SUMMARY
Ampullary carcinomas are highly malignant neoplasms that can have either intestinal or pancreatobiliary differentiation. To characterize somatic alterations in ampullary carcinomas, we performed whole-exome sequencing and DNA copy number analysis on 60 ampullary carcinomas resected from clinically well-characterized Japanese and American patients. We next selected 92 genes and performed targeted-sequencing to validate significantly mutated genes in additional 112 cancers. The prevalence of driver gene mutations in carcinomas with the intestinal phenotype is different from those with the pancreatobiliary phenotype. We identified a characteristic significantly mutated driver gene (ELF3) as well as previously known driver genes (TP53, KRAS, APC and others). Functional studies demonstrated that ELF3 silencing in normal human epithelial cells enhances their motility and invasion.
INTRODUCTION
Carcinoma of the ampulla of Vater is a highly malignant neoplasm. Ampullary carcinomas localized to the ampulla have a significantly better 5-year survival rate (45%) than those with regional extension beyond the ampulla directly into surrounding tissues (31%), or distant disease (4%) (metastasis in distant lymph nodes or in visceral organs) (Winter et al., 2010).
Three distinct epithelial linings (duodenal, biliary, and pancreatic) converge at the ampulla of Vater, with pancreatic and biliary epithelium merging within the ampulla of Vater to form the epithelium of the ampulla. Ampullary carcinomas can be separated into two histological phenotypes, intestinal-type or pancreatobiliary-type (Ang et al., 2014; Kimura and Ohtsubo, 1988). These phenotypes have different pathogenic and clinical characteristics (Okano et al., 2014). In most studies, carcinomas of the pancreatobiliary subtype are found to be more aggressive than are those of the intestinal subtype (Kim et al., 2012). Several studies suggest that ampullary carcinomas with intestinal differentiation respond to different chemotherapeutic regimens than do ampullary carcinomas with biliary differentiation (Neoptolemos et al., 2012; Shoji et al., 2014).
Because the genomic biology of ampullary carcinomas is currently poorly defined, we have conducted an in-depth analysis of the genomic abnormalities of these carcinomas through an international multicenter collaboration to establish a potential basis for treatments of this disease.
RESULTS
Clinicopathological Features
To provide comprehensive data on the landscape of genomic aberrations that contribute to ampullary carcinoma, we investigated a large cohort of clinically and pathologically well-characterized patients with ampullary cancer that included 70 patients from Japan and 102 patients from USA (Table S1). Since ampullary carcinomas can have either intestinal or pancreatobiliary differentiation (Figure S1), we also clarified the difference of genomic alterations based on the histological phenotypes (Ang et al., 2014). Cases not fitting the categories in which we classified ampullary carcinomas immunohistochemically into intestinal or pancreatobiliary differentiation are regarded as “ambiguous” according to the previous report (Ang et al., 2014). In the present study, pancreatobiliary–type carcinomas were significantly (p = 0.0387) smaller than intestinal-type carcinomas, but pancreatobiliary–type carcinomas were significantly associated with more advanced T-stage (p < 0.0001) and lymph node metastasis (p < 0.0001) (Table 1). We also included 18 separate duodenal carcinomas to compare the genomic characteristics with those of ampullary carcinomas. Three of 18 (16.7%) duodenal carcinomas were immunohistochemically classified into pancreatobiliary-type carcinomas, which is in accordance with the previous report (Ushiku et al., 2014).
Table 1.
Clinocopathlogical Features of Intestinal-type Carcinomas and Pancreatobiliary-type Carcinomas
| Variable | Intestinal-type (n = 93) | Pancreatobiliary-type (n = 66) | p value |
|---|---|---|---|
| Gender | 0.6004 | ||
| Female | 37 (39.8%) | 29 (43.9%) | |
| Male | 56 (60.2%) | 37 (56.1%) | |
| Age (years) | 0.7843 | ||
| Mean | 67.8 | 67.4 | |
| Median | 69 | 68 | |
| Range | 20–88 | 34–84 | |
| T-factor | < 0.0001 | ||
| Tis | 19 (20.4%) | 0 | |
| 1 | 15 (16.1%) | 1 (5.5%) | |
| 2 | 34 (36.6%) | 17 (25.6%) | |
| 3 | 16 (17.2%) | 41 (62.1%) | |
| 4 | 9 (9.7%) | 7 (10.6%) | |
| N-factor | < 0.0001 | ||
| 0 | 59 (63.4%) | 19 (28.8%) | |
| 1 | 34 (36.6%) | 47 (71.2%) | |
| Tumor size (mm) | 0.0387 | ||
| Mean | 27.4 | 22.5 | |
| Range | 5–165 | 8–45 | |
| Tumor differentiation | |||
| Well | 34 (36.6%) | 8 (12.1%) | |
| Moderately | 31 (33.3%) | 35 (53.0%) | |
| Poorly | 21 (22.6%) | 22 (33.3%) | |
| Papillary | 4 (3.3%) | 1 (1.5%) | |
| Others | 3 (3.2%) | 0 | |
| Nationality | |||
| Japanese | 43 (46.2%) | 24 (36.4%) | |
| American | 50 (53.8%) | 42 (63.6%) | |
We macrodissected frozen tissue samples to enrich the tumor fraction relative to the dominant non-neoplastic stromal component and other normal cells. The flowchart of the entire analysis can be found in Figure S1.
Exome-Sequence Analysis
To gain insight into the genetic basis of this tumor type, we determined the exomic sequence of ~20,965 protein coding genes in a discovery set of 60 ampullary carcinomas and 10 duodenal carcinomas. DNA from the enriched neoplastic samples and from matched non-neoplastic tissues was used to prepare fragment libraries (SureSelect Human All Exon v4.0, Agilent Technologies) suitable for massively parallel sequencing (HiSeq2000, Illumina). Sequencing depths were 188X on average (range 117–271). We identified a total of 14,720 somatic non-silent mutations across the entire data set, including 12,564 missense, 718 nonsense, 312 splice-site and 23 read-through mutations, as well as 710 deletions and 393 insertions. Cases with mutation rates < 4.89/Mb (93.1% of all cases; median number of non-silent mutations = 61) were separated from those with mutation rates of > 27.89/Mb. The four cases in the latter class (6.9% of the total cases) were classified as hypermutated (number of non-silent mutations: 928, 1,384, 2,297 and 5,955) and harbored inactivating (nonsense, frameshift or splice-site) mutations in mismatch-repair complex components (Table S2). The significantly (q value < 0.1) mutated driver genes identified in the 60 ampullary carcinomas included KRAS, TP53, APC, ELF3, SMAD4, CTNNB1 and MUC4 (Table S3 and Figure S2). Among them, we identified the significantly mutated driver gene, ELF3, which is characteristic of ampullary carcinomas. The eight (13.3%) somatic mutations in ELF3 among the 60 ampullary carcinomas included six frameshift, one nonsense and one missense mutations. The seven of eight ELF3 mutations were estimated to be heterozygous based on the analysis of adjusted variant allele frequencies for ELF3 mutations by comparison to tumor percent/average variant allele frequencies (Table S4). The remaining case was estimated to be homozygous. Sequencing data demonstrated that ELF3 mutations are present at relatively high allele frequencies, implying that ELF3 mutation may represent an early event (founder mutation) in ampullary carcinomas. OncoPlot summary of significantly mutated, amplified or deleted genes in the discovery screen is shown in Figure S2.
Targeted Deep Sequence Analysis
We next selected 92 genes (genes are listed in Supplemental Experimental Procedures) that were recurrently altered in the discovery screen, or which were well-documented components of a pathway or potentially therapeutic targetable, since alterations in these genes are most likely to be clinically relevant. The sequences of these 92 genes were determined using a target enrichment system (HaloPlex, Agilent Technologies) that differed from that in the exome sequencing. The validation screen consisted 60 ampullary carcinomas in the discovery screen and 112 separate ampullary carcinomas (n = 172 in total), and 10 duodenal carcinomas in the discovery screen and 8 additional duodenal carcinomas (n = 18 in total). Sequencing depths were 2,535X on average (range 1,470–5,814). Validation of candidate mutations with HaloPlex platform showed that 95% of candidate mutations in the discovery set was detected in the validation set. Twenty-four genes were significantly (q < 0.1) mutated driver genes in the 172 ampullary carcinomas (Table 2, Figure 1 and Table S5). The number of significantly (q < 0.1) mutated driver genes in intestinal-type carcinomas (n = 93) was 20 genes (Table S6). On the other hand, the number of significantly (q < 0.1) mutated driver genes in pancreatobiliary-type carcinomas (n = 66) was nine genes (Table S6). There were differences between the genomic landscapes of intestinal phenotype and those of pancreatobiliary phenotype. Among the significantly mutated genes, genes high-ranking based on the prevalence of mutations (Table 3) were similar between intestinal-type ampullary carcinomas and colorectal carcinomas (APC, TP53, KRAS, and SMAD4; p < 0.00001 in a permutation test) and between pancreatobiliary-type carcinomas and pancreatic carcinomas (KRAS, TP53 and SMAD4; p < 0.00001 in a permutation test). The APC, ACVR2A, SOX9 and EPHA6 genes were significantly mutated in intestinal-type carcinomas as compared with pancreatobiliary-type carcinomas (p < 0.05, fisher’s exact test), whereas the KRAS, TP53 and CDH10 genes were significantly mutated in pancreatobiliary-type carcinomas (p < 0.05) as compared to intestinal-type carcinomas.
Table 2.
Significantly (q < 0.1) Mutated Genes in Ampullary Carcinomas (n = 172)
| Gene | Frequency (%) | Patients | Number of inactivating alteration | p value | q value | |
|---|---|---|---|---|---|---|
| 1 | KRAS | 47.7 | 82 | 0 | 1.14E-118 | 1.06E-116 |
| 2 | TP53 | 55.8 | 96 | 30 | 5.88E-117 | 2.73E-115 |
| 3 | CTNNB1 | 23.3 | 40 | 1 | 2.71E-22 | 8.40E-21 |
| 4 | SMAD4 | 16.3 | 28 | 10 | 4.37E-16 | 1.02E-14 |
| 5 | APC | 33.7 | 58 | 54 | 1.89E-14 | 3.52E-13 |
| 6 | ELF3 | 12.2 | 21 | 17 | 1.15E-11 | 1.78E-10 |
| 7 | GNAS | 11.6 | 20 | 1 | 5.58E-08 | 7.41E-07 |
| 8 | ACVR2A | 7.6 | 13 | 9 | 1.34E-05 | 0.000147192 |
| 9 | ERBB3 | 10.5 | 18 | 0 | 1.42E-05 | 0.000147192 |
| 10 | ACVR1B | 8.1 | 14 | 5 | 4.60E-05 | 0.000428017 |
| 11 | ARID2 | 15.7 | 27 | 21 | 5.68E-05 | 0.000479964 |
| 12 | BRAF | 9.3 | 16 | 0 | 0.00013007 | 0.00100804 |
| 13 | CDKN2A | 4.7 | 8 | 5 | 0.000227549 | 0.00162785 |
| 14 | TGFBR1 | 6.4 | 11 | 3 | 0.000609132 | 0.00404637 |
| 15 | SOX9 | 8.1 | 14 | 12 | 0.000876341 | 0.00543331 |
| 16 | LOXHD1 | 11 | 19 | 0 | 0.00128913 | 0.00749306 |
| 17 | ERBB2 | 11.6 | 20 | 0 | 0.00165826 | 0.00907168 |
| 18 | FBXW7 | 5.8 | 10 | 3 | 0.00254902 | 0.01317 |
| 19 | EPHA3 | 7 | 12 | 2 | 0.005577 | 0.027298 |
| 20 | EPHA6 | 5.8 | 10 | 0 | 0.0059163 | 0.0275108 |
| 21 | TGFBR2 | 5.8 | 10 | 3 | 0.0110298 | 0.0466613 |
| 22 | RNF43 | 7 | 12 | 9 | 0.0110381 | 0.0466613 |
| 23 | CNTN4 | 7 | 12 | 2 | 0.0154074 | 0.0622994 |
| 24 | SLITRK5 | 8.1 | 14 | 2 | 0.0255168 | 0.0988775 |
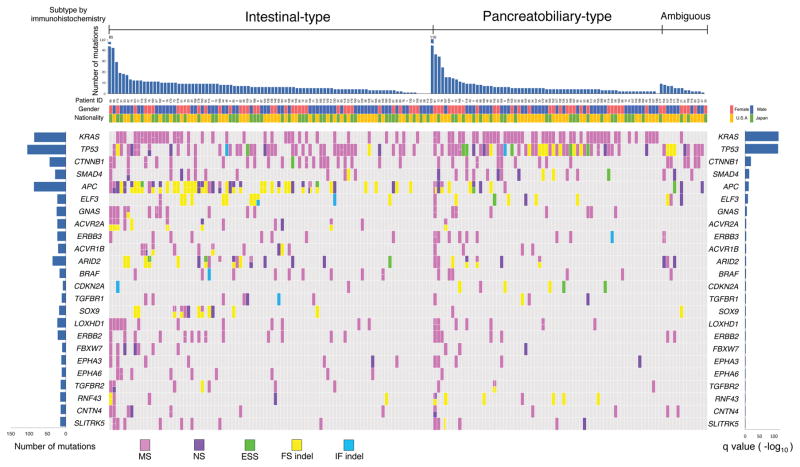
Figure 1. OncoPlot Summary of Significantly (q < 0.1) Mutated Genes in 172 Ampullary Carcinomas (Validation Screen).
Ampullary carcinomas are immunohistchemically classified into intestinal-type or pancreatobiliary-type (or ambiguous-type). The top bar plot shows the number of somatic mutations among 92 genes for tumors from each case. The bottom left plot shows the mutation count for each individual gene. The bottom right bar plot shows the significance of each gene on –log10 (q) values. MS, missense mutation; NS, nonsense mutation; ESS, essential splice-site mutation (the first or last 2 bp of an intron); FS indel, frameshift insertion or deletion; IF indel, in-frame insertion or deletion.
See also Figure S2 and Tables S2, S5, and S10.
Table 3.
Mutation Frequencies of Significantly Mutated Genes in Intestinal-type Ampullary Carcinomas versus Colorectal Carcinomas and Pancreatobiliary-type Carcinomas versus Pancreatic Carcinomas
| Intestinal-type ampullary carcinomas | Colorectal carcinomas (TCGA) | Pancreatobiliary-type ampullary carcinomas | Pancreatic carcinomas (Biankin et al., 2012) | |
|---|---|---|---|---|
| 1 | APC (50%) | APC (81%) | KRAS (68%) | KRAS (99%) |
| 2 | TP53 (46%) | TP53 (60%) | TP53 (67%) | TP53 (33%) |
| 3 | KRAS (39%) | KRAS (43%) | SMAD4 (20%) | SMAD4 (16%) |
| 4 | CTNNB1 (26%) | TTN (31%) | CTNNB1 (15%) | MLL3 (7%) |
| 5 | ARID2 (18%) | PIK3CA (18%) | ERBB3 (14%) | ATM (5%) |
| 6 | ERBB2 (14%) | FBXW7 (11%) | GNAS (12%) | NALCN (5%) |
| 7 | ACVR2A (13%) | SMAD4 (10%) | CDH10 (12%) | ARID1A (4%) |
| 8 | SMAD4 (13%) | NRAS (9%) | ELF3 (11%) | SF3B1 (4%) |
| 9 | GNAS (13%) | TCF7L2 (9%) | CDKN2A (9%) | TGFBR2 (4%) |
| 10 | SOX9 (13%) | FAM123B (7%) | ARID2 (3%) |
The 25 somatic mutations in ELF3 among the 172 ampullary carcinomas included 14 frameshift, 6 missense, 2 nonsense, 2 in-frame, and 1 splice-site mutations (Table S7). We performed the concurrence and mutual exclusion analysis among the significantly mutated genes in ampullary carcinomas, based on the permutation test. Interestingly, ELF3 mutations were mutually exclusive of mutations in CDKN2A (p < 0.0001), but overlapped with mutations in ERBB2 (p = 0.0055), SLITRK5 (p = 0.0204), PIK3CA (p = 0.0232) and FLG (p = 0.0323). The immunohistochemical analysis demonstrated that ELF3 mutations were significantly (p = 0.0006, chi-squared test) correlated with loss of Elf3 protein expression (Table S8 and Figure S5).
Potentially therapeutic targetable mutations, including in ERBB2, ERBB3, BRAF, BRCA2, PIK3CA and others, were identified in 51% (88/172) of patients with ampullary carcinomas (Table S9). Fifteen of 24 (62.5%) ERBB2 mutations were known oncogenic driver mutations and/or in vitro therapeutic targets in several cancers (Greulich et al., 2012; Herter-Sprie et al., 2013; Yamamoto et al., 2014). Among significantly mutated genes, there were no genes which show significant difference between Japanese and American patients with ampullary carcinomas (Tables S10).
We also investigated a smaller subset of 18 duodenal carcinomas. The duodenal carcinomas had two significantly (q < 0.1) mutated driver genes (KRAS and TP53).
Somatic Copy-Number Alterations (SCNAs)
Fifty-eight ampullary carcinomas and 10 duodenal carcinomas which were included in the whole exome sequencing in the discovery screen were profiled for SCNAs with Agilent CGH array (SurePrint G3 CGH Microarray, 1x1M). Matched non-tumor tissues were used as a copy number reference. We applied the GISTIC2.0 algorithm (Beroukhim et al., 2007) to identify SCNAs that might be responsible for driver tumorigenesis and identified 13 focal events (Figure S3). There were four regions of significant (q < 0.1) focal amplification and two regions of significant (q < 0.1) focal deletion. Significantly amplified chromosome arms were 8q24.21 (including MYC), 12q15 (including MDM2), 3q26.2 (including PRKCI and SKIL) (Hagerstrand et al., 2013) and 1p31.1 (including NEGR1) (Takita et al., 2011). Significantly deleted chromosome arms were 9p21.3 (including CDKN2A) and 18q21.2 (including SMAD4). In general, intestinal-type carcinomas tended to have more SCNAs than those of pancreatobiliary-type carcinomas. Unexpectedly, the deletion in 9p21.3 (including CDKN2A) was frequently observed in intestinal-type carcinomas (33.3%, 11/33) (Figure S2), which is uncommon in colorectal carcinomas (Cancer Genome Atlas, 2012). The deletion in 18q21.2 (including SMAD4) was detected in both phenotypes: intestinal phenotype, 24.2% (8/33); pancreatobiliary phenotype, 25.0% (6/24).
Altered Pathways
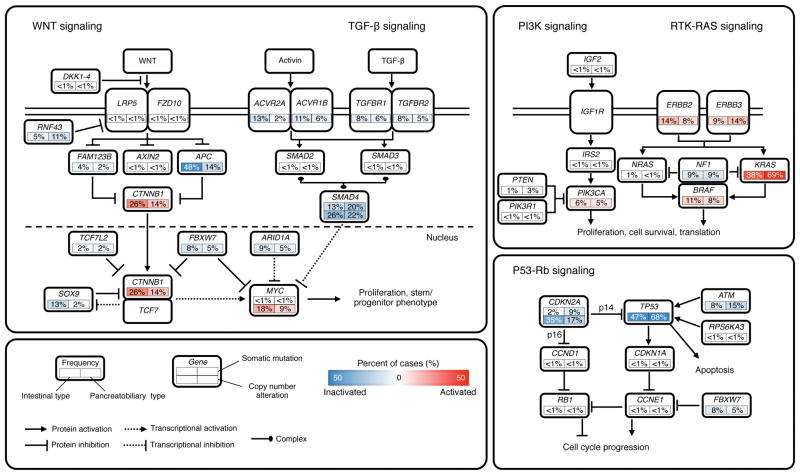
Integrated analysis of mutations (n = 172) and copy number changes (n = 58) enriched our understanding of how some well-defined pathways are deregulated. We grouped samples by histological phenotype and identified alterations in the WNT, TGF-β, PI3K, RTK-RAS and P53-Rb signaling pathways (Figure 2).
Figure 2. Diversity and Frequency of Genetic Changes Leading to Deregulation of Signaling Pathways in Ampullary Carcinomas.
Mutation frequencies are expressed as a percentage of cases analyzed in the validation screen: intestinal-type, n = 93; pancreatobiliary-type, n = 66. Frequencies of high-level focal amplifications and deletions are expressed as a percentage of cases analyzed in the discovery screen: intestinal-type, n = 32; pancreatobiliary-type, n = 23. Red denotes activated genes and blue denotes inactivated genes.
We found that the WNT signaling pathway was more commonly (p < 0.001) altered in the intestinal-type carcinomas (76% (71/93)), mainly by APC and CTNNB1 alterations, as compared to pancreatobiliary-type carcinomas (38% (25/66)), based on the somatic mutation data analysis. On the other hand, RTK-RAS signaling and P53-Rb signaling alterations were significantly more common in pancreatobiliary-type carcinomas than in intestinal-type carcinomas: RTK-RAS signaling, 81.8% (54/66) in pancreatobiliary versus 63.4% (59/93) in intestinal, p = 0.006; P53-Rb signaling, 74.2% (49/66) in pancreatobiliary versus 54.8% (51/93) in intestinal, p = 0.008. These findings are consistent with the recent report sequencing of 279 cancer genes in 32 ampullary carcinomas (Hechtman et al., 2015).
Mutation Pattern and Signature Analyses
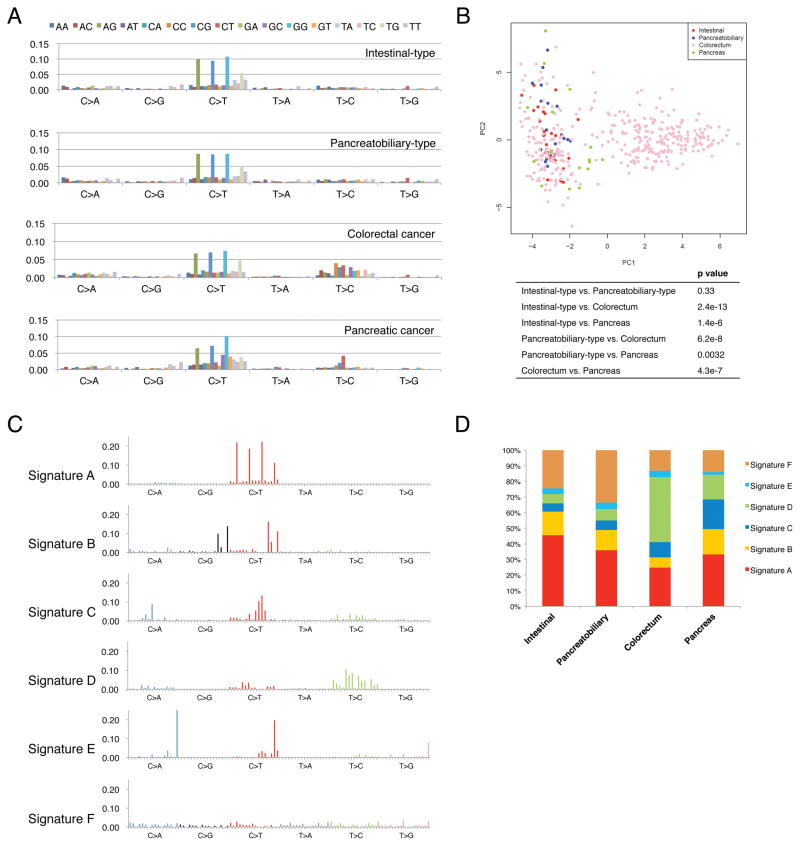
We applied the 96-substitution classification to ampullary carcinomas, colorectal carcinomas and pancreatic carcinomas. Mutational data of individual patients with colorectal carcinoma or pancreatic carcinoma were obtained from mutational catalogues of ICGC (Alexandrov et al., 2013). They were mixed with mutational data of patients with ampullary carcinoma and we re-analyzed all of these data together. In ampullary carcinomas, CpG>TpG substitutions were specifically increased compared with other substitutions regardless of histological phenotypes and nationalities (Figure 3A). Principal-component analysis (PCA) based on frequency in the 96-substituion matrix demonstrated that substitution patterns of intestinal-type and pancreatobiliary-type carcinomas were similar, but significantly different from colorectal carcinomas and pancreatic carcinomas (Figure 3B). We next applied non-negative matrix factorization (NMF) analysis to the 96-substitution patterns of ampullary carcinomas, colorectal carcinomas and pancreatic carcinomas. We excluded hypermutated tumors in further NMF analyses since there is a very significant difference in the mutational signatures between hypermutated and non-hypermutated tumors (Figure S4). Based on the work of Alexandrov et al (Alexandrov et al., 2013), and the updated COSMIC database (http://cancer.sanger.ac.uk/cosmic/signatures), these four hypermutated tumors belong to Signature 6, one of four mutational signatures associated with defective DNA mismatch repair. After excluding these hypermutated tumors, we identified six mutation signatures (Figure 3C). Signature A is characterized by prominence of C>T substitutions at NpCpG trinucleotides, and Signature B is characterized by C>T and C>G mutations at TpCpN trinucleotides. Signature B is consistent with Signature 2 in the previous report (Alexandrov et al., 2013), which is considered to be associated with over activity of members of the APOBEC family of cytidine deaminases (Nik-Zainal et al., 2012). On the other hand, Signature A is consistent with Signature 1 (Alexandrov et al., 2013), and was significantly associated with age, tumor size and intestinal-type in the present study (Figure S4). There was a significant correlation (Pearson’s correlation coefficient = 0.930, p = 0.0072) only between the mutational signatures of intestinal-type and pancreatobiliary-type across four cancer types (Figure 3D). These findings suggest that even though ampullary carcinomas can be classified into two histological phenotypes (intestinal-type or pancreatobiliary-type) based on the prevalence of driver gene mutations, they can be considered as the same category of disease from the point of view of mutational patterns and signatures.
Figure 3. Mutational Patterns and Signatures.
(A) Each signature is displayed according to the 96-substitution classification defined by the substitution class and sequence context immediately 5′ and 3′ to the mutated base. The top legend shows the bases immediately 5′ and 3′ to each substitution. The y axis indicates the frequency of the 96-substitution patterns. Mutational data of individual patients with colorectal carcinomas or pancreatic carcinomas were obtained from mutational catalogues of ICGC (International Cancer Genome Consortium). They were mixed with mutational data of patients with ampullary carcinomas and we re-analyzed all of these data together.
(B) Principal component analysis (PCA) of somatic substitution patterns in individual patients with intestinal-type ampullary carcinoma, pancreatobiliary-type ampullary carcinoma, colorectal cancer or pancreatic cancer. The composition of substitution patterns was statistically significantly different among types of cancer excluding intestinal-type ampullary carcinoma versus pancreatobiliary-type ampullary carcinoma. P values were shown in the box.
(C) Non-negative matrix factorization (NMF) analysis to the 96-substitution patterns of ampullary carcinomas, colorectal carcinomas and pancreatic carcinomas identifies six mutational signatures.
(D) Contribution of the six mutational signatures to intestinal-type ampullary carcinoma, pancreatobiliary-type ampullary carcinoma, colorectal cancer and pancreatic cancer. The y axis indicates the percentage of mutations comprised in each signature.
See also Figure S4.
Recurrent ELF3 Mutations and Functional Assays
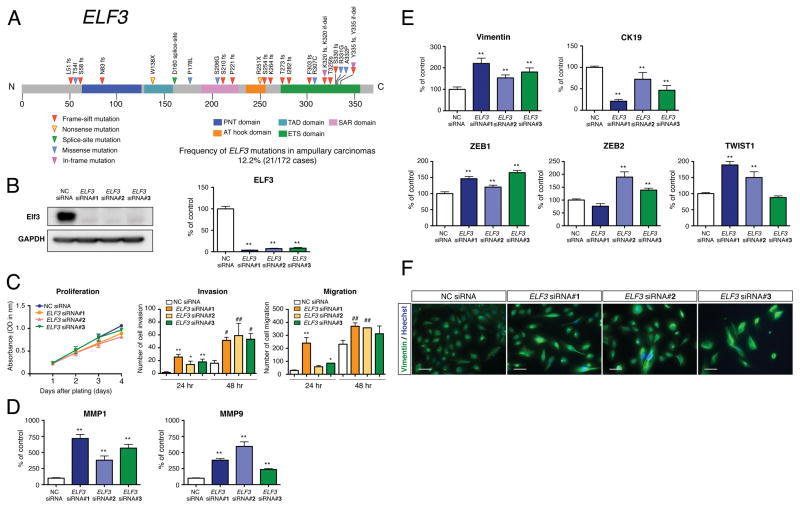
In the validation screen of 172 ampullary cancers, most of the ELF3 mutations were deleterious (16 insertions or deletions, 2 nonsense and 1 splice-site mutations), while 6 were missense (Figure 4A and Table S7). The ELF3 mutations were validated by Sanger sequencing. These mutations were observed in intestinal-type carcinoma, 11 patients (11.8%, 11/93); pancreatobiliary-type carcinoma, 7 patients (10.6%, 7/66); ambiguous-type carcinoma, 3 patients (23.1%, 3/13); duodenal carcinoma, 1 patient (5.6%, 1/18) and across racial differences: Japanese, 8 patients (11.4%, 8/70); American, 13 patients (12.7%, 13/102).
Figure 4. Recurrent ELF3 Mutations and Functional Assays.
(A) Recurrent somatic mutations of ELF3 in 21 ampullary carcinomas and one duodenal carcinoma. ELF3 is shown in the context of the protein domain model derived from UniProt and Pfam annotations. Numbers refer to amino acid residues. Each inverted triangle represents an individual mutated tumor sample: frameshift, nonsense, splice-site, missense and in-flame mutations are represented by filled red, yellow, green, blue and pink inverted triangles, respectively. Domains are depicted with various colors with an appropriate key located below the domain model.
(B) Representative western blot for the expression of Elf3 in HBDEC2-3H10 cells treated with the specific siRNA (ELF3 siRNA #1, #2 and #3) and negative control siRNA (NC siRNA). Quantitative RT-PCR analysis for the expression of ELF3 in control and ELF3 knockdown cells. Total RNA was prepared 72 hr after transfection (mean ± SEM n = 4 per group, **p < 0.01 versus NC siRNA).
(C) Cellular proliferation and invasion/migration assays with control and ELF3 knockdown cells. Cell growth was measured for 1, 2, 3 and 4 days and performed in triplicate (mean ± SEM). Cell invasion and migration were measured for 24 and 48 hr and performed in three times (mean ± SD, *p < 0.05 versus 24 hr NC siRNA, **p < 0.01 versus 24 hr NC siRNA, #p < 0.05 versus 48 hr NC siRNA, ##p < 0.01 versus 48 hr NC siRNA).
(D) Quantitative RT-PCR analysis for the expression of MMP1 and MMP9 in control and ELF3 knockdown cells. Total RNA was prepared 72 hr after transfection (mean ± SEM, n = 4 per group, **p < 0.01 versus NC siRNA).
(E) Expression of vimentin, CK19, ZEB1, ZEB2 and TWIST1 in control and ELF3 knockdown cells determined by quantitative RT-PCR analysis (mean ± SEM, n = 4 per group, **p < 0.01 versus NC siRNA).
(F) Immunofluorescence analysis for the expression of vimentin in control and ELF3 knockdown cells. Hoechst staining (blue) identifies the nuclei of all cells in the field. Scale bars = 50 μm.
See also Figure S5 and Tables S4, S7 and S8.
Since an immortalized normal epithelial cell line has not been established from ampullary cells, we used an immortalized normal epithelial cell line of common bile duct origin, designated HBDEC2-3H10 and an immortalized normal epithelial cell line of duodenal mucosa origin, designated HDuodEC3. These lines were selected for functional analyses because ELF3 mutations have also been observed in 7/74 (9.5%) common bile duct carcinomas in our recent study (Nakamura et al., 2015) and 1/18 (5.6%) duodenal carcinomas in the present study. To investigate the consequences of the loss-of-function mutation in ELF3, three human ELF3-specific small interfering RNA (siRNA) oligonucleotides were utilized to knockdown ELF3 expression in the HBDEC2-3H10 cells. Efficient loss of ELF3 was detected at both the mRNA and protein levels (Figure 4B). Compared to HBDEC2-3H10 cells treated with negative control siRNA (NC siRNA), ELF3 siRNA-transfected cells (ELF3 siRNA #1, #2 and #3) didn’t show any significant difference in cellular proliferation (Figure 4C). Invasion/Migration assay using Matrigel invasion chambers and control inserts demonstrated that invasive activities and motilities in ELF3 knockdown cells were significantly increased compared to control cells (Figure 4C). HDuodEC3 cells treated with ELF3 siRNAs showed similar phenotypic changes in terms of cell proliferation, invasion and motility (Figure S5). Consistent with the present data, aggressive invasion phenotype (extended cell bodies into the Matrigel matrix) of ELF3 knockdown cells was observed in time-lapse images of 3D cell invasion assay. Quantitative RT-PCR analysis for the expression of matrix metalloproteinase-1 and -9 (MMP1 and MMP9) further supported this observation, showing higher expression levels of MMP1 and MMP9 in ELF3 knockdown cells compared with control cells (Figure 4D). Knockdown of ELF3 is associated with epithelial-to-mesenchymal transition (EMT): immunofluorescence and quantitative RT-PCR analysis showed that the expression of vimentin, which is a mesenchymal marker of EMT and a regulator of cell migration, was increased in ELF3 knockdown cells compared with the control cells (Figures 4E and 4F). By contrast, the expression of the epithelial marker, cytokeratin 19 (CK19), was decreased in cells with ELF3 deficiency. In addition, key regulators of EMT, such as ZEB1, ZEB2 and TWIST1, were upregulated in ELF3 knockdown cells (Figure 4E), while E-cadherin expression was variable depending on individual siRNAs. We have performed the microarray-based expression analysis on ELF3-knockout HBDEC2-3H10 cell lines using the CRISPR/Cas9 system, which demonstrated that ELF3-knockout modulated key pathways typically activated in cancer, including WNT and RTK-RAS signaling pathways (data not shown).
Multi-region Whole Exome Sequencing
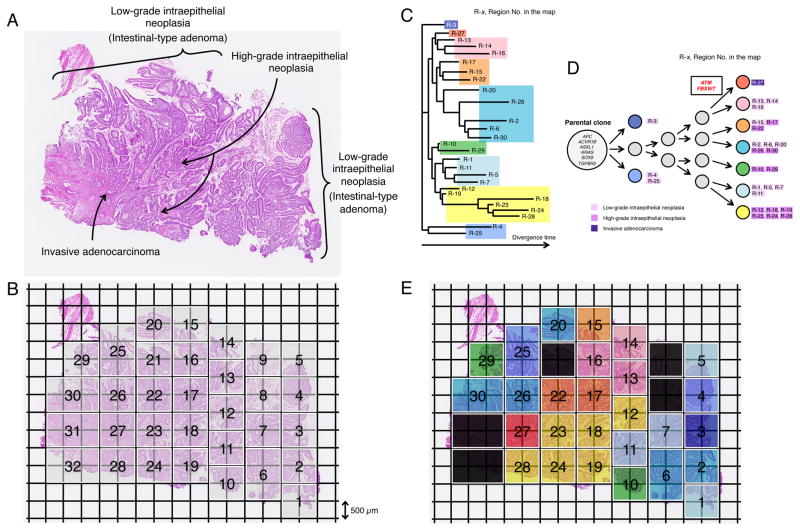
We also performed multi-region whole exome sequencing in an ampullary carcinoma using the new technology (Glass Chip Macrodissection, GCM, Olympus) to understand tumor heterogeneity and carcinogenesis of this disease. The classic intestinal ampullary carcinoma consists of the components of low-grade intraepithelial neoplasia (intestinal-type adenoma), high-grade intraepithelial neoplasia and invasive adenocarcinoma (Figure 5A). In this study, five serial frozen sections were prepared and we dissected the tissue from 32 squares of 1 mm2 units (thickness of 18 μm) (Figure 5B) and collected the same region from all five sections into the same tube. The amount of gDNA was 155.0 ng per region on average (range 69.6–242.7 ng). We excluded the two samples since the amount of DNA was less than 100 ng (regions No.8 and No.9 in Figure 5B). We next performed whole exome sequencing on 30 regions obtained from 30 units of the GCM, as well as from bulk tumor tissue and the matched bulk normal duodenal tissue. We prepared the DNA libraries for sequencing using Illumina pair-read platform, according to the manufacture’s protocol for the preparation of gDNA libraries from 200 ng DNA samples (SureSelect Human All Exon v4.0). Whole-exome sequencing of 30 regions yielded an average of 134X coverage with 95.4% of targeted bases covered by ≥ 20X, while the depth in bulk tumor tissue and bulk normal tissue was 192X and 179X, respectively. We excluded the samples from three units since the sequencing errors or low tumor purities were suspected (regions No.21, No.31 and No.32 in Figure 5B), leaving 27 final regions of the tumor. A total of 7,139 synonymous and non-synonymous somatic mutations were identified in 27 regions. The number of non-synonymous somatic mutations ranged from 95 to 113 with an average of 100.7, while the bulk tumor tissue had 95 mutations (Table S11). We next drew a phylogenetic tree based on the sequencing data of synonymous mutations (Figure 5C) (Saitou and Nei, 1987), which showed widely spreading branches. Interestingly, although the regions No.2, No.6, No.20, No.26 and No.30 were far apart on the section, they belong to the same group, probably since these regions might be connected three-dimensionally (Figures 5C and 5E).
Figure 5. Geographic Mapping of Subclones Based on Multi-region Exome Sequencing and Proposed Clonal Evolution of an Ampullary Carinoma with Low-grade and High-grade Intraepithelial Neoplasias.
(A) Histology of a sample consisting of the components of low-grade intraepithelial neoplasia (Intestinal-type adenoma), high-grade intraepithelial neoplasia and invasive carcinoma (hematoxylin and eosin stain).
(B) A whole section was broken up into 32 squares of 1 mm2 units (thickness, 18 μm) by Glass Chip Macrodissection (GCM) technology and was dissected. Genomic DNA (gDNA) was individually extracted from the same compartment using the five serial sections and subjected to Agilent SureSelect Human All Exon v4.0 based hybrid selection followed by exome library construction for Illumina sequencing.
(C) Phylogenetic tree was drawn based on the sequencing data of synonymous mutations.
(D) Proposed clonal evolution model based on the phylogenetic tree during tumor progression. Cancer driver genes reported in COSMIC are represented. The mutations of the cancer driver genes, ATM and FBXW7 are observed only in region No.27 where cancer cells start to be invasive morphologically. Gray circles indicate predicted subclones based on the phylogenetic tree.
(E) Sections are colored, corresponding to the colors of the phylogenetic tree in (C) and (D).
See also Table S11.
A proposed clonal evolution model based on the phylogenetic tree is shown in Figure 5D. In all clones, six driver genes (‘founder’ mutations: APC, ACVR1B, ASXL1, KRAS, SOX9 and TGFBR2) are mutated. Interestingly, mutations in ATM and FBXW7, which are well-known driver genes, were observed only in region No.27 where cancer cells start to be invasive morphologically. These findings confirmed that parental clones have superimposed ‘progressor’ mutations associated with clonal evolution (Yachida et al., 2010). We didn’t detect ELF3 mutations in any of the regions studied in the ampullary carcinoma which was selected for multi-region whole exome sequencing. Although the whole-exome sequencing using the bulk tumor tissue found all of the ‘founder’ driver mutations (APC, ACVR1B, ASXL1, KRAS, SOX9 and TGFBR2), ‘progressor’ mutations (ATM and FBXW7) observed in subclones couldn’t be detected, indicating the advantage of this equipment for detecting the subclones with different genomic features.
DISCUSSION
This is the study of whole exome sequencing and targeted-sequencing in adenocarcinomas of the ampulla of Vater from a large series of Japanese and American patients. We found that the prevalence of driver gene mutations in carcinomas with the intestinal phenotype was different from those with the pancreatobiliary phenotype; the former is similar to colorectal carcinomas, the latter to pancreatic carcinomas. However, intestinal-type ampullary carcinomas have some alterations that are distinct from those observed in colorectal cancers. Furthermore, PCA and NMF analysis suggest that even though ampullary carcinomas can be classified into two histological phenotypes based on the prevalence of driver gene mutations, the mutational patterns and signatures are similar in intestinal- and pancreatobiliary-types, differing from those in colorectal carcinomas and pancreatic carcinomas. These findings suggest that the cancers were exposed to similar mutagens, or that the affected cells have similar DNA repair processes, or both.
Genes frequently mutated in intrahepatic biliary cancers (BAP1, ARID1A, PBRM1, IDH1 and IDH2) were only rarely targeted in ampullary carcinomas (Nakamura et al., 2015; Simbolo et al., 2014). These findings indicate that ampullary carcinoma is genetically distinct from intrahepatic biliary carcinoma although until now the patients with ampullary carcinoma are often treated with regimens (gemcitabine and cisplatin) designed for biliary tract adenocarcinomas without distinction (Malka et al., 2014; Valle et al., 2010). Our findings support both the biological and clinical significance of classifying ampullary adenocarcinomas into two distinct phenotypes, and suggest potential subtype-specific therapeutic strategies.
We identified the significantly mutated driver gene, ELF3, characteristic of ampullary carcinomas. ELF3 (E74-like factor-3)/ESE1 encodes a member of the ETS transcription factor family that is expressed and upregulated in epithelial cancers (Chang et al., 1997). Recently, ELF3 mutations were reported in 4 out of 100 patients of gastric cancers (Wang et al., 2014) and 3 out of 24 cervical adenocarcinomas (Ojesina et al., 2014). In the present study, almost all of ELF3 mutations were truncating, suggesting that inactivation of this gene could drive ampullary tumorigenesis. The ampulla of Vater sits at the intersection of pancreatobiliary and intestinal differentiation (Gürbüz and Klöppel, 2004), and it is therefore of interest to note that, in the ELF3-null mice model, inactivation of ELF3 results in dysmorphogenesis and altered differentiation of small intestinal epithelium (Ng et al., 2002). Elf3 is critical for proper epithelial differentiation in pancreas as well as intestine (Kobberup et al., 2007). Our functional analyses in normal epithelial cells demonstrated that ELF3 silencing induced EMT and enhanced motility and invasion, concomitant with upregulation of vimentin, MMP1 and MMP9. The mechanisms of tumorigenesis by loss of ELF3 are still unclear. However, it has been reported that knockdown of EHF/ESE3, which encodes a protein with similar structural elements to Elf3 (Hollenhorst et al., 2011), induced EMT, stem-like features, and tumor-initiating and metastatic properties in prostate epithelial cells (Albino et al., 2012; Kar and Gutierrez-Hartmann, 2013). This suggests common tumor suppressive properties in these structurally similar proteins. In the present study, we focused on the effects of loss-of-functions since ELF3 was mainly mutated through protein-truncating alterations, but the possibility of gain-of-function by some missense mutations should be clarified in the future.
Through whole exome sequencing and subsequent targeted deep sequencing, we identified potentially therapeutically targetable mutations in approximately half of patients with ampullary carcinomas. The genes coding for members of the Fanconi anemia pathway included ATM (n = 17), BRCA2 (n = 10) and PALB2 (n = 2). It has been reported that adenocarcinomas with inactivating mutations in genes coding for members of this pathway can be exquisitely sensitive to DNA cross-linking agents (Villarroel et al., 2011) and Poly (ADP-ribose) polymerase (PARP) inhibitors (Fogelman et al., 2011). PIK3CA (n = 10), PTEN (n = 4), PIK3C2A (n = 3) and PIK3C2G (n = 3) are attractive molecular targets for anti-cancer molecules, such as PI3K/AKT/mTOR pathway inhibitors (Beaver et al., 2013; Lopez-Chavez et al., 2015). In addition, BRAF (n = 16), JAK3 (n = 8) and NF1 (n = 15) were currently in human clinical trials for BRAF inhibitors, JNK inhibitors and Hsp90 inhibitors, respectively, with promising results (Fedorenko et al., 2015; Hu-Lieskovan et al., 2015; Springuel et al., 2014). We can speculate that ampullary carcinomas will be good candidate for a personalized approach to therapy based on the genetic changes in these patients’ cancers.
Recently a “big bang” model of human colorectal tumor growth was proposed (Sottoriva et al., 2015) and it is clarified that tumor heterogeneity and clonal evolution in cancers are complex (Waclaw et al., 2015). We conducted a multi-region exome sequencing of an ampullary carcinoma using the new technology (GCM). This analysis demonstrated clonal evolution during ampullary cancer progression. The clones which are located in the regions of low-grade intraepithelial neoplasia (intestinal-type adenoma) and high-grade intraepithelial neoplasia harbored alterations in the same six driver genes. Although the phylogenic tree based on synonymous mutations shows wide divergence, the subclone in region No.27 is superimposed on an accumulation of driver mutations (ATM and FBXW7), which is genetically evolved from the parental clone, and presumably acquired the capacity to invade.
In summary, whole exome sequencing and subsequent targeted deep sequencing has led to the identification of the tumor suppressor gene, ELF3, characteristic of ampullary carcinomas. ELF3 mutations are present in both histological phenotypes and across racial differences.
EXPERIMENTAL PROCEDURES
Patients and Tissue Samples
The tissues and clinical information used in this study were obtained under informed consent and approval of the institutional review boards of each institute. All tumors and corresponding non-tumor tissues were frozen after surgical resection. We performed macrodissection to enrich the tumor fraction relative to the dominant stromal component and other normal cells.
Immunohistochemistry
Paraffin-embedded samples of the primary carcinomas from 172 patients were immunostained for MUC1, MUC2, CDX2 and Cytokeratin 20 (CK20). Immunohistochemical labeling was carried out using a Bond Max instrument (Leica Microsystems). According to the previous paper (Ang et al., 2014), “intestinal-type” is defined as having (1) positive staining for CK20 or CDX2 or MUC2 and negative staining for MUC1, or (2) positive staining for CK20, CDX2, and MUC2, irrespective of the MUC1 result; and “pancreatobiliary-type” was defined as having positive staining for MUC1 and negative staining for CDX2 and MUC2, irrespective of CK20 results. Cases not fitting one of these three categories are regarded as “ambiguous”.
Genome Analysis
Massively parallel sequencing exome capture was performed using Agilent SureSelect Human All Exon Kit v4.0 according to the manufacturer’s instructions. Whole exome sequencing was performed on the Illumina HiSeq platforms. We next selected 92 genes and performed using a target enrichment system (HaloPlex) that differed from that in the exome sequencing to validate significantly mutated genes. Mutation calling, processing the significantly mutated genes and further analyses on mutational patterns and signatures are described in Supplemental Experimental Procedures.
Somatic Copy Number Analysis
We profiled for SCNAs with Agilent CGH array, using their matched non-tumor tissues as a copy number reference. We applied the GISTIC2.0 algorithm (Beroukhim et al., 2007) to identify SCNAs that might be responsible for driver tumorigenesis.
Functional Analysis
We used an immortalized normal epithelial cell line of common bile duct origin, designated HBDEC2-3H10, and an immortalized normal epithelial cell line of duodenal mucosa origin, designated HDuodEC3. To investigate the consequences of the loss-of-function mutation in ELF3, three human ELF3-specific siRNA oligonucleotides were utilized to knockdown ELF3 expression in the HBDEC2-3H10 cells and HDuodEC3 cells. Biochemical assays were performed as provided in Supplemental Experimental Procedures.
Statistical Analyses
Statistical analyses were conducted with IBM SPSS soft ware version 20. Gene with q (false detection rate) < 0.1 was considered to be significantly mutated. Frequency distributions were compared by χ2 test. Continuous variables were compared using the Student’s t-test when the data were normally distributed or Wilcoxon signed-rank test when the data were not normally distributed. A p value of < 0.05 was considered statistically significant. We performed a permutation test to evaluate the similarity between genes mutated in intestinal-type ampullary cancers and those of colorectal cancer (Comparison A) and between genes mutated in pancreatobiliary-type ampullary cancers and those of pancreatic cancers (Comparison B). We focused on high-ranking significantly mutated genes of APC, TP53, KRAS and SMAD4 in Comparison A, and KRAS, TP53 and SMAD4 in Comparison B. The permutation test was based on the sum of absolute differences (SAD) in the ranks of those genes between the two datasets in each comparison. In one permutation, we randomly assigned ranks across significantly mutated genes, and calculated a SAD value across the genes of interest. We repeated this permutation process for 100,000 times. We then calculated the frequency when SAD values in permutated data were less or equal to an observed SAD value, which was used as an empirical p value.
Supplementary Material
SIGNIFICANCE.
The genetic landscape of ampullary carcinomas is currently poorly defined. We established an international multicenter collaboration and conducted an in-depth analysis of the genomic abnormalities of these carcinomas. Whole exome sequencing and subsequent targeted deep sequencing led to the identification of a tumor suppressor gene, ELF3, characteristic of ampullary carcinomas. In addition, potentially therapeutic targetable mutations were identified in 51% (88/172) of the carcinomas. A difference in the prevalence of driver gene mutations between the intestinal and pancreatobiliary phenotypes was found. Our findings form a foundation for a personalized approach to the treatment of patients with carcinoma of the ampulla of Vater.
Acknowledgments
We wish to thank all patients and families who contributed to this study. We are grateful to Ms. Kaho Minoura (Agilent Technologies), Ms. Chika Shima, Ms. Keiko Igarashi, Ms. Risa Usui, Ms. Shoko Ohashi, Ms. Tomoko Urushidate, and Ms. Naoko Okada (National Cancer Center Research Institute) for technical assistance. The authors thank Dr. Hiroyuki Miyoshi (RIKEN, BioResource Center) for lentiviral constructs. This work was supported by the following grants: Grant-in Aid for Scientific Research from the Ministry of Education, Culture, Science and Technology of Japan (25134719 to S.Y.; 25134721 to M.K.; 25134720 to T.S.); the Takeda Science Foundation (S.Y.); the Mochida Memorial Foundation for Medical and Pharmaceutical Research (S.Y.); Princess Takamatsu Cancer Research Fund (S.Y.); the National Cancer Center Research and Development Fund (25-A-3 to S.Y.); the Ministry of Health Labor and Welfare (Health and Labor Sciences Research Expenses for Commission and Applied Research for Innovative Treatment of Cancer (S.Y., E.T., M.K., C.M., T.O., and T.S.); AIRC grant 12182 (C.L.); Italian Cancer Genome Project (FIRB RBAP10AHJB) (C.L.), and the US National Cancer Institutes SPORE grant CA62924 (R.H.H.). The super-computing resource SHIROKANE was provided by the Human Genome Center, The University of Tokyo (http://sc.hgc.jp/shirokane.html). Bert Vogelstein is one of the founders of Personal Genome Diagnostics, Inc. and PapGene, Inc., and a member of the Scientific Advisory Board of Sysmex-Inostics. These companies are focused on the identification of genetic alterations in human cancer for diagnostic or therapeutic purposes. These companies and others have licensed patent applications on genetic technologies from Johns Hopkins, some of which result in royalty payments. The terms of these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies.
Footnotes
ACCESSION NUMBERS
The accession number for the raw sequencing data reported in this paper is NBDC: DRR050378-DRR050437.
Supplemental information includes Supplemental Experimental Procedures, 5 figures and 11 tables.
AUTHOR CONTRIBUTIONS
Designed experiments: S.Y., L.D.W., Hitoshi.N., B.V., R.H.H., and T.S.; performed experiments: S.Y., M.S., E.T., Y.A., N.H., F.H., N.M., K.H., J.F., H.T., and T.K.; DNA sequence analysis: S.Y., E.T., Y.T., M.K., Hiromi.N., N.H., and A.E.; statistical analysis: E.T., Y.T., M.K., Hiromi.N., N.H., and A.E.; sample acquisition and clinical data collection: S.Y., L.D.W., C.L., T.S., C.M., T.O., S.N., K.S., N.H., H.T., R.H., M.O., K.O., S.H., M.M., K.A., M.Y., M.U., H.Y., M.J.W., C.L.W., and T.F.; Manuscript writing: S.Y., L.D.W., M.S., Y.T., N.M., T.K., and R.H.H.; project oversight: S.Y., L.D.W., Hitoshi.N., B.V., T.K., R.H.H., and T.S.
References
- Albino D, Longoni N, Curti L, Mello-Grand M, Pinton S, Civenni G, Thalmann G, D’Ambrosio G, Sarti M, Sessa F, et al. ESE3/EHF controls epithelial cell differentiation and its loss leads to prostate tumors with mesenchymal and stem-like features. Cancer research. 2012;72:2889–2900. doi: 10.1158/0008-5472.CAN-12-0212. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang DC, Shia J, Tang LH, Katabi N, Klimstra DS. The utility of immunohistochemistry in subtyping adenocarcinoma of the ampulla of vater. The American journal of surgical pathology. 2014;38:1371–1379. doi: 10.1097/PAS.0000000000000230. [DOI] [PubMed] [Google Scholar]
- Beaver JA, Gustin JP, Yi KH, Rajpurohit A, Thomas M, Gilbert SF, Rosen DM, Ho Park B, Lauring J. PIK3CA and AKT1 mutations have distinct effects on sensitivity to targeted pathway inhibitors in an isogenic luminal breast cancer model system. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:5413–5422. doi: 10.1158/1078-0432.CCR-13-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Scott GK, Kuo WL, Xiong X, Suzdaltseva Y, Park JW, Sayre P, Erny K, Collins C, Gray JW, Benz CC. ESX: a structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene. 1997;14:1617–1622. doi: 10.1038/sj.onc.1200978. [DOI] [PubMed] [Google Scholar]
- Fedorenko IV, Gibney GT, Sondak VK, Smalley KS. Beyond BRAF: where next for melanoma therapy? British journal of cancer. 2015;112:217–226. doi: 10.1038/bjc.2014.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogelman DR, Wolff RA, Kopetz S, Javle M, Bradley C, Mok I, Cabanillas F, Abbruzzese JL. Evidence for the efficacy of Iniparib, a PARP-1 inhibitor, in BRCA2-associated pancreatic cancer. Anticancer research. 2011;31:1417–1420. [PubMed] [Google Scholar]
- Greulich H, Kaplan B, Mertins P, Chen TH, Tanaka KE, Yun CH, Zhang X, Lee SH, Cho J, Ambrogio L, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14476–14481. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürbüz Y, Klöppel G. Differentiation pathways in duodenal and ampullary carcinomas: a comparative study on mucin and trefoil peptide expression, including gastric and colon carcinomas. Virchows Arch. 2004;444:536–541. doi: 10.1007/s00428-004-1008-2. [DOI] [PubMed] [Google Scholar]
- Hagerstrand D, Tong A, Schumacher SE, Ilic N, Shen RR, Cheung HW, Vazquez F, Shrestha Y, Kim SY, Giacomelli AO, et al. Systematic interrogation of 3q26 identifies TLOC1 and SKIL as cancer drivers. Cancer Discov. 2013;3:1044–1057. doi: 10.1158/2159-8290.CD-12-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hechtman JF, Liu W, Sadowska J, Zhen L, Borsu L, Arcila ME, Won HH, Shah RH, Berger MF, Vakiani E, et al. Sequencing of 279 cancer genes in ampullary carcinoma reveals trends relating to histologic subtypes and frequent amplification and overexpression of ERBB2 (HER2) Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2015;28:1123–1129. doi: 10.1038/modpathol.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herter-Sprie GS, Greulich H, Wong KK. Activating Mutations in ERBB2 and Their Impact on Diagnostics and Treatment. Frontiers in oncology. 2013;3:86. doi: 10.3389/fonc.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annual review of biochemistry. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu-Lieskovan S, Mok S, Homet Moreno B, Tsoi J, Robert L, Goedert L, Pinheiro EM, Koya RC, Graeber TG, Comin-Anduix B, Ribas A. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAFV600E melanoma. Science translational medicine. 2015;7:279ra241. doi: 10.1126/scitranslmed.aaa4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A, Gutierrez-Hartmann A. Molecular mechanisms of ETS transcription factor-mediated tumorigenesis. Critical reviews in biochemistry and molecular biology. 2013;48:522–543. doi: 10.3109/10409238.2013.838202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WS, Choi DW, Choi SH, Heo JS, You DD, Lee HG. Clinical significance of pathologic subtype in curatively resected ampulla of vater cancer. Journal of surgical oncology. 2012;105:266–272. doi: 10.1002/jso.22090. [DOI] [PubMed] [Google Scholar]
- Kimura W, Ohtsubo K. Incidence, sites of origin, and immunohistochemical and histochemical characteristics of atypical epithelium and minute carcinoma of the papilla of Vater. Cancer. 1988;61:1394–1402. doi: 10.1002/1097-0142(19880401)61:7<1394::aid-cncr2820610720>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Kobberup S, Nyeng P, Juhl K, Hutton J, Jensen J. ETS-family genes in pancreatic development. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236:3100–3110. doi: 10.1002/dvdy.21292. [DOI] [PubMed] [Google Scholar]
- Lopez-Chavez A, Thomas A, Rajan A, Raffeld M, Morrow B, Kelly R, Carter CA, Guha U, Killian K, Lau CC, et al. Molecular Profiling and Targeted Therapy for Advanced Thoracic Malignancies: A Biomarker-Derived, Multiarm, Multihistology Phase II Basket Trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:1000–1007. doi: 10.1200/JCO.2014.58.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malka D, Cervera P, Foulon S, Trarbach T, de la Fouchardiere C, Boucher E, Fartoux L, Faivre S, Blanc JF, Viret F, et al. Gemcitabine and oxaliplatin with or without cetuximab in advanced biliary-tract cancer (BINGO): a randomised, open-label, non-comparative phase 2 trial. The Lancet Oncology. 2014;15:819–828. doi: 10.1016/S1470-2045(14)70212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, et al. Genomic spectra of biliary tract cancer. Nature genetics. 2015;47:1003–1010. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- Neoptolemos JP, Moore MJ, Cox TF, Valle JW, Palmer DH, McDonald AC, Carter R, Tebbutt NC, Dervenis C, Smith D, et al. Effect of adjuvant chemotherapy with fluorouracil plus folinic acid or gemcitabine vs observation on survival in patients with resected periampullary adenocarcinoma: the ESPAC-3 periampullary cancer randomized trial. Jama. 2012;308:147–156. doi: 10.1001/jama.2012.7352. [DOI] [PubMed] [Google Scholar]
- Ng AY, Waring P, Ristevski S, Wang C, Wilson T, Pritchard M, Hertzog P, Kola I. Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology. 2002;122:1455–1466. doi: 10.1053/gast.2002.32990. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano K, Oshima M, Yachida S, Kushida Y, Kato K, Kamada H, Wato M, Nishihira T, Fukuda Y, Maeba T, et al. Factors predicting survival and pathological subtype in patients with ampullary adenocarcinoma. Journal of surgical oncology. 2014;110:156–162. doi: 10.1002/jso.23600. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shoji H, Morizane C, Hiraoka N, Kondo S, Ueno H, Ohno I, Shimizu S, Mitsunaga S, Ikeda M, Okusaka T. Twenty-six cases of advanced ampullary adenocarcinoma treated with systemic chemotherapy. Japanese journal of clinical oncology. 2014;44:324–330. doi: 10.1093/jjco/hyt237. [DOI] [PubMed] [Google Scholar]
- Simbolo M, Fassan M, Ruzzenente A, Mafficini A, Wood LD, Corbo V, Melisi D, Malleo G, Vicentini C, Malpeli G, et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget. 2014;5:2839–2852. doi: 10.18632/oncotarget.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D, Curtis C. A Big Bang model of human colorectal tumor growth. Nature genetics. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springuel L, Hornakova T, Losdyck E, Lambert F, Leroy E, Constantinescu SN, Flex E, Tartaglia M, Knoops L, Renauld JC. Cooperating JAK1 and JAK3 mutants increase resistance to JAK inhibitors. Blood. 2014;124:3924–3931. doi: 10.1182/blood-2014-05-576652. [DOI] [PubMed] [Google Scholar]
- Takita J, Chen Y, Okubo J, Sanada M, Adachi M, Ohki K, Nishimura R, Hanada R, Igarashi T, Hayashi Y, Ogawa S. Aberrations of NEGR1 on 1p31 and MYEOV on 11q13 in neuroblastoma. Cancer Sci. 2011;102:1645–1650. doi: 10.1111/j.1349-7006.2011.01995.x. [DOI] [PubMed] [Google Scholar]
- Ushiku T, Arnason T, Fukayama M, Lauwers GY. Extra-ampullary duodenal adenocarcinoma. The American journal of surgical pathology. 2014;38:1484–1493. doi: 10.1097/PAS.0000000000000278. [DOI] [PubMed] [Google Scholar]
- Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, Madhusudan S, Iveson T, Hughes S, Pereira SP, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. The New England journal of medicine. 2010;362:1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- Villarroel MC, Rajeshkumar NV, Garrido-Laguna I, De Jesus-Acosta A, Jones S, Maitra A, Hruban RH, Eshleman JR, Klein A, Laheru D, et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Molecular cancer therapeutics. 2011;10:3–8. doi: 10.1158/1535-7163.MCT-10-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waclaw B, Bozic I, Pittman ME, Hruban RH, Vogelstein B, Nowak MA. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525:261–264. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nature genetics. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- Winter JM, Cameron JL, Olino K, Herman JM, de Jong MC, Hruban RH, Wolfgang CL, Eckhauser F, Edil BH, Choti MA, et al. Clinicopathologic analysis of ampullary neoplasms in 450 patients: implications for surgical strategy and long-term prognosis. Journal of gastrointestinal surgery : official journal of the Society for Surgery of the Alimentary Tract. 2010;14:379–387. doi: 10.1007/s11605-009-1080-7. [DOI] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Higasa K, Sakaguchi M, Shien K, Soh J, Ichimura K, Furukawa M, Hashida S, Tsukuda K, Takigawa N, et al. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. Journal of the National Cancer Institute. 2014;106:djt338. doi: 10.1093/jnci/djt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.