Abstract
Healthy aging has become a major goal of public health. Many studies have provided evidence and theories to explain molecular mechanisms of the aging process. Recent studies suggest that epigenetic mechanisms are responsible for life span and the progression of aging. Epigenetics is a fascinating field of molecular biology, which studies heritable modifications of DNA and histones that regulate gene expression without altering the DNA sequence. DNA methylation is a major epigenetic mark that shows progressive changes during aging. Recent studies have investigated aging-related DNA methylation as a biomarker that predicts cellular age. Interestingly, growing evidence proposes that nutrients play a crucial role in the regulation of epigenetic modifiers. Because various nutrients and their metabolites function as substrates or cofactors for epigenetic modifiers, nutrition can modulate or reverse epigenetic marks in the genome as well as expression patterns. Here, we will review the results on aging-associated epigenetic modifications and the possible mechanisms by which nutrition, including nutrient availability and bioactive compounds, regulate epigenetic changes and affect aging physiology.
Keywords: aging, epigenetics, nutrition, bioactive compounds
INTRODUCTION
Aging is a complex process that impairs an organism’s biological functions and increases its susceptibility to disease and death. The aging process is affected by a combination of genetic and environmental factors, which means that aging is both genetically determined and influenced by the environment. For example, longevity clusters within families. The parents and siblings of centenarians have a greater likelihood of longevity, and the offspring of centenarians appear to have a delay in age-related disease (1). These results indicate that genetic components affect aging. Genetic variants related to aging have been investigated in genome-wide association studies in centenarian populations (2–5).
Diet is a key environmental factor that has profound effects on human development and health. Chronic exposure to certain dietary patterns, such as unbalanced energy or deficiencies in essential nutrients, creates metabolic stress. This type of stress is closely related to the incidence and progression of various chronic, non-communicable diseases and aging. Metabolic stress affects the regulation of gene expression, resulting in cellular and physiological changes (6), but it rarely causes acute and deleterious damage, such as mutations to the genome.
Nutrigenomics is an emerging science that aims to understand how dietary components and metabolites affect gene expression and interact with the genome. It provides a mechanistic link between dietary factors and the genomic response. It is important to understand gene-nutrient interactions to learn how to regulate metabolic processes that contribute to age-related disease risk factors, such as obesity, cardiovascular disease, and inflammation (7). Intriguingly, dietary and nutritional effects can be passed on to the next generation. The effects of nutrition on the body are also mediated by epigenetic mechanisms (8). Epigenetic mechanisms can mediate between nutrient availability and phenotype throughout life; however, little is known about how nutrition plays a role in longevity and aging via epigenetic mechanisms. The aim of this review is to summarize the growing evidence that numerous dietary factors, such as calorie restriction and polyphenols, can modify epigenetic marks influencing longevity and aging.
MOLECULAR FACTORS IN AGING
Adult or somatic stem cells progressively decline with aging. At present, the molecular mechanisms of cellular aging are partially understood. For instance, defects in genes involved in the DNA repair system result in the accumulation of unrepaired DNA and chromosomal damage. In aging populations, variations in exonuclease 1 (EXO1) and postmeiotic segregation increased 2 (PMS2) have been shown to contribute to human longevity (4,5). These candidates mainly function in the DNA mismatch repair system (9). The role of DNA repair in the aging process has been documented in previous studies in both human patients and mouse models. Patients with Werner syndrome, an autosomal recessive disease characterized by premature aging, have defects in DNA helicase, which is important in DNA repair pathways (10). A trichothiodystrophy mouse model with a genetic mutation in a DNA repair pathway exhibited accelerated aging and a shortened life span (11). These results suggest that both defects in DNA repair systems and risk factors that induce DNA damage are associated with the aging process. Intrinsic cellular sources, such as replication errors and chemical changes to DNA, and external sources, such as ionizing radiation and genotoxic drugs, are the main factors that cause DNA damage (1,12). Reactive oxygen species (ROS), such as superoxide anions, hydroxyl radicals, hydrogen peroxide, and nitric oxide, are the major molecules that impair cellular function. If ROS exceeds cellular antioxidant defenses, cells experience oxidative stress, which contributes to DNA damage and aging (13). In addition to DNA damage, telomere length is associated with age-related diseases in humans. Somatic stem cell function may be impaired by telomere shortening. Moreover, telomerase-deficient mice have short telomeres and age prematurely, whereas cancer-resistant mice that overexpress telomerase have long telomeres and delayed aging (14,15).
EPIGENETIC AND AGING
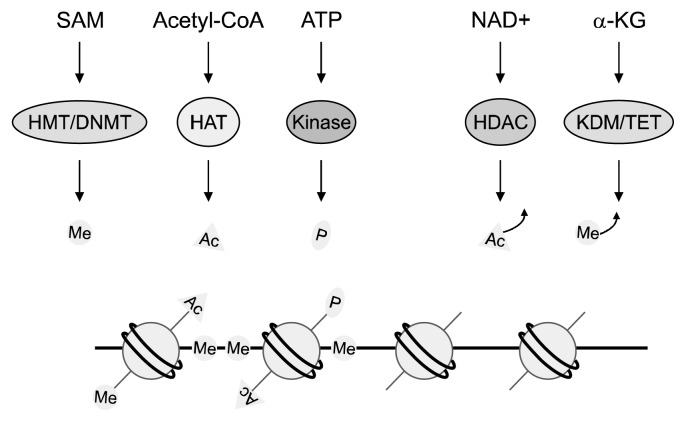
Since the late 1980s, epigenetic changes found in cancer have been connected with aging (16). Epigenetics is associated with changes in the genome that are passed on from one generation to the next that alter gene expression without changing the primary DNA sequence. DNA methylation and histone modification are two well-known epigenetic mechanisms that affect chromatin structure and eventually regulate gene expression patterns (Fig. 1) (17). The combination of these marks is responsible for cell development, not only during cellular differentiation in embryonic development but also throughout life (18). A recent study highlights that epigenetic patterns, especially DNA methylation, are critical biomarkers for aging that can predict the age of human tissue and cell type (19). In this study, 353 markers of DNA methylation were selected and found to predict various tissue types, including blood cells, and brain, breast, kidney, liver, and lung tissue (19). Intriguingly, the DNA methylation status of the markers in embryonic and induced pluripotent stem cells was close to zero (19). Methylation markers were also shown to reflect cell passage number and could predict the age of chimpanzees (19). These findings indicate that epigenetic changes are directly linked to the aging process.
Fig. 1.
Epigenetic modifiers require nutrients and their metabolites. The availability of nutrients and metabolites regulates the activities of various epigenetic enzymes, such as histone methyltransferase (HMT), DNA methyltransferase (DNMT), histone acetyltransferase (HAT), kinase, histone deacetylase (HDAC), histone demethylase (KDM), and ten-eleven translocation (TET, a DNA demethylation enzyme). These epigenetic enzymes utilize S-adenosylmethionine (SAM), acetyl-CoA, ATP, NAD+, and α-ketoglutarate (α-KG) to regulate the status of methylation (Me), acetylation (Ac), and phosphorylation (P) in chromatin.
DNA methylation
DNA methylation occurs when a methyl group is added to position 5 of the cytosine residue in 5-methylcytosine (20), preferentially in CG dinucleotides. The methyl groups are added by DNA methyltransferases (DNMTs). DNMT3A and DNMT3B catalyze the establishment of DNA methylation, and DNMT1 serves to maintain DNA methylation during DNA replication. DNA demethylation occurs either passively by blocking DNMT1 maintenance methyltransferase activity during DNA replication or due to an active demethylation process. Several studies have revealed that altered DNA methylation during aging seems to occur both at a global level and at a locus-specific level (Table 1). At the global level, genome-wide hypomethylation and reduced methylation of repetitive elements, such as intracisternal A-type particle (IAP), have been observed in aging mouse models (21–23). Maintenance of methylation at repetitive elements, which exists in multiple copy sequences in the genome, is crucial for genome stability, and in particular for centromeres and telomeres (24). Research using whole-genome bisulfate sequencing (25) revealed a distinct pattern in the DNA methylome of centenarians, compared to that of newborns. Centenarian DNA shows relatively lower DNA methylation at several regions, whereas newborn DNA has more homogeneous methylation patterns. The loss of DNA methylation in centenarians mainly occurred at CpG poor promoters whereas hypermethylated sequences were observed at CpG island promoters (25). Another epigenome-wide study suggested that the DNA methylation status of 155 CpG sites in whole blood DNA were significantly associated with age (26). Among the age-associated sites, methylation levels were positively associated with aging, and these sites were mainly within CpG islands. These global methylation changes may cause genomic instability (21) and loss of telomere integrity (27), which may consequently contribute to the aging cellular phenotype.
Table 1.
Epigenetic biomarkers observed in aging
| Epigenetic mark | Changes with aging | Genomic loci | Tissue | Species | Reference |
|---|---|---|---|---|---|
| DNA methylation | Hypomethylation | Intracisternal A-type particle | Liver | Mouse | 23 |
| Hypermethylation | Promoter and exon 1 of ESR1 | Colon | Human | 28 | |
| Intron 1 to exon 4 of c-fos | Liver | Human | 29 | ||
| Promoter of DAPK1 and E-cadherin | Stomach, intestines | Human | 30 | ||
| Histone modification | Acetylation of H4 lysine 16 | Subtelomeric regions | – | Yeast | 37 |
| Trimethylation of H4-K20 | Global level | Liver | Rat | 45 | |
| Dephosphorylation of H1.4 and H1.5 | Global level | Peripheral blood lymphocytes | Human | 46 |
Age-associated methylation also occurs in a site-specific manner. A well-known locus-specific alteration in methylation is aberrant hypermethylation of CpG islands, including the promoter and exon 1 of the estrogen receptor (ESR1) gene in normal human colon (28). Aging-associated locus-specific hypermethylation was also observed in intron 1 to exon 4 of c-fos in human liver (29). Moreover, aging-associated hypermethylation was observed in various tumor suppressor genes, such as death-associated protein kinase 1 (DAPK1), E-cadherin, glutathione S-transferase P1 (GSTP1), human mutL homologue 1 (hMLH1), p16, and runt-related transcription factor 3 (RUNX3), with tissue-specificity (30). In addition, in a cross-sectional and longitudinal epigenome-wide study, methylation levels at CpG sites in the fatty acid elongase 2 (ELOVL2), four and a half LIM domains 2 (FHL2), zyg-11 homolog A (ZYG11A), and neurofilament medium polypeptide (NEFM) genes were significant predictors of aging (26). This aging-associated methylation might be due to changes in expression and/or activity of DNMTs or other epigenetic modifiers, which are responsible for DNA demethylation with aging. Indeed, there are several studies demonstrating that aging-related global DNA hypomethylation results from a decrease in expression of DNMT1, which is known for ‘copying’ the DNA methylation pattern to the nascent strand during DNA replication (31,32). In contrast, aging-related DNA hypermethylation results from an increase in expression of DNMT3, which is known for de novo methylation of DNA (32). Moreover, a recent study suggests that aging-related hypomethylation is associated with several histone markers (33). McClay et al. (33) discovered aging-related differentially methylated regions (DMRs) using whole blood DNA from people aged 25 to 92 years. Among the DMRs, hypomethylated regions are associated with histone modifications, such as acetylation and methylation. These results suggest that during the aging process, coordination between DNA methylation and histone modifications might regulate aging-related epigenetic changes and chromatin structure.
Histone modification
Histones are proteins that form a core particle around in which double-stranded genomic DNA is wrapped. They are susceptible to covalent modifications, such as acetylation, methylation, and phosphorylation. These modifications regulate chromatin structure and protein-protein interactions, and they play a role in transcriptional regulation. In addition to DNA methylation, histone modifications are recognized as aging-associated epigenetic markers (Table 1). For example, aging-related hypomethylated regions are associated with various histone modifications, including H3K27ac, H3K4m1, H3K4m2, H3K4m3, and H3K9ac (33). Histone acetylation is a well-understood histone modification associated with aging. Histone acetylation occurs mainly at the lysine residues of the N-terminal tail. The acetylation of histones loosens the connection between DNA and histone proteins, leading to transcriptional activation, whereas the deacetylation of histones results in a compact nucleosome and transcriptional repression.
Of the epigenetic modifiers regulating acetylation status, sirtuins (SIRTs) are known to affect the aging process and aging-related diseases. SIRTs are a family of nicotinamide adenine dinucleotide (NAD)-dependent proteins that catalyze deacetylation, and they are distributed in a variety of organisms from yeast to mammals. In yeast, Sir2 is a SIRT protein that regulates longevity (34). Down-regulation of Sir2 in yeast led to a shortening of the replicative life span, whereas activation of Sir2 significantly extended life span (35). Sir2 modulates aging by translocating its complex from telomeres to ribosomal DNA repeats and avoiding the formation of extrachromosomal ribosomal DNA circles (ERCs), which could result in genomic instability (36). Sir2 protein abundance declines with aging and results in an increase in H4 lysine 16 acetylation and a loss of histones at specific subtelomeric regions in replicative old yeast cells. Antagonizing activities of Sir2 and Sas2, a histone acetyltransferase, regulate the replicative life span through acetylation of histone H4 lysine 16 at subtelomeric regions, indicating that SIRTs have an evolutionarily conserved role in maintaining intact telomeric chromatin (37). Genomic stabilization may also be regulated by Sir2-associated proteins (Hst3 and Hst4), resulting in H3K56 deacetylation and heterochromatinization at ERCs (38).
In mammals, seven SIRTs (SIRT1 to SIRT7) have been identified, and SIRT1 is the closet homolog of yeast Sir2. Interestingly, unlike Sir2 in yeast and worms, the mammalian homolog of Sir2 mainly acts as a deacetylase of other proteins, such as p53, rather than histones (39). To date, little is known about the direct role of the mammalian SIRT family in aging. Instead, studies have investigated the function of SIRT proteins in aging-related diseases, such as metabolic disease and cancer. For example, in mutant mice, a SIRT1 point mutation that abolished its catalytic activity induced susceptibility to a high-fat diet and exhibited accumulation of hepatic lipid and insulin resistance (40). SIRT6-deficient mice display a degenerative phenotype with premature aging traits, such as cachexia, kyphosis, and osteopenia, due to a malfunction in the base-excision repair system (41). In addition, activation of the deacetylase SIRT6 on the telomeric H3K9 and H3K56 regions is linked with genomic instability and aging (42,43). SIRT6 is also proposed to attenuate signaling of the stress-responsive transcription factor, nuclear factor-κB, via H3K9 deacetylation (44).
Besides acetylation, other histone modifications have been scarcely investigated with regard to aging. Aside from SIRT-related histone modifications, increased H4-K20 trimethylation has been reported in aged rat liver (45). Progressive dephosphorylation of histones H1.4 and H1.5 is another histone modification mark in aging that was detected in peripheral blood lymphocytes (46).
EPIGENETIC LINK BETWEEN NUTRITION AND LONGEVITY
Metabolism is important in aging because energy regulation changes during the aging process. Metabolic challenges are related to age-related chronic diseases. Aging-associated changes in epigenetic modifications and epigenetic enzymatic activity have mainly been shown to influence metabolic pathways for insulin signaling. For example, the histone demethylase activity of ubiquitously transcribed TPR on X (Utx-1) was shown to regulate the expression of key components of the insulin/insulin-like growth factor (IGF) pathway in Caenorhabditis elegans (47). RNA interference of Utx-1 increases H3K27 trimethylation of the Daf-2 gene, which leads to nuclear accumulation of the Daf-16, an ortholog of the forkhead box O (FOXO) transcription factor, and results in the activation of anti-aging genes (47). Furthermore, increased levels of Sir2 extend life span through the insulin/IGF-1 pathway (48), in which a cascade of phosphorylation occurs with inactivation of transport of FOXO transcription factors to the nucleus and inhibition of anti-aging genes. These metabolism-associated epigenetic modulations in cellular signaling pathways are mediated by nutrient availability or bioactive compounds (Fig. 1).
Longevity-related epigenetic changes in response to nutrient availability and calorie restriction
Calorie restriction has been shown to extend life span and delay the incidence of age-related chronic diseases (49). In primate studies, calorie restriction was shown to modulate longevity positively. A study of monkeys at the Wisconsin National Primate Research Center reported that moderate calorie restriction improved both overall mortality and age-related mortality, and it reduced the incidence of cancer, cardiovascular disease, and diabetes (50). Consistent with these findings, clinical research has shown beneficial effects of calorie restriction on age-related phenomena (51,52). Calorie restriction reduced the risk factors for atherosclerosis, including lipid parameters and blood pressure (51), and it improved two types of aging biomarkers, fasting insulin levels and core body temperature (52).
The calorie restriction-driven reduction of the metabolic rate involves deactivation of the nutrient-sensing mammalian target of rapamycin (mTOR), an evolutionarily conserved serine/threonine protein kinase that is strongly involved in most cellular functions and cell growth (53,54). Recent experimental work implies that the mTOR signaling pathway plays an important role in the cell’s aging process (55). Protein synthesis is modulated by target of rapamycin (TOR) on the ribosomal protein S6 kinase and on the translation initiation factor 4E-binding protein (56). Therefore, the inhibition of TOR may reduce protein synthesis and result in a prolonged life span because inhibition of translation could stabilize the cell’s metabolic state. TOR is also involved in regulating autophagy, which contributes to cell longevity (57). Autophagy provides a metabolic buffering capacity to resist stress, and it is induced by stress and caloric restriction. It serves to clean damaged organelles and cell components and to prevent their accumulation. Reduced autophagy may be an oncogenic event related to tumor progression, whereas increased autophagy in tumor cells provides an adaptive survival mechanism to overcome stress and drug cytotoxicity (58,59). mTOR negatively regulates autophagy, so down-regulation of mTOR by caloric restriction could enhance autophagy in cells.
A number of studies have demonstrated that Sir2 and its related genes determine life span (36,48), and Sir2 might act as a mediator of longevity via calorie restriction (60). The mammalian deacetylase SIRT1 detects the increase in NAD+ concentrations from NADH oxidation, which allows SIRT1 to respond to calorie restriction and up-regulate its activity. Therefore, stimulated SIRT1 activity affects transcriptional regulators involved in aging-related metabolism, such as fat mobilization, insulin secretion, and stress resistance (61). In SIRT1 knock-out mice, which displayed alterations in metabolic rate and physical activity, calorie restriction did not affect survival (62). Caloric restriction stimulates autophagy through the activation of the mammalian SIRT1, which leads to deacetylation of essential autophagy modulators and affects several glucose homeostasis-related pathways (63, 64).
Calorie restriction increased the expression and/or activity of the histone deacetylase SIRT1, leading to increased histone deacetylation and increased DNA methylation (65). Based on the above results that indicate that SIRT1 plays a critical role in the longevity response to calorie restriction or compounds that mimic calorie restriction like resveratrol, we hypothesize that SIRT1 promotes longevity in part through epigenetic effects, including maintenance of DNA methylation integrity.
Bioactive compounds and longevity
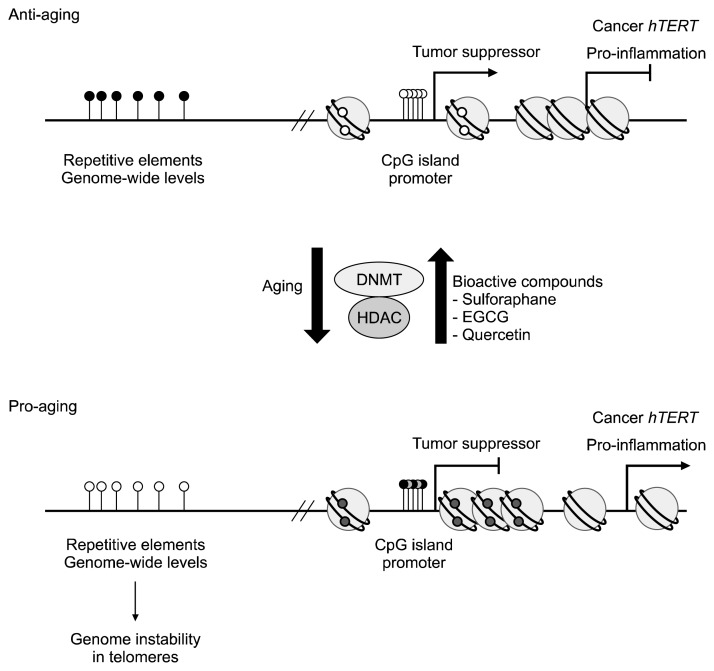
Although calorie restriction has been shown to have a beneficial role in aging, calorie restriction therapy has several limitations or potential side effects, such as infertility, menstrual irregularities, hypertension, and depression (66). For these reasons, recent studies have aimed to identify bioactive compounds that may mimic calorie restriction and provide therapeutic anti-aging effects. Unfortunately, direct evidence showing linkage between bioactive compound consumption and longevity is scarce. However, several studies have reported beneficial effects of natural compounds on age-related phenomena, mainly in providing anticancer and anti-inflammatory effects (Fig. 2).
Fig. 2.
Epigenetic changes in anti-aging and pro-aging status. Relative to anti-aged normal cells, aged cells exhibit DNA hypomethylation in the repetitive elements and at genome-wide levels. Pro-aging cells, which are mainly cancer cells, exhibit DNA hypermethylation at CpG islands within the promoters of tumor suppressor. Pro-aging cells also show induction of pro-inflammatory genes and human telomerase reverse transcriptase (hTERT) expression, possibly due to the loosened chromatin structure. These pro-aging related epigenetic marks can be reversed by treatment with bioactive compounds, such as sulforaphane, epigallocatechin-3-gallate (EGCT), and quercetin. White and black dots indicate unmethylated and methylated DNA status, respectively.
Resveratrol is the most characterized bioactive polyphenolic compound in anti-aging diets. Dietary polyphenols have antioxidant capacity and protect against age-related degenerative diseases. They can activate endogenous defense systems and modulate cellular signaling processes (60). Resveratrol is found in grapes and grape-based red wine, as well as strawberries and blueberries. Resveratrol and other polyphenols have low bioavailability in humans. However, resveratrol and its metabolites accumulate in human cells in vivo in a tissue-specific and dose-dependent manner (67). The possible role of resveratrol in extending life span recently gained worldwide attention. Resveratrol has been identified as a potent SIRT1 activator (68,69) that mimics the effects of calorie restriction and regulates longevity from lower organisms to humans (70). In 2003, Howitz and colleagues showed that resveratrol increases the deacetylase activity of SIRT1 (70). A number of studies showed that resveratrol induced SIRT1 activity in several species (71). Resveratrol mimics calorie restriction effects, which may result in increased life span (68–70).
Although the effects of resveratrol and SIRT1 on longevity are still debated, resveratrol clearly appears to improve metabolism and attenuate the risk of age-related chronic diseases in animal models (72). For example, increased SIRT1 activation from resveratrol improves energy expenditure and prevents diet-induced obesity and other metabolic diseases (69). In addition, middle-aged mice on a high-calorie diet that were treated with resveratrol showed healthy and longevity benefits (68). In humans, resveratrol supplementation induces metabolic changes in obese humans, mimicking the effects of calorie restriction (73). In addition to metabolic regulation, resveratrol has an intrinsic antioxidant capacity and induces the expression of antioxidant enzymes, which reduces oxidative stress (74). To date, little is known about the underlying epigenetic mechanism by which resveratrol improves longevity and aging-related metabolism. Research suggests that resveratrol may target metabolism-related pathways, such as AMP-activated protein kinase and peroxisome proliferator-activated receptor gamma coactivator 1 α (69,75,76).
In addition to resveratrol, accumulating evidence suggests that various bioactive compounds may improve life span and aging-related diseases, mainly cancer. We reviewed research on several bioactive components of which beneficial effects are mediated by epigenetic modifications, namely, sulforaphane, epigallocatechin-3-gallate (EGCG), quercetin, and genistein. Sulforaphane is a well-studied dietary inhibitor of histone deacetylase, and it is found in cruciferous vegetables. It is an isothiocyanate that exhibits antioxidative and anti-inflammatory effects mainly by transcriptional regulation. McWalter et al. (77) reported that broccoli seed extract containing sulforaphane activates nuclear factor E2-related factor 2, which is involved in an antioxidative pathway. In addition, sulforaphane has been shown to have inhibitory effects on DNMT in human breast cancer cells. In MCF-7 and MDA-MB-231 cells, sulforaphane treatment decreased expression of human telomerase reverse transcriptase (hTERT) and increased occupancy of histone acetylation levels at the hTERT promoter. Additionally, sulforaphane decreased the expression levels of DNMT1 and DNMT3a, although there were only slight changes in DNA methylation in the hTERT promoter region (78).
EGCG is an abundant catechin compound in green tea. This catechin has been shown to have anticancer effects by acting as a DNMT inhibitor. According to research by Fang et al. (79), EGCG treatment reversed hypermethylation at promoters of tumor suppressor genes, including p16INK4a, retinoic acid receptor β (RARβ), O6-methylguanine methyltransferase (MGMT), and hMLH1, in human esophageal cancer KYSE 510 cells. These cells showed re-expression of tumor suppressor genes, mainly for RARβ and hMLH1. These results indicate that by reversing silenced tumor suppressors with DNMT inhibition, EGCG treatment may have an anticancer effect. In line with sulforaphane, EGCG treatment exhibits anticancer effects by reducing cellular proliferation and increasing apoptosis (80). In addition, hTERT expression was down-regulated by EGCG treatment in MCF-7 breast cancer cells (80).
The anti-inflammatory and anticancer properties of quercetin and genistein have also been investigated. Quercetin is a polyphenolic substance found in onions and citrus fruits. Quercetin treatment inhibited pro-inflammatory gene expression, such as interferon-gamma-inducible protein 10 and macrophage inflammatory protein 2 expression (81). It has been proposed to regulate transcription factor recruitment and decrease histone acetylation levels (81). Genistein is an isoflavone, or phytoestrogen, found in soybeans and soy products. Similar to EGCG, genistein treatment led to the re-expression of tumor suppressor genes, such as RARβ, p16INK4a, and MGMT, in KYSE 510 esophageal squamous cancer cells (82). Genistein treatment led to demethylation of promoter regions of the tumor suppressor genes, indicating that genistein functions as a DNMT inhibitor (82).
CONCLUSIONS
With a growing aging population, healthy aging has become an important aim for public health. For decades, aging has been associated with genetics and environmental factors. In addition, here we suggest that epigenetic modifications also play a role in the aging process. It is clear that epigenetic marks, including DNA methylation and histone modifications, are associated with aging. Several studies have reported epigenetic biomarkers, mainly involving DNA methylation, which can predict the cellular age of various tissues. These results indicate that epigenetic approaches may be applied to develop effective anti-aging interventions. Nutrients and their metabolites are crucial substrates for epigenetic modifiers. There is mounting evidence demonstrating that dietary interventions influence epigenetic status, and furthermore that altered epigenetic changes are inherited by offspring. These data suggest that physiological changes induced by nutritional intervention may be responsible for epigenetic changes and that the modified epigenetic patterns are long lasting. Some reports have suggested that changes in nutrient availability, such as energy restriction and direct supplementation of bioactive compounds, are associated with aging via epigenetic alterations. However, in many cases, the epigenetic changes are associated with aging-related diseases. Therefore, in the future, it is important to find direct evidence of nutritional effects on longevity and to investigate epigenetic markers that can predict individual responses.
ACKNOWLEDGEMENTS
This study was supported by ILSI Korea and the National Research Foundation of Korea (NRF 2016R1D1A1A 02937546). J.H.P and Y.Y are supported by Brain Korea 21 project (22A20130012143).
Footnotes
AUTHOR DISCLOSURE STATEMENT
The authors declare no conflict of interest.
REFERENCES
- 1.Karasik D, Demissie S, Cupples LA, Kiel DP. Disentangling the genetic determinants of human aging: biological age as an alternative to the use of survival measures. J Gerontol A Biol Sci Med Sci. 2005;60:574–587. doi: 10.1093/gerona/60.5.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deelen J, Beekman M, Uh HW, Broer L, Ayers KL, Tan Q, Kamatani Y, Bennet AM, Tamm R, Trompet S, Guðbjartsson DF, Flachsbart F, Rose G, Viktorin A, Fischer K, Nygaard M, Cordell HJ, Crocco P, van den Akker EB, Böhringer S, Helmer Q, Nelson CP, Saunders GI, Alver M, Andersen-Ranberg K, Breen ME, van der Breggen R, Caliebe A, Capri M, Cevenini E, Collerton JC, Dato S, Davies K, Ford I, Gampe J, Garagnani P, de Geus EJ, Harrow J, van Heemst D, Heijmans BT, Heinsen FA, Hottenga JJ, Hofman A, Jeune B, Jonsson PV, Lathrop M, Lechner D, Martin-Ruiz C, Mcnerlan SE, Mihailov E, Montesanto A, Mooijaart SP, Murphy A, Nohr EA, Paternoster L, Postmus I, Rivadeneira F, Ross OA, Salvioli S, Sattar N, Schreiber S, Stefánsson H, Stott DJ, Tiemeier H, Uitterlinden AG, Westendorp RG, Willemsen G, Samani NJ, Galan P, Sørensen TI, Boomsma DI, Jukema JW, Rea IM, Passarino G, de Craen AJ, Christensen K, Nebel A, Stefánsson K, Metspalu A, Magnusson P, Blanché H, Christiansen L, Kirkwood TB, van Duijn CM, Franceschi C, Houwing-Duistermaat JJ, Slagboom PE. Genomewide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum Mol Genet. 2014;23:4420–4432. doi: 10.1093/hmg/ddu139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nebel A, Flachsbart F, Till A, Caliebe A, Blanché H, Arlt A, Häsler R, Jacobs G, Kleindorp R, Franke A, Shen B, Nikolaus S, Krawczak M, Rosenstiel P, Schreiber S. A functional EXO1 promoter variant is associated with prolonged life expectancy in centenarians. Mech Ageing Dev. 2009;130:691–699. doi: 10.1016/j.mad.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Ryu S, Moskowitz DM, Rothenberg D, Leahy DJ, Atzmon G, Barzilai N, Suh Y. Discovery of novel non-synonymous SNP variants in 988 candidate genes from 6 centenarians by target capture and next-generation sequencing. Mech Ageing Dev. 2013;134:478–485. doi: 10.1016/j.mad.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell. 2010;40:323–332. doi: 10.1016/j.molcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips CM. Nutrigenetics and metabolic disease: current status and implications for personalised nutrition. Nutrients. 2013;5:32–57. doi: 10.3390/nu5010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKay JA, Mathers JC. Diet induced epigenetic changes and their implications for health. Acta Physiol. 2011;202:103–118. doi: 10.1111/j.1748-1716.2011.02278.x. [DOI] [PubMed] [Google Scholar]
- 9.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Huang S, Lee L, Davalos A, Schiestl RH, Campisi J, Oshima J. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell. 2003;2:191–199. doi: 10.1046/j.1474-9728.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- 11.de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, Weeda G, van der Horst GT, van Leeuwen W, Themmen AP, Meradji M, Hoeijmakers JH. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296:1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- 12.Fraga MF. Genetic and epigenetic regulation of aging. Curr Opin Immunol. 2009;21:446–453. doi: 10.1016/j.coi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 14.Tomás-Loba A, Flores I, Fernández-Marcos PJ, Cayuela ML, Maraver A, Tejera A, Borrás C, Matheu A, Klatt P, Flores JM, Viña J, Serrano M, Blasco MA. Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell. 2008;135:609–622. doi: 10.1016/j.cell.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 15.Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/S0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 16.Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 17.Niculescu MD, Lupu DS. Nutritional influence on epigenetics and effects on longevity. Curr Opin Clin Nutr Metab Care. 2011;14:35–40. doi: 10.1097/MCO.0b013e328340ff7c. [DOI] [PubMed] [Google Scholar]
- 18.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 19.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doskočil J, Šorm F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- 21.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 22.Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- 23.Barbot W, Dupressoir A, Lazar V, Heidmann T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 2002;30:2365–2373. doi: 10.1093/nar/30.11.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Putiri EL, Robertson KD. Epigenetic mechanisms and genome stability. Clin Epigenetics. 2011;2:299–314. doi: 10.1007/s13148-010-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ, Puca AA, Sayols S, Pujana MA, Serra-Musach J, Iglesias-Platas I, Formiga F, Fernandez AF, Fraga MF, Heath SC, Valencia A, Gut IG, Wang J, Esteller M. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Florath I, Butterbach K, Müller H, Bewerunge-Hudler M, Brenner H. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet. 2014;23:1186–1201. doi: 10.1093/hmg/ddt531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 28.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 29.Choi EK, Uyeno S, Nishida N, Okumoto T, Fujimura S, Aoki Y, Nata M, Sagisaka K, Fukuda Y, Nakao K, Yoshimoto T, Kim YS, Ono T. Alterations of c-fos gene methylation in the processes of aging and tumorigenesis in human liver. Mutat Res. 1996;354:123–128. doi: 10.1016/0027-5107(96)00056-5. [DOI] [PubMed] [Google Scholar]
- 30.Waki T, Tamura G, Sato M, Motoyama T. Age-related methylation of tumor suppressor and tumor-related genes: an analysis of autopsy samples. Oncogene. 2003;22:4128–4133. doi: 10.1038/sj.onc.1206651. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Deng C, Lu Q, Richardson B. Age-dependent DNA methylation changes in the ITGAL (CD11a) promoter. Mech Ageing Dev. 2002;123:1257–1268. doi: 10.1016/S0047-6374(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 32.Casillas MA, Jr, Lopatina N, Andrews LG, Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. doi: 10.1023/A:1025548623524. [DOI] [PubMed] [Google Scholar]
- 33.McClay JL, Aberg KA, Clark SL, Nerella S, Kumar G, Xie LY, Hudson AD, Harada A, Hultman CM, Magnusson PK, Sullivan PF, Van Den Oord EJ. A methylome-wide study of aging using massively parallel sequencing of the methyl-CpG-enriched genomic fraction from blood in over 700 subjects. Hum Mol Genet. 2014;23:1175–1185. doi: 10.1093/hmg/ddt511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell SJ, Kahn CR. Endocrine regulation of ageing. Nat Rev Mol Cell Biol. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- 35.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hachinohe M, Hanaoka F, Masumoto H. Hst3 and Hst4 histone deacetylases regulate replicative lifespan by preventing genome instability in Saccharomyces cerevisiae. Genes Cells. 2011;16:467–477. doi: 10.1111/j.1365-2443.2011.01493.x. [DOI] [PubMed] [Google Scholar]
- 39.Sommer M, Poliak N, Upadhyay S, Ratovitski E, Nelkin BD, Donehower LA, Sidransky D. ΔNp63&alpha overexpression induces downregulation of Sirt1 and an accelerated aging phenotype in the mouse. Cell Cycle. 2006;5:2005–2011. doi: 10.4161/cc.5.17.3194. [DOI] [PubMed] [Google Scholar]
- 40.Caron AZ, He X, Mottawea W, Seifert EL, Jardine K, Dewar-Darch D, Cron GO, Harper ME, Stintzi A, McBurney MW. The SIRT1 deacetylase protects mice against the symptoms of metabolic syndrome. FASEB J. 2014;28:1306–1316. doi: 10.1096/fj.13-243568. [DOI] [PubMed] [Google Scholar]
- 41.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 42.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarg B, Koutzamani E, Helliger W, Rundquist I, Lindner HH. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J Biol Chem. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 46.Happel N, Doenecke D, Sekeri-Pataryas KE, Sourlingas TG. H1 histone subtype constitution and phosphorylation state of the ageing cell system of human peripheral blood lymphocytes. Exp Gerontol. 2008;43:184–199. doi: 10.1016/j.exger.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Jin C, Li J, Green CD, Yu X, Tang X, Han D, Xian B, Wang D, Huang X, Cao X, Yan Z, Hou L, Liu J, Shukeir N, Khaitovich P, Chen CD, Zhang H, Jenuwein T, Han JD. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 49.Masoro EJ. Subfield history: caloric restriction, slowing aging, and extending life. Sci Aging Knowl Environ. 2003;2003:re2. doi: 10.1126/sageke.2003.8.re2. [DOI] [PubMed] [Google Scholar]
- 50.Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fontana L, Meyer TE, Klein S, Holloszy JO. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci USA. 2004;101:6659–6663. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heilbronn LK, de Jonge L, Frisard MI, DeLany JP, Larson-Meyer DE, Rood J, Nguyen T, Martin CK, Volaufova J, Most MM, Greenway FL, Smith SR, Deutsch WA, Williamson DA, Ravussin E Pennington CALERIE Team. Effect of 6-month calorie restriction on biomarkers of longevity, metabolic adaptation, and oxidative stress in overweight individuals: a randomized controlled trial. JAMA. 2006;295:1539–1548. doi: 10.1001/jama.295.13.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10:2305–2316. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan L, Mieulet V, Lamb RF. Nutrient regulation of mTORC1 and cell growth. Cell Cycle. 2010;9:2473–2474. doi: 10.4161/cc.9.13.12124. [DOI] [PubMed] [Google Scholar]
- 55.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 57.Silver N, Proctor GB, Arno M, Carpenter GH. Activation of mTOR coincides with autophagy during ligation-induced atrophy in the rat submandibular gland. Cell Death Dis. 2010;1:e14. doi: 10.1038/cddis.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bennett HL, Fleming JT, O’Prey J, Ryan KM, Leung HY. Androgens modulate autophagy and cell death via regulation of the endoplasmic reticulum chaperone glucose-regulated protein 78/BiP in prostate cancer cells. Cell Death Dis. 2010;1:e72. doi: 10.1038/cddis.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 60.Chen C, Yu R, Owuor ED, Kong AN. Activation of antioxidant-response element (ARE), mitogen-activated protein kinases (MAPKs) and caspases by major green tea polyphenol components during cell survival and death. Arch Pharm Res. 2000;23:605–612. doi: 10.1007/BF02975249. [DOI] [PubMed] [Google Scholar]
- 61.Chen D, Guarente L. SIR2: a potential target for calorie restriction mimetics. Trends Mol Med. 2007;13:64–71. doi: 10.1016/j.molmed.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 62.Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blagosklonny MV. Linking calorie restriction to longevity through sirtuins and autophagy: any role for TOR. Cell Death Dis. 2010;1:e12. doi: 10.1038/cddis.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stein S, Matter CM. Protective roles of SIRT1 in atherosclerosis. Cell Cycle. 2011;10:640–647. doi: 10.4161/cc.10.4.14863. [DOI] [PubMed] [Google Scholar]
- 65.Wakeling LA, Ions LJ, Ford D. Could Sirt1-mediated epigenetic effects contribute to the longevity response to dietary restriction and be mimicked by other dietary interventions? AGE. 2009;31:327–341. doi: 10.1007/s11357-009-9104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dirks AJ, Leeuwenburgh C. Tumor necrosis factor alpha signaling in skeletal muscle: effects of age and caloric restriction. J Nutr Biochem. 2006;17:501–508. doi: 10.1016/j.jnutbio.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 67.Smoliga JM, Baur JA, Hausenblas HA. Resveratrol and health–a comprehensive review of human clinical trials. Mol Nutr Food Res. 2011;55:1129–1141. doi: 10.1002/mnfr.201100143. [DOI] [PubMed] [Google Scholar]
- 68.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 70.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 71.Baur JA. Resveratrol, sirtuins, and the promise of a DR mimetic. Mech Ageing Dev. 2010;131:261–269. doi: 10.1016/j.mad.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cantó C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD+? Pharmacol Rev. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Timmers S, Konings E, Bilet L, Houtkooper RH, van de Weijer T, Goossens GH, Hoeks J, van der Krieken S, Ryu D, Kersten S, Moonen-Kornips E, Hesselink MK, Kunz I, Schrauwen-Hinderling VB, Blaak EE, Auwerx J, Schrauwen P. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011;14:612–622. doi: 10.1016/j.cmet.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Halliwell B. Dietary polyphenols: good, bad, or indifferent for your health? Cardiovasc Res. 2007;73:341–347. doi: 10.1016/j.cardiores.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 75.Um JH, Park SJ, Kang H, Yang S, Foretz M, McBurney MW, Kim MK, Viollet B, Chung JH. AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes. 2010;59:554–563. doi: 10.2337/db09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cantó C, Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci. 2010;67:3407–3423. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McWalter GK, Higgins LG, McLellan LI, Henderson CJ, Song L, Thornalley PJ, Itoh K, Yamamoto M, Hayes JD. Transcription factor Nrf2 is essential for induction of NAD(P)H: quinone oxidoreductase 1, glutathione S-transferases, and glutamate cysteine ligase by broccoli seeds and isothiocyanates. J Nutr. 2004;134:3499S–3506S. doi: 10.1093/jn/134.12.3499S. [DOI] [PubMed] [Google Scholar]
- 78.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;6:e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 80.Berletch JB, Liu C, Love WK, Andrews LG, Katiyar SK, Tollefsbol TO. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008;103:509–519. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruiz PA, Braune A, Hölzlwimmer G, Quintanilla-Fend L, Haller D. Quercetin inhibits TNF-induced NF-κB transcription factor recruitment to proinflammatory gene promoters in murine intestinal epithelial cells. J Nutr. 2007;137:1208–1215. doi: 10.1093/jn/137.5.1208. [DOI] [PubMed] [Google Scholar]
- 82.Fang MZ, Chen D, Sun Y, Jin Z, Christman JK, Yang CS. Reversal of hypermethylation and reactivation of p16INK4a, RARβ, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res. 2005;11:7033–7041. doi: 10.1158/1078-0432.CCR-05-0406. [DOI] [PubMed] [Google Scholar]