential flotation and density gradient procedures. Clinica chimica acta; international journal of clinical chemistry. 1994;230:215–220. doi: 10.1016/0009-8981(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 40.Reeder BJ. The redox activity of hemoglobins: from physiologic functions to pathologic mechanisms. Antioxidants & redox signaling. 2010;13:1087–1123. doi: 10.1089/ars.2009.2974. [DOI] [PubMed] [Google Scholar]
- 41.Malygin AA, Babaylova ES, Loktev VB, Karpova GG. A region in the C-terminal domain of ribosomal protein SA required for binding of SA to the human 40S ribosomal subunit. Biochimie. 2011;93:612–617. doi: 10.1016/j.biochi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Minarik P, Tomaskova N, Kollarova M, Antalik M. Malate dehydrogenases--structure and function. General physiology and biophysics. 2002;21:257–265. [PubMed] [Google Scholar]
- 43.Kimura R, Takahashi N, Lin S, Goto T, Murota K, Nakata R, Inoue H, Kawada T. DHA attenuates postprandial hyperlipidemia via activating PPARalpha in intestinal epithelial cells. J Lipid Res. 2013;54:3258–3268. doi: 10.1194/jlr.M034942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchida A, Slipchenko MN, Cheng JX, Buhman KK. Fenofibrate, a peroxisome proliferator-activated receptor alpha agonist, alters triglyceride metabolism in enterocytes of mice. Biochim Biophys Acta. 2011;1811:170–176. doi: 10.1016/j.bbalip.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura R, Takahashi N, Murota K, Yamada Y, Niiya S, Kanzaki N, Murakami Y, Moriyama T, Goto T, Kawada T. Activation of peroxisome proliferator-activated receptor-alpha (PPARalpha) suppresses postprandial lipidemia through fatty acid oxidation in enterocytes. Biochem Biophys Res Commun. 2011;410:1–6. doi: 10.1016/j.bbrc.2011.05.057. [DOI] [PubMed] [Google Scholar]
- 46.Cartwright IJ, Higgins JA. Direct evidence for a two-step assembly of ApoB48-containing lipoproteins in the lumen of the smooth endoplasmic reticulum of rabbit enterocytes. J Biol Chem. 2001;276:48048–48057. doi: 10.1074/jbc.M104229200. [DOI] [PubMed] [Google Scholar]
- 47.Kumar NS, Mansbach CM. Determinants of triacylglycerol transport from the endoplasmic reticulum to the Golgi in intestine. Am J Physiol. 1997;273:G18–30. doi: 10.1152/ajpgi.1997.273.1.G18. [DOI] [PubMed] [Google Scholar]
- 48.Mansbach CM, Siddiqi SA. The biogenesis of chylomicrons. Annual review of physiology. 2010;72:315–333. doi: 10.1146/annurev-physiol-021909-135801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexander CA, Hamilton RL, Havel RJ. Subcellular localization of B apoprotein of plasma lipoproteins in rat liver. The Journal of cell biology. 1976;69:241–263. doi: 10.1083/jcb.69.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McFie PJ, Banman SL, Kary S, Stone SJ. Murine diacylglycerol acyltransferase-2 (DGAT2) can catalyze triacylglycerol synthesis and promote lipid droplet formation independent of its localization to the endoplasmic reticulum. The Journal of biological chemistry. 2011;286:28235–28246. doi: 10.1074/jbc.M111.256008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuerschner L, Moessinger C, Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 2008;9:338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- 52.Ding Y, Wu Y, Zeng R, Liao K. Proteomic profiling of lipid droplet-associated proteins in primary adipocytes of normal and obese mouse. Acta biochimica et biophysica Sinica. 2012;44:394–406. doi: 10.1093/abbs/gms008. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H, Wang Y, Li J, Yu J, Pu J, Li L, Zhang H, Zhang S, Peng G, Yang F, Liu P. Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein a-I. Journal of proteome research. 2011;10:4757–4768. doi: 10.1021/pr200553c. [DOI] [PubMed] [Google Scholar]
- 54.Crunk AE, Monks J, Murakami A, Jackman M, Maclean PS, Ladinsky M, Bales ES, Cain S, Orlicky DJ, McManaman JL. Dynamic regulation of hepatic lipid droplet properties by diet. PloS one. 2013;8:e67631. doi: 10.1371/journal.pone.0067631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan SA, Wollaston-Hayden EE, Markowski TW, Higgins L, Mashek DG. Quantitative analysis of the murine lipid droplet-associated proteome during diet-induced hepatic steatosis. Journal of lipid research. 2015;56:2260–2272. doi: 10.1194/jlr.M056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saka HA, Thompson JW, Chen YS, Dubois LG, Haas JT, Moseley A, Valdivia RH. Chlamydia trachomatis Infection Leads to Defined Alterations to the Lipid Droplet Proteome in Epithelial Cells. PloS one. 2015;10:e0124630. doi: 10.1371/journal.pone.0124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye J, Li JZ, Liu Y, Li X, Yang T, Ma X, Li Q, Yao Z, Li P. Cideb, an ER- and lipid droplet-associated protein, mediates VLDL lipidation and maturation by interacting with apolipoprotein B. Cell metabolism. 2009;9:177–190. doi: 10.1016/j.cmet.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 58.Zhang LJ, Wang C, Yuan Y, Wang H, Wu J, Liu F, Li L, Gao X, Zhao YL, Hu PZ, Li P, Ye J. Cideb facilitates the lipidation of chylomicrons in the small intestine. Journal of lipid research. 2014;55:1279–1287. doi: 10.1194/jlr.M046482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilfling F, Haas JT, Walther TC, Farese RV., Jr Lipid droplet biogenesis. Current opinion in cell biology. 2014;29:39–45. doi: 10.1016/j.ceb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan JS, Seow CJ, Goh VJ, Silver DL. Recent advances in understanding proteins involved in lipid droplet formation, growth and fusion. Journal of genetics and genomics = Yi chuan xue bao. 2014;41:251–259. doi: 10.1016/j.jgg.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Liu Q, Siloto RM, Lehner R, Stone SJ, Weselake RJ. Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Progress in lipid research. 2012;51:350–377. doi: 10.1016/j.plipres.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Molecular Biology of the Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim JJ, Battaile KP. Burning fat: the structural basis of fatty acid beta-oxidation. Current opinion in structural biology. 2002;12:721–728. doi: 10.1016/s0959-440x(02)00390-1. [DOI] [PubMed] [Google Scholar]
- 64.Chen J, Yu L, Li D, Gao Q, Wang J, Huang X, Bi G, Wu H, Zhao S. Human CRYL1, a novel enzyme-crystallin overexpressed in liver and kidney and downregulated in 58% of liver cancer tissues from 60 Chinese patients, and four new homologs from other mammalians. Gene. 2003;302:103–113. doi: 10.1016/s0378-1119(02)01095-8. [DOI] [PubMed] [Google Scholar]
- 65.Cadel S, Foulon T, Viron A, Balogh A, Midol-Monnet S, Noel N, Cohen P. Aminopeptidase B from the rat testis is a bifunctional enzyme structurally related to leukotriene-A4 hydrolase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:2963–2968. doi: 10.1073/pnas.94.7.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haeggstrom JZ. Leukotriene A4 hydrolase/aminopeptidase, the gatekeeper of chemotactic leukotriene B4 biosynthesis. The Journal of biological chemistry. 2004;279:50639–50642. doi: 10.1074/jbc.R400027200. [DOI] [PubMed] [Google Scholar]
- 67.Caviglia JM, Gayet C, Ota T, Hernandez-Ono A, Conlon DM, Jiang H, Fisher EA, Ginsberg HN. Different fatty acids inhibit apoB100 secretion by different pathways: unique roles for ER stress, ceramide, and autophagy. J Lipid Res. 2011;52:1636–1651. doi: 10.1194/jlr.M016931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haas JT, Winter HS, Lim E, Kirby A, Blumenstiel B, DeFelice M, Gabriel S, Jalas C, Branski D, Grueter CA, Toporovski MS, Walther TC, Daly MJ, Farese RV., Jr DGAT1 mutation is linked to a congenital diarrheal disorder. The Journal of clinical investigation. 2012;122:4680–4684. doi: 10.1172/JCI64873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stephen J, Vilboux T, Haberman Y, Pri-Chen H, Pode-Shakked B, Mazaheri S, Marek-Yagel D, Barel O, Di Segni A, Eyal E, Hout-Siloni G, Lahad A, Shalem T, Rechavi G, Malicdan MC, Weiss B, Gahl WA, Anikster Y. Congenital protein losing enteropathy: an inborn error of lipid metabolism due to DGAT1 mutations. Eur J Hum Genet. 2016;24:1268–1273. doi: 10.1038/ejhg.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyers CD, Amer A, Majumdar T, Chen J. Pharmacokinetics, pharmacodynamics, safety, and tolerability of pradigastat, a novel diacylglycerol acyltransferase 1 inhibitor in overweight or obese, but otherwise healthy human subjects. Journal of clinical pharmacology. 2015 doi: 10.1002/jcph.509. [DOI] [PubMed] [Google Scholar]
- 71.Meyers CD, Tremblay K, Amer A, Chen J, Jiang L, Gaudet D. Effect of the DGAT1 inhibitor pradigastat on triglyceride and apoB48 levels in patients with familial chylomicronemia syndrome. Lipids in health and disease. 2015;14:8. doi: 10.1186/s12944-015-0006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dahlhoff M, Frohlich T, Arnold GJ, Muller U, Leonhardt H, Zouboulis CC, Schneider MR. Characterization of the sebocyte lipid droplet proteome reveals novel potential regulators of sebaceous lipogenesis. Exp Cell Res. 2015;332:146–155. doi: 10.1016/j.yexcr.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 73.Li L, Zhang H, Wang W, Hong Y, Wang J, Zhang S, Xu S, Shu Q, Li J, Yang F, Zheng M, Qian Z, Liu P. Comparative proteomics reveals abnormal binding of ATGL and dysferlin on lipid droplets from pressure overload-induced dysfunctional rat hearts. Scientific reports. 2016;6:19782. doi: 10.1038/srep19782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 75.Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R. Association of stomatin with lipid bodies. J Biol Chem. 2004;279:23699–23709. doi: 10.1074/jbc.M310546200. [DOI] [PubMed] [Google Scholar]
- 76.Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 77.Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. Journal of proteome research. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 78.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 79.Sato S, Fukasawa M, Yamakawa Y, Natsume T, Suzuki T, Shoji I, Aizaki H, Miyamura T, Nishijima M. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. Journal of biochemistry. 2006;139:921–930. doi: 10.1093/jb/mvj104. [DOI] [PubMed] [Google Scholar]
- 80.Orban T, Palczewska G, Palczewski K. Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues. The Journal of biological chemistry. 2011;286:17248–17258. doi: 10.1074/jbc.M110.195198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turro S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernandez MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A. Identification and characterization of associated with lipid droplet protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic (Copenhagen, Denmark) 2006;7:1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.