Abstract
The pathogenic bacterium Staphylococcus aureus has evolved to actively evade many aspects of the human innate immune system by expressing a series of secreted inhibitory proteins. Among these, the extracellular adherence protein (Eap) has been shown to inhibit the classical and lectin pathways of the complement system. By binding to complement component C4b, Eap is able to inhibit formation of the CP/LP C3 pro-convertase. Secreted full-length, mature Eap consists of four ~98 residue domains, all of which adopt a similar beta-grasp fold, and are connected through a short linker region. Through multiple biochemical approaches, it has been determined that the third and fourth domains of Eap are responsible for C4b binding. Here we report the backbone and side-chain resonance assignments of the 11.3 kDa fourth domain of Eap. The assignment data has been deposited in the BMRB database under the accession number 26726.
Keywords: Staphylococcus aureus, Virulence factor, Extracellular adherence protein, NMR, Resonance assignment
Biological context
Staphylococcus aureus is one the leading causes of health care-associated and community-acquired infections (Boucher et al. 2010). One reason for this trend is the increasing number of drug resistant strains of Staph that are insensitive to standard antibiotic treatments, and which have been shown to account for upwards of 50 % of all S. aureus strains causing disease within the healthcare system (Drago et al. 2007). While antibiotic resistance is a significant threat to public health on its own, the enhanced virulence of widespread clinical strains is not simply due to their resistance phenotypes (Nimmo 2012, Thurlow et al. 2012). Indeed, the ability of S. aureus to thrive in many tissues within its host can be attributed to its array of secreted virulence factors that interfere with the host’s immune response. A number of these virulence factors have been shown to specifically bind to and inhibit components of the complement system, which serves as a critical aspect of innate immunity responsible for opsonizing and killing invading pathogens. Recent reviews have highlighted many of the underlying mechanisms by which these virulence factors are able to inhibit various facets of the innate immune system, including complement (Thammavongsa et al. 2015, Serruto et al. 2010).
The extracellular adherence protein (Eap) has been shown to affect multiple aspects of the innate immune system, and contributes to the overall virulence of Staphylococcus aureus (Stapels et al. 2014, Woehl et al. 2014). Eap is secreted as a 50- to 70-kDa protein depending upon the specific strain in question, and consists of multiple ~97 residue repeating EAP domains (Geisbrecht et al. 2005). Each EAP domain is characterized by a beta-grasp fold and is connected to the adjacent domain through a short (~10 residue) linker region. Through a multi-disciplinary approach involving analytical ultracentrifugation and small-angle X-ray scattering, Hammel et al. determined that Eap adopts an elongated, but structured conformation in solution (Hammel et al. 2007). Subsequent experimentally-constrained structural modeling on a four-domain isoform of Eap suggested a potential for transient interactions between adjacent pairs of EAP domains, most notably Eap1 with Eap2, and Eap3 with Eap4. Indeed, Raman spectroscopy studies of samples comprised of full-length Eap versus a stoichiometric mixture of all four individual domains revealed that interdomain interactions are detectable, but only in the context of the full-length protein (Hammel et al. 2007). This dependence on the linker regions that connect the domains argues that any interdomain interactions are very low affinity on their own, and EAP domains are unlikely associate with one another at physiologically-relevant concentrations.
Whereas several S. aureus complement inhibitors block the alternative complement pathway by binding C3b and disrupting the multi-subunit alternative pathway C3 convertase complex (C3bBb) which processes additional C3 into C3b, Eap instead targets the classical and lectin complement pathways. Though they are initiated by distinct stimuli, the classical and lectin pathways of complement converge at the point of C4, which is then cleaved to form C4b. C4b binds to complement component C2, forming the C3 pro-convertase (C4bC2). Cleavage of C4b-bound C2 forms the fully active C3 convertase of the classical and lectin pathways (C4bC2a), which is also able to proteolyze C3 into C3b. Eap has been shown to bind with a nanomolar affinity to C4b, and competes out the interaction between C4b and C2. In this manner, Eap blocks activity of the classical and lectin pathways by inhibiting formation of their C3 pro-convertase (Woehl et al. 2014).
Neutrophils also play a critical role in the innate immune response against invading bacterial pathogens. Upon activation, neutrophils release a number of chymotrypsin-like serine proteases into their phagocytic compartment or extracellular environment, including neutrophil elastase, cathepsin G, and proteinase 3 (Amulic et al. 2012). These so-called neutrophil-serine proteases (NSPs) have been shown to degrade outer membrane components of bacteria as well facilitating in the removal of secreted bacterial virulence factors (Stapels et al. 2015). Recently, Eap as well as two homologs comprised of a single EAP repeat, called EapH1 and EapH2, have been shown to bind and inhibit NSPs (Stapels et al. 2014). Mechanistically, Eap, EapH1, and EapH2 bind in a non-covalent manner with nanomolar affinity to each NSP and appear to block substrate access to the protease catalytic cleft (Stapels et al. 2014).
Although the mechanisms of classical and lectin pathway complement inhibition and NSP inhibition have been identified, the structural basis for these diverse activities of EAP domain proteins remain poorly understood. This is particularly intriguing because while all EAP domains appear to be equally potent inhibitors of NSPs, only an Eap fragment containing domains 3 and 4 retained high affinity C4b binding and CP/LP inhibitory activity similar to the full-length Eap protein. Here, we report the backbone 1H, 15N, and 13C resonance assignments for the fourth domain of the extracellular adherence protein (i.e. Eap 4). These assignments and the structure calculations will be invaluable in identifying specific binding-site interactions between Eap4 and its partners and will promote further NMR-based solution characterization of EAP proteins.
Materials and experiments
A DNA fragment encoding the linker-free Eap4 sequence (residues 374-468 of accession code WP_001557458.1) was subcloned into the Sal1 and Not1 sites of the prokaryotic expression vector pT7HMT (Geisbrecht et al. 2006). The recombinant protein expressed from this plasmid retains a “Gly-Ser-Thr” artifact at its N-terminus following digestion by tobacco etch virus (TEV) protease when removing the polyhistidine purification tag. Uniformly 15N- and/or 13C-labeled Eap4 was expressed in E. coli BL21(DE3) cells grown in M9 minimal media supplemented with 1 g/L 15NH4Cl and 2 g/L 13C-D-glucose for 15N-labeling and 13C-labeling, respectively. The cells containing the expression plasmid were grown in a starter culture consisting of 30 mL LB media supplemented with kanamycin at 37 °C overnight. The starter was centrifuged and re-suspended twice in sterile PBS to wash out excess LB. The pellet was then re-suspended in M9 minimal media, grown at 37 °C to an OD600 value of .8–1.0, induced with 1.0 mM isopropyl β-D-thiogalactoside, and further grown at 18 °C overnight. The cells were harvested by centrifugation (7000 RCF for 15 min, 4 °C), and subsequently re-suspended in native binding buffer and lysed using a microfluidizer. After a second centrifugation (30,000 RCF for 35 min, 4 °C), the cell lysate supernatant was purified using a gravity flow Ni–NTA affinity column. Overnight dialysis into native binding buffer and TEV-protease cleavage was performed to remove the hexahistidine and c-myc epitope tags. The protein was re-purified over a reverse Ni–NTA affinity column to remove the cleaved hexahistidine and c-myc epitope tags. Size exclusion chromatography on a HiLoad 26/600 Superdex 75 prep grade (GE healthcare) was employed as a final purification step. The final protein yield was 7–10 mg from 1 L of growing culture and all NMR samples contained 0.5–1.0 mM uniformly 15N- or 13C/15N-labeled Eap4 in NMR buffer (50 mM sodium phosphate pH 6.5 and 0.1 % NaN3) in 95 % H2O/5 % D2O.
NMR measurements were carried out at 25 °C on a Varian 500VNMR System (Agilent Technologies) equipped with a 5 mm triple-resonance inverse detection pulse gradient cold probe operating at 499.84 MHz for 1H frequency, and on a Bruker 800 MHz Avance III spectrometer equipped with a TCI cryo-probe. Backbone resonance assignments were achieved using the following NMR spectra: 2D 1H–15N HSQC and 3D HNCA, HN(CO)CA, HNCACB, CBCA(-CO)NH, and HNCO. The following NMR spectra were collected for side chain assignments: 2D 13C HSQC and 3D (H)C(CCO)NH-TOCSY, H(CCCO)NH-TOCSY, HCCH-TOCSY, and 15N-edited NOESY (mixing time 100 ms). Validation of the assignments has also been conducted with the 13C-NOESY-HSQC spectrum. All NMR spectra were processed using NMRPipe (Delaglio et al. 1995), and analyzed with CARA (http://www.nmr.ch) (Keller 2004) and UNIO (http://perso.ens-lyon.fr/torsten.herrmann/Herrmann/Software.html) (Serrano et al. 2012). The 1H chemical shift assignment was referenced by using 2,2-dimethy-2-silapentane-5-sulphonic acid (DSS) at 25 °C as a standard. The 13C and 15N chemical shift were referenced indirectly.
Extent of assignments and data deposition
2D 1H–15N HSQC measurement of Eap4 resulted in a well-dispersed spectrum (Fig. 1). Amino acid numbering is based upon the authentic Eap4 sequence, excluding the N-terminal His tag, c-myc tag, and TEV-protease cleavage recognition site. A total of 97 % of backbone 1H and 15N resonances of 95 non-proline residues, and 100 % of all 13Cα resonances, 13Cβ resonances, and 13C′ resonances have been unambiguously assigned based on a standard set of triple resonance spectra described above. The backbone amide residues that could not be assigned include G1, S2, and S24. G1 and S2 lie in a loop region of the artifactual N-terminus as a result of the subcloning procedure, while S24 lies in the C-terminal region of a short β-strand as judged by comparison to the secondary structure prediction and alignment to the crystal structures of three EAP domains (Geisbrecht et al. 2005). Side chain 1H and 13C resonance assignments were 90 % completed excluding aromatic rings. The secondary structure elements of Eap4 were predicted by the TALOS+ program (Shen et al. 2009) using the resonance assignments of 13Cα, 13Cβ, and 13C′ (Fig. 2). This prediction is in excellent agreement with the secondary structures observed in EapH1, EapH2, and Eap2, as determined by X-ray crystallography (Geisbrecht et al. 2005). The chemical shift assignments have been deposited in BioMagResBank (http://www.bmrb.wisc.edu) under the accession number 26726.
Fig. 1.
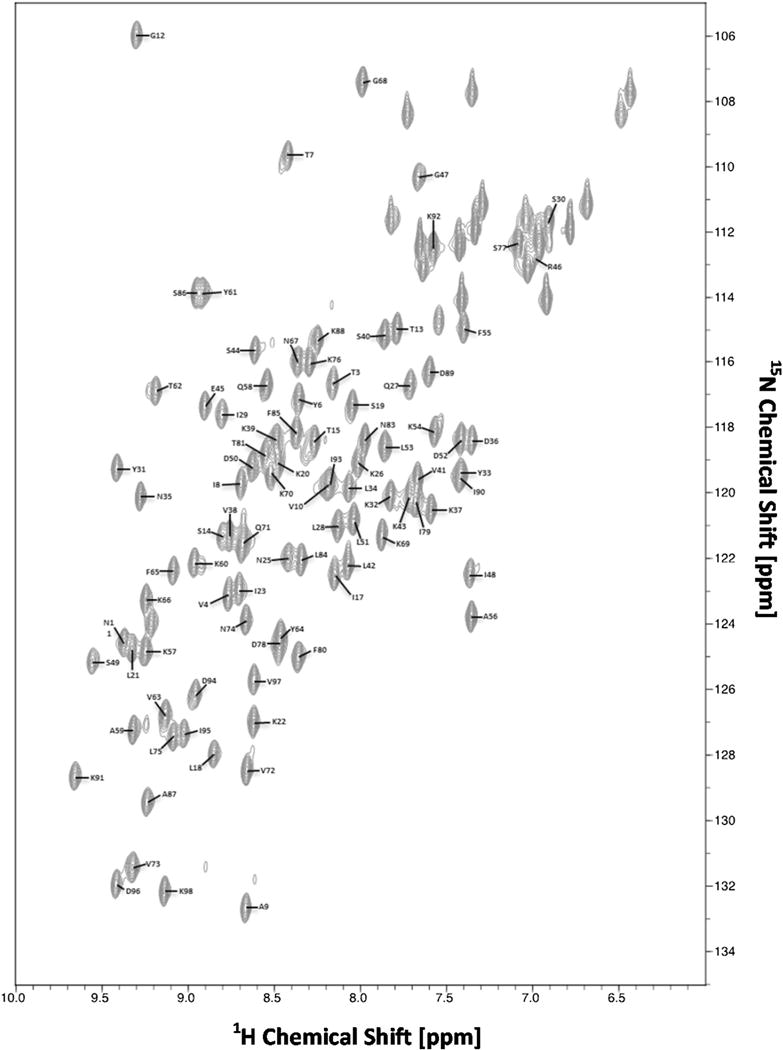
2D 1H–15N HSQC spectrum of 0.7 mM 13C/15N-labeled Eap4► recorded at 298 K on a Bruker 800 MHz Avance III spectrometer equipped with a TCI cryoprobe. Sequence specific assignments are indicated by single letter residue name and sequence number
Fig. 2.
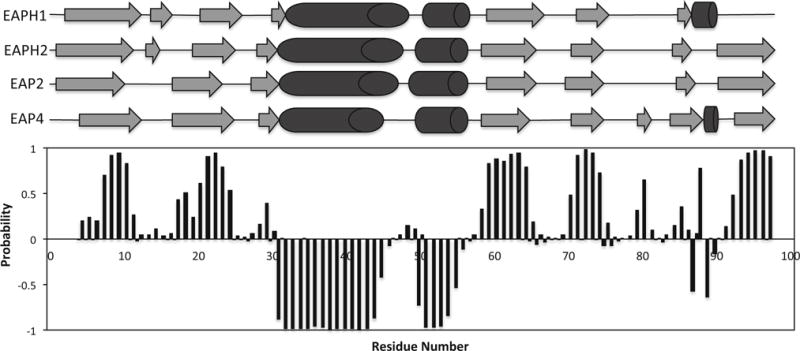
Secondary structure prediction for the Eap4 domain based on the TALOS+ program using obtained chemical shift values. β-strand probabilities are given by positive values, α-helices are given by negative values, and loop regions are given by values approximately from −0.3 to 0.3. Shown in the top portion are the secondary structure topology obtained from the published crystal structure of Eap2 (Geisbrecht et al. 2005) and the TALOS+ prediction of Eap4 with α-helices shown as cylinders and β-sheets shown as arrows. The predicted secondary structure of Eap4 shows very similar topology to the other three EAP domains whose crystal structures are available
Acknowledgments
This work was supported by the American Heart Association Midwest Affiliate Pre-doctoral Research Fellowship 15PRE25750013 and by National Institutes of Health (NIH) Grant AI111203.
References
- Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- Boucher H, Miller LG, Razonable RR. Serious infections caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2010;51(Suppl 2):S183–S197. doi: 10.1086/653519. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Drago L, De Vecchi E, Nicola L, Gismondo MR. In vitro evaluation of antibiotic combinations for empirical therapy of suspected methicillin resistant Staphylococcus aureus severe respiratory infections. BMC Infect Dis. 2007;7:111. doi: 10.1186/1471-2334-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht BV, Hamaoka BY, Perman B, Zemla A, Leahy DJ. The crystal structures of EAP domains from Staphyloccocus aureus reveal an unexpected homology to bacterial superantigens. J Biol Chem. 2005;280(17):17243–17250. doi: 10.1074/jbc.M412311200. [DOI] [PubMed] [Google Scholar]
- Geisbrecht BV, Bouyain S, Pop M. An optimized system for expression and purification of secreted bacterial proteins. Protein Expr Purif. 2006;46:23–32. doi: 10.1016/j.pep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Hammel M, Nemecek D, Keightley JA, Thomas GJ, Jr, Geisbrecht BV. The Staphylococcus aureus extracellular adherence protein (Eap) adopts an elongated but structured conformation in solution. Protein Sci. 2007;16(12):2605–2617. doi: 10.1110/ps.073170807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. The computer aided resonance assignment tutorial. Verlag Goldau; Cantina: 2004. [Google Scholar]
- Nimmo GR. USA300 abroad: global spread of a virulent strain of community-associated methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2012;18:725–734. doi: 10.1111/j.1469-0691.2012.03822.x. [DOI] [PubMed] [Google Scholar]
- Serrano P, Pedrini B, Mohanty B, Geralt M, Herrmann T, Wüthrich K. The J-UNIO protocol for automated protein structure determination by NMR in solution. J Biomol NMR. 2012;53(4):341–354. doi: 10.1007/s10858-012-9645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto D, Rappuoli R, Scarselli M, Gros P, van Strijp JA. Molecular mechanisms of complement evasion: learning from staphylococci and meningococci. Nat Rev Microbiol. 2010;8(6):393–399. doi: 10.1038/nrmicro2366. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS?: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapels DA, Ramyar KX, Bischoff M, von Köckritz-Blickwede M, Milder FJ, Ruyken M, Eisenbeis J, McWhorter WJ, Herrmann M, van Kessel KP, Geisbrecht BV, Rooijakkers SH. Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors. Proc Natl Acad Sci USA. 2014;111(36):13187–13192. doi: 10.1073/pnas.1407616111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapels DA, Geisbrecht BV, Rooijakkers SH. Neutrophil serine proteases in antibacterial defense. Curr Opin Microbiol. 2015;23:42–48. doi: 10.1016/j.mib.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thammavongsa V, Kim HK, Missiakas D, Schneewind O. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol. 2015;13(9):529–543. doi: 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow LR, Joshi GS, Richardson AR. Virulence strategies of the dominant USA300 lineage of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) FEMS Immunol Med Microbiol. 2012;65:5–22. doi: 10.1111/j.1574-695X.2012.00937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehl JL, Stapels DA, Garcia BL, Ramyar KX, Keightley A, Ruyken M, Syriga M, Sfyroera G, Weber AB, Zolkiewski M, Ricklin D, Lambris JD, Rooijakkers SH, Geisbrecht BV. The extracellular adherence protein from Stapylococcus aureus inhibits the classical and lectin pathways of complement by blocking formation of the C3 proconvertase. J Immunol. 2014;193(12):6161–6171. doi: 10.4049/jimmunol.1401600. [DOI] [PMC free article] [PubMed] [Google Scholar]


