Abstract
Infections can cause a multitude of stresses on the host and microbe. To detect potential infections, the mammalian immune system utilizes several families of pattern recognition receptors, which survey the intracellular and extracellular environments for microbial products. Members of each receptor family induce antimicrobial effector responses, which include inflammatory cytokine or interferon expression, downregulation of protein synthesis or host cell death. In this Review, we discuss the benefits of each of these innate immune responses. We highlight how non-infectious bacteria and viruses typically activate a single family of receptors, which results in a predictable host response. Infections with virulent pathogens, in contrast, may activate receptors from distinct families. As each receptor family may induce responses that antagonize or synergize with the activities of another family, cell fate decisions during pathogenic encounters are unpredictable. Understanding the antagonistic antimicrobial activities of the innate immune system should provide insight into how cell fate decisions are made during infections, and potentially during other environmental stresses.
eTOC Blurb
Franz et al describe how innate immune receptors control host cell fate decisions during various types of microbial encounters. During infections, multiple receptors recognize the same microbial ligand, but these receptors can induce cellular responses that antagonize each other. This competition among receptors determines host cell fate decisions during infection.
Introduction
The ability to recognize and adapt to stress is a necessity of life, as organisms that cannot accomplish these seemingly simple tasks are less fit than those that can (Ades, 2008; Torres and Dangl, 2005; Walter and Ron, 2011). The mammalian immune system provides a useful model to study host responses that are designed to prevent or eliminate stress. The best-defined potential stress that is detected by the immune system is the presence of microbes (Janeway, 1989). Microbes that can survive under conditions that are similar to mammalian cells, and can consume similar energy sources, are potentially dangerous to the host. The reason for this is that most microbes replicate faster than mammalian cells. With a fixed food supply, microbial cells will overtake this supply and cause catastrophic consequences to the host. For this reason, at the most basic level, the task of the mammalian immune system is to eradicate microbial infections in order to preserve the host food supply. Due to the rapid pace of bacterial, viral and fungal replication, two selective pressures may have been placed on the immune system to fight infection. The first pressure is to identify the presence of the microbe, and the second pressure is to elicit defense responses rapidly. The inability to detect and rapidly fight infection places a life-threatening stress on the host (Pandey et al., 2014). As such, much genetic information has been dedicated to create rapid response pathways dedicated to host defense (Daugherty and Malik, 2012). However, different microbes pose different threats to the host, with some being avirulent, and others being highly virulent. It therefore stands to reason that the immediate (innate) immune response systems can gauge the threat to the host.
In recent years, several ideas of the means by which infectious threat can be gauged have been discussed, with the dominant view being that virulent pathogens can activate a greater inflammatory response than their avirulent counterparts (Blander and Sander, 2012; Vance et al., 2009). This greater inflammatory response is thought to result from the combined actions of pathogenic activities that promote infection, and the ability of some pathogens to thrive in an inflammatory tissue environment (Faber et al., 2016; Winter et al., 2010). Pathogen replication results in a larger abundance and wider range of microbial ligands present at sites of virulent infections than avirulent ones. These microbial ligands are referred to as pathogen associated molecular patterns (PAMPs) (Janeway, 1989). This strategy of gauging the threat of virulence can be considered microbe-centric, in that the innate immune system would recognize specific types and amounts of PAMPs to determine its state of activation. However, host molecules can also influence the activation state of immune cells. These host molecules are called damage associated molecular patterns (DAMPs), and are released from dying cells at the sites of tissue damage and infection (Newton and Dixit, 2012). Thus, in an infected tissue, distinct sources of immunomodulatory molecules exist, which raises the question of how the host interprets this plethora of information.
In this Review, we will discuss the various cell fate decisions that mammalian cells must make during microbial encounters. Particular focus is placed on macrophages and dendritic cells, as these phagocytes survey all tissues of the body, and are therefore positioned to detect infections rapidly. Attention is also given to tissue resident cells (fibroblasts, for example), as much of our knowledge has derived from the study of these cell types. We discuss the common activation state achieved when cells detect and respond to innocuous microbial encounters, and then focus on the various types of activation that occur upon encounters with pathogens. In particular, we discuss the means by which the receptors of the innate immune system influence cell fate decisions through the detection of PAMPs, DAMPs or both, and highlight how pathogens can activate receptors that elicit activities that oppose one another in the responding cell. These opposing activities of PRRs pose interesting challenges to the host in terms of which activation state to achieve.
PAMPs from non-infectious microbes induce a generic phagocyte activation state
All mammalian cells detect microbes through the actions of receptors that recognize specific PAMPs. There are numerous examples of these host sensors of infection, which are collectively known as pattern recognition receptors (PRRs). The best-characterized PRRs include the Toll-like Receptor (TLR) family, the Dectin family of C-type lectin receptors, the nucleotide binding domain leucine rich repeat containing proteins (NLRs), the AIM2 like Receptors (ALRs), and the RIG-I like Receptors (RLRs) (Medzhitov, 2009). Other PRRs also exist that do not fall into these families, and will be discussed as needed throughout this manuscript. PRRs do not contain a common protein fold or primary amino acid sequence, yet they operate by the common principle of detecting microbes and inducing host-defense responses (Pandey et al., 2014). From a cell biological perspective, PRRs can be classified into two groups—those that survey the extracelluar space for PAMPs, and those that survey the intracellular (usually cytosolic) space for these microbial products (Brubaker et al., 2015).
The mechanisms by which PRRs detect their ligands and the means by which specific PRRs are localized within cells have been the focus of several reviews in recent years (Kagan, 2012; Pandey et al., 2014), and will therefore only be discussed briefly. PRRs can operate as high-affinity PAMP-binding proteins (Chuenchor et al., 2014), or they can operate as low-affinity receptors whose activation depends on the functions of other high-affinity ligand-binding proteins (Gioannini et al., 2004). It is unclear why some PRRs evolved to detect PAMPs with high-affinity directly, and others achieve high-affinity interactions through the aid of accessory factors. One possible advantage of the latter is that accessory proteins may help amplify the number of cells activated during a microbial encounter. An example of this concept derives from studies of the mammalian receptors for extracellular bacterial lipopolysaccharides (LPS). TLR4 is classically viewed as the LPS receptor, as this protein is solely responsible for the inflammatory gene expression program that is induced by this microbial product (Meng et al., 2011; Poltorak et al., 1998). However, TLR4 forms few direct contacts with LPS (Park et al., 2009). High-affinity LPS interactions depend first on interactions with several LPS-binding proteins, including LPS binding protein (LBP), CD14 and MD-2 (Gioannini et al., 2004; Gioannini and Weiss, 2007). LBP binds directly to the bacterial cell wall (or micelles of free LPS), and this process facilitates the capture of a monomer of LPS by CD14. CD14 then transfers LPS to a heterodimer of MD-2 and TLR4 to promote inflammatory gene expression. It has been estimated that CD14 can extract 1000 molecules of LPS from a single bacterium, each of which can possibly activate a different macrophage to promote inflammation (Gioannini and Weiss, 2007). Thus, the process of using accessory proteins to facilitate PRR signaling may benefit the host in terms of the number of phagocytes participating in the inflammatory response. At present, it is unclear if similar mechanisms exist to promote an amplified response to other microbial encounters, but the studies of TLR4 signaling justify such an inquiry.
Regardless of the mechanism by which a PRR detects its microbial ligand, a general consequence of PRR activation is the initiation of an inflammatory response. PRRs that detect extracellular PAMPs are commonly activated by microbes, regardless of their virulence potential, and include the TLRs and Dectin families (Kagan, 2012). TLRs, in particular, survey the extracellular space and the contiguous luminal compartments of endosomal vesicles for PAMPs (Majer et al., 2016). These microbial products range from bacterial cell surface components (e.g. LPS and flagellin subunits) to viral nucleic acids (Pandey et al., 2014). Once these ligands are detected, signaling pathways are induced that culminate in the activation of inflammatory transcription factors of the AP-1 and NF-κB families, among others. These factors then induce the expression of a variety of inflammatory and immunomodulatory genes that shape the subsequent immune response.
Mechanistically, TLRs, Dectins (and most other PRRs) induce thematically similar signaling pathways to promote cell activation. Once these receptors detect their microbial ligands, they induce the formation of supramolecular organizing centers (SMOCs) that consist of several cytosolic signaling proteins (Kagan et al., 2014). SMOCs function as organelles that are assembled on-demand, or as needed, by the host cell, and serve as the principal source of inflammatory signals that emanate from active PRRs (Kagan et al., 2014). These structures are therefore the subcellular sites where cell fate decisions are induced after microbial detection by PRRs.
In the TLR pathway, the best studied SMOCs are the myddosome (Lin et al., 2010; Motshwene et al., 2009) and the putative triffosome (Gay et al., 2014) whose effector functions are to promote the activation of inflammatory and immunoregulatory transcription factors. Dectins induce the CARD9-MALT1-BCL10 complex that can be also be considered a SMOC, and links active receptors a similar set of inflammatory transcription factors (Gross et al., 2006). In several of these PRR-induced signaling events, the strong upregulation of inflammatory gene expression is coupled with a shift in the metabolic state of the cell from aerobic respiration to glycolysis (O’Neill and Pearce, 2016). This shift correlates with an increase in the translational and secretory capacities of the cells, thereby ensuring efficient synthesis and release of the induced cytokines, chemokines and other immune mediators that are important to achieve a general inflammatory activation state.
In addition to the local inflammatory responses induced by TLR and Dectin mediated responses, signaling of these receptors within phagocytes promotes T- and B-cell mediated immune responses that provide long-term host defense (Iwasaki and Medzhitov, 2010; LeibundGut-Landmann et al., 2007). These responses have been best-defined in dendritic cells, where PRR-dependent signaling events induce the expression of major histocompatibility complex (MHC) and costimulatory molecules (Schnare et al., 2001), the processing and loading of microbial antigens on MHC (Blander and Medzhitov, 2006), the transport of peptide-loaded MHC to the cell surface (Chow et al., 2002), and the trafficking of dendritic cells from the sites of microbial encounter to the draining lymph node where naïve T-cells reside (Lee and Iwasaki, 2007). Importantly, the PRR-dependent responses described above can be considered generic, in that any microbe should activate these responses.
Much of the discussion offered above is based on the idea that a simple “if-then” decision determines the activation states of mammalian cells that encounter microbes. In the case of microbial encounters where a single PRR family is activated (e.g. activation of TLRs, but not other PRRs by non-pathogenic E. coli), this if-then decision can accurately predict the activation state of the host cell. However, the cell fate decision becomes more complex during encounters with virulent pathogens, as under these conditions a wider range of PRRs may be engaged. As will be described below, different PRR families exhibit different effector functions, and some of these functions antagonize the activities of other PRRs. Understanding how competing signals are interpreted by the host to commit to a specific activation state is a challenge faced by the community today.
PAMPs from virulent microbes induce diverse activation states of infected cells
While the above-described sequence of events summarizes much of our knowledge of how the immune system links microbial detection to inflammatory and adaptive immune responses, there is another variable to consider. This variable is virulence. Not all microbes pose the same threat to the host, as some are avirulent and some are highly pathogenic. In principle, it would be beneficial to the host if systems would be in place to create a higher state of immune activation when encounters with virulent pathogens are detected (Blander and Sander, 2012; Vance et al., 2009). For this discussion, it is important to note that there are two types of pathogenic encounters. One type involves encounters with microbes that display complex activities to manipulate and replicate in a mammalian host (Cossart and Roy, 2010; Diamond and Pierson, 2015; Orzalli and Knipe, 2014). Encounters with these microbes are commonly considered virulent encounters, and a successful host response should eliminate the host-microbe interaction entirely. The second type of pathogenic encounter occurs with microbes that naturally inhabit our bodies (Chow and Mazmanian, 2010). Interactions with these microbes are normally innocuous, but can convert to a pathology under conditions of immune-dysregulation, stress, or a change in the composition of commensal microbiota (e.g. during antibiotic treatment). Unlike the sterilizing immunity that results from successful host responses to virulent pathogens, host responses to these opportunistic commensals may not eliminate the microbe. Rather, the host responses may be designed to restore a state of commensalism between the host and microbe. As the distinction between microbe and pathogen is blurred by potentially virulent commensals, we will focus our discussion on encounters with avirulent environmental microbes or virulent pathogens.
Work in recent years has provided support to the idea that a heightened state of immune activation can be achieved during virulent infections. The means by which infection (as opposed to microbial encounters) is detected by the host is not via TLRs and Dectins, as these receptors may detect any extracellular microbe. Rather, receptors of the NLR, RLR and ALR family are responsible. The central difference between these receptors and the TLRs and Dectins is that the former group surveys the intracellular (usually cytosolic) space for microbial products (Kagan and Barton, 2014).
The cytosol is a highly unusual environment, in that it is perhaps the only germ-free location on the planet. In contrast, all extracellular environments have the possibility of being occupied by one or more microbes. During many infectious encounters, the germ-free state of the cytosol is disrupted, as most virulent pathogens must access the cytosol in some way. Cytosolic access can come in the form of actual pathogen occupancy, such as is the case for Listeria monocytogenes, Shigella flexneri, and numerous viral pathogens. Cytosolic access can also occur indirectly, as is the case during infectious encounters with extracellular or vacuolar bacteria. Like their aforementioned cytosolic counterparts, these microbes must manipulate the host in order become pathogenic. This task is most commonly accomplished by the injection of virulence factors (classically called toxins or effectors) into the host cytosol (Cambronne and Roy, 2006; Galan, 2009). Thus, many pathogens, regardless of their infectious strategies, must access the cytosol in some way in order to manipulate the host. Because the uninfected cytosol is so definitively germ-free, cytosolic PRRs are in a unique position to operate as pathogen sensory proteins. An excellent example of this principle comes from studies of Listeria monocytogenes infections of macrophages (Leber et al., 2008). Wild type (virulent) bacteria enter cells via phagocytosis and then escape phagosomes to replicate in the cytosol (Tilney and Portnoy, 1989). These bacteria activate signaling events that are mediated by TLRs and NLRs (Leber et al., 2008). In contrast, avirulent bacteria that are unable to escape phagosomes only activate TLR-dependent gene expression (Leber et al., 2008). NLRs cannot be activated because these bacteria never enter the cytosol, where these receptors are located. As only cytosolic Listeria are virulent, the activation of NLRs upon entry into this intracellular location can be considered an indicator of a virulent encounter.
In the next sections, we describe the various cellular responses induced by cytosolic PRRs, with a focus on activities that appear to be incompatible with the effector functions of other PRRs that may be activated simultaneously during pathogenic encounters.
Infected cells must decide between immune activation and pyroptosis
If cytosolic PRRs serve to distinguish microbe from pathogen, detection of PAMPs by these receptors should elicit different cell activation states than PRRs that survey the extracellular space. Work over the last decade has supported this idea. For example, several members of the NLR and ALR families detect microbial products in the cytosol of infected cells (Hornung et al., 2009; Zhao et al., 2011). Once detected, these proteins promote a lytic form of cell death called pyroptosis, which prevents the host cell from supporting pathogen replication (Man and Kanneganti, 2015). It is likely that during natural infections of mammalian hosts, some cells will be exposed to PAMPs, but will not be infected. In these cells, TLRs and Dectins may be activated to promote general inflammatory and adaptive immune responses. Other cells in the same tissue, in contrast, will be infected by the pathogen. In these cells, TLRs/Dectins will be activated, along with cytosolic PRRs that promote pyroptosis.
Mechanistically, the best understood PRRs that induce pyroptosis are members of the NLR and ALR family. These proteins detect a variety of molecules, but for the purpose of this discussion, PAMP detection will be highlighted. The ALR AIM2, in particular, detects viral double stranded DNA (dsDNA) (Hornung et al., 2009), and the NLRs NAIP5 and NAIP6 detect subunits of bacterial flagellin (Rauch et al., 2016; Zhao et al., 2016). Once these receptors detect their microbial ligands, they induce the formation a SMOC called an inflammasome, whose principal effector functions are to cleave and activate cytosolic IL-1 family cytokines and the protein gasdermin D (GSDMD) (Kayagaki et al., 2015; Schroder and Tschopp, 2010; Shi et al., 2015). Once cleaved, GSDMD forms pores in the plasma membrane of the infected cell that results in lysis (pyroptosis), and the release of the aforementioned IL-1 family members that amplify the inflammatory response (Kayagaki et al., 2015; Shi et al., 2015). Pyroptosis will also amplify the antimicrobial response directly, as several intracellular pathogens can become trapped in pyroptotic corpses, rendering these microbes sensitive to capture by other phagocytes (Jorgensen et al., 2016). Thus, pyroptosis offers many benefits to the host in terms of amplifying the inflammatory and antimicrobial activities of the host.
While the induction of pyroptosis will amplify host defenses in an infected tissue, there are likely to be tradeoffs associated with this cell fate decision. The most obvious tradeoff is that the infected cell will die and therefore its ability to participate in subsequent immune responses will end. These immunomodulatory activities include, at the minimum, the ability of macrophages to promote additional antimicrobial activities in the infected tissue and the ability to resolve the local inflammatory response after the infection has been controlled (Okabe and Medzhitov, 2016). In addition, in the case of dendritic cells, the pyroptotic death of these cells will render them incapable of activating naïve antigen specific T-cells, as these events require long-distance cell migration to draining lymph nodes and many hours (or perhaps days) of residence in the lymph node (Mempel et al., 2004). Thus, it is possible that the decision of a cell to undergo pyroptosis has costs and benefits to the coordination of innate and adaptive immune responses.
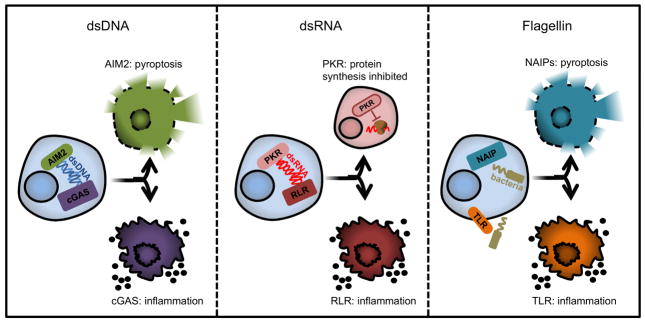
How a cell determines whether to initiate pyroptotic death pathways or to promote inflammation while retaining viability is unknown. This question becomes even more interesting when considering the fact that different PRRs recognize the same PAMP and induce these different responses (Figure 1). For example, whereas cytosolic AIM2 detects dsDNA and promotes pyroptosis (Hornung et al., 2009), the cytosolic PRR cGAS also binds dsDNA, but subsequently stimulates interferon (IFN) and chemokine production (Sun et al., 2013). Mechanistically, detection of dsDNA by cGAS activates its enzymatic activity to synthesize a cyclic dinucleotide called 2′-3′ cyclic GMP-AMP (cGAMP) (Sun et al., 2013; Wu et al., 2013). cGAMP then binds with high-affinity to the endoplasmic reticulum (ER) localized protein STING, which functions to activate various immunoregulatory transcription factors (Liu et al., 2015) that promote IFN and inflammatory chemokine expression.
Figure 1. One ligand drives opposing cell fate decisions by unknown mechanisms.
PRRs can bind the same ligand but induce antagonistic cell fates. The left panel depicts the decision between activation of inflammatory cytokines by cGAS or pyroptotic death by AIM2. The middle panel illustrates activation of the cell by RLR or translation shutdown and possibly apoptosis by PKR. The rightmost panel demonstrates the decision between pyroptotic death after flagellin sensing by NAIP5 and NAIP6 or TLR-mediated activation of inflammatory cytokines. The context and mechanism for how each ligand drives one decision over another is not known.
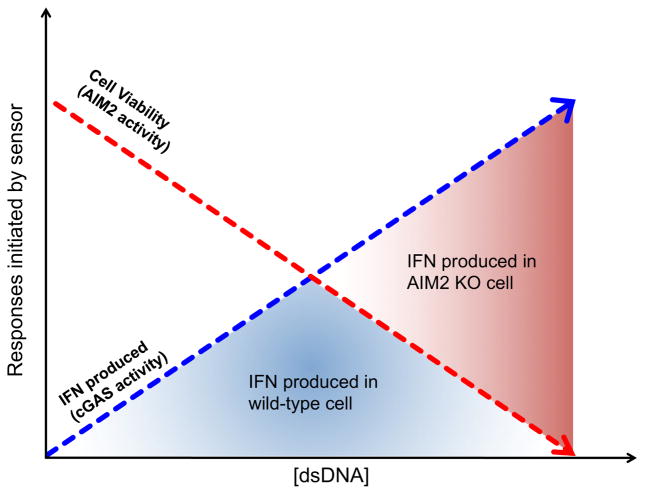
A common means of studying AIM2 and cGAS signaling events has been to use transfection of dsDNA as a model for viral entry into the cytosol. Performing these experiments in macrophages and dendritic cells revealed that increasing amounts of transfected dsDNA yields a proportional increase in IFN expression, before a maximum IFN response is reached (Corrales et al., 2016). Transfecting higher concentrations of DNA past that of the peak induces decreasing amounts of IFN expression. Interestingly, if this same experiment is conducted in the absence of AIM2 or downstream inflammasome components, IFN production continues to increase with increased concentration of transfected dsDNA (Corrales et al., 2016). One interpretation of this result is that in the absence of AIM2, cells do not enter pyroptosis after DNA transfection and are thus capable of producing more IFN. This idea suggests an antagonistic relationship between AIM2 and cGAS, despite both receptors binding the same PAMP and being important for host defense against the same viral pathogens. A possible explanation for the antagonistic relationship between AIM2 and cGAS is that cGAS may be activated at lower concentrations of cytosolic dsDNA than AIM2. Thus, during an initial viral infection, IFN expression may be the dominant host defense response. When a particular threshold of cytosolic dsDNA is passed, such as under conditions of uncontrolled viral replication, AIM2 is activated and pyroptosis ensues (Figure 2). One prediction of this model is that cGAS should have a higher affinity for dsDNA than AIM2, such that low concentrations of cytosolic dsDNA would only activate cGAS. However, in vitro studies estimate the affinity of AIM2 for dsDNA to be similar to that cGAS (Jin et al., 2012; Kranzusch et al., 2013). This finding suggests a more complex means of regulating the antagonistic activities of AIM2 and cGAS. Additional work is necessary to understand the basis for these important regulatory mechanisms.
Figure 2. AIM2 limits cGAS-activation of phagocytes.
cGAS-dependent IFN production increases with increasing concentration of transfected dsDNA (blue dashed line). Cell viability declines as AIM2-dependent death is activated with increasing concentration of dsDNA (red dashed curves). The area under the intersection of both curves (blue shaded area) defines the maximum amount of IFN produced in a cell with functional cGAS and AIM2 responses. Since AIM2 negatively regulates cGAS responses, more IFN is produced in the absence of AIM2 and is represented by the total area under the blue curve (blue shaded area + red shaded area). We hypothesize that the greatest competition for ligand between AIM2 and cGAS occurs at the intersection of these two curves, as at low concentrations of dsDNA, cell death is negligible and cGAS responses dominate and at the highest concentrations of dsDNA, AIM2-dependent death dominates and no IFN is made.
Cytosolic PAMPs can bind PRRs that induce transcription and reduce translation
Life verses death decisions are not the only antagonistic activities of the innate immune system. Even within populations of viable cells, different PRRs can induce signals that appear to counteract each other. An example of this idea derives from studies of PRRs that detect viral RNA. One set of PRRs that detect RNA viruses are the RLRs, which survey the cytosol for the presence of viral genomes, replication intermediates or transcripts (Kato et al., 2006). Upon detection of these RNAs, RLRs promote an antiviral cell activation state, which is characterized by the strong upregulation of inflammatory chemokines, cytokines and the expression of type I and III IFNs (Gitlin et al., 2006; Kato et al., 2006; Odendall et al., 2014). However, viral RNA can also detected by another cytosolic PRR called PKR (Levin and London, 1978), which acts to inhibit the protein synthesis machinery of the infected cell (Garcia et al., 2006). These virus-triggered activities are seemingly at odds with one another, as RLR-induced IFN transcripts need to be translated, but PKR downregulates protein synthesis.
Recent studies have highlighted that RLR and PKR activities may not be occurring independent of one another, but rather RLRs promote PKR activity. During viral infections, free viral RNA is rare, as these nucleic acids are often coated with viral proteins that have diverse functions (Garcia et al., 2006). One of the functions of viral nucleoproteins is to prevent access of the RNA to PKR. Interestingly, while PKR cannot access coated viral RNA, the RLRs can. RLRs have the ability to displace viral RNA binding proteins, thereby exposing these PAMPs to PKR and promoting translation shutdown (Yao et al., 2015). Thus, a PRR family that promotes gene expression (RLRs) is important to activate a PRR that limits protein synthesis (PKR).
While the above-described sequence of events provides an example of seemingly antagonistic innate immune responses, the receptors that regulate these responses are important for controlling viral infection. Thus, some means of coordinating RLR and PKR activity must exist. One possibility is that the ability of RLRs to displace viral proteins from RNA may only occur at high concentration of RLRs. Since RLRs are upregulated after IFN induction (Kang et al., 2004; Yoneyama et al., 2004), such a concentration may require an initial round of RLR-dependent IFN and ISG expression. This idea would ensure that PKR does not prevent the translation of IFN-regulated genes during the initial viral encounter, but would do so once an IFN response has been initiated. Consistent with this idea are observations that some viruses induce PKR activity late in infection (Mulvey et al., 2003; Stojdl et al., 2000). A notable aspect of this model would be that the concentration of RLRs in the cell would determine effector response induced during infection. Low RLR abundance in resting cells would be sufficient to induce IFN expression, whereas high RLR abundance could be necessary to activate PKR and shutdown translation. Another possibility to consider is that there is some means by which mRNAs encoding IFNs and other antiviral factors are insensitive to the translation-inhibitory actions of PKR. This scenario would allow RLR-induced transcripts to be translated, even under conditions of PKR activation. Support for this possibility comes from the findings that, during some viral infections, PKR is required for the polyadenylation and translation of mRNAs encoding various IFNs (Schulz et al., 2010). Kinetic analysis of multiple virus encounters in multiple cell types may be necessary before a clear understanding of how RLR and PKR activities can be provided.
Altering PRR-induced cell fate decisions by sensors of cellular stress
In the example described above, one PAMP, dsRNA, was capable of downregulating protein synthesis via PKR while also activating IFN expression via RLRs. Not all PAMPs are able to induce these dual activities. However, the stress of a replicating pathogen on the host can be sensed simultaneously with a PAMP, through the use of different receptors. Under these conditions, a cell may receive an activating signal from a PAMP and at the same time a signal to inhibit protein synthesis via the integrated stress response (ISR). This scenario provides two contexts for the detection of PAMPs: with ISR activation and without. Concurrent activation of PRRs and ISR may serve as a sensor of virulence of a pathogen and as such, downstream signaling from either pathway may be modified.
The ISR is regulated by a group of kinases consists of the aforementioned PKR, general control nonderepressible 2 (GCN2), Heme-Regulated Inhibitor (HRI), and PKR-like ER-localized Kinase (PERK). Respectively, these kinases sense the presence of dsRNA, amino acid starvation, heme deficiency, oxidative stress, and ER stress (Berlanga et al., 1999; Chen and London, 1995; Harding et al., 2003; Levin and London, 1978). PERK is also one of three ER-localized transmembrane sensors that make up the unfolded protein response (UPR) that are activated during ER stress (Walter and Ron, 2011). While each of these kinases detect a unique cellular stress, they phosphorylate the same target protein to induce the same cellular response—downregulation of protein synthesis and activating transcriptional programs to return to protein homeostasis (Walter and Ron, 2011).
When the ISR kinases are active, they phosphorylate a key translation initiation protein, eIF2α. eIF2α is part of the heterotrimeric complex, eIF2, that binds GTP and delivers the initiating methionyl tRNA to the assembling ribosome (Hinnebusch and Lorsch, 2012). However, when eIF2α is phosphorylated it binds eIF2B and competitively inhibits its GEF activity (Krishnamoorthy et al., 2001). GEF inhibition prevents the recycling of GDP to GTP, inhibiting ternary complex formation and ultimately turning off canonical translation initiation. When canonical eIF2-dependent translation is inhibited, selective translation of a group of mRNAs is upregulated (Morris and Geballe, 2000). These mRNAs include the transcription factors ATF4 and CHOP, which help relieve inhibitory stress responses, but long-term induction will lead to apoptosis (Walter and Ron, 2011).
Diverse immunological consequences of ISR activation exist. The most obvious consequence of ISR activation is that virus replication may be restricted, as all pathogens of this class require host protein synthesis to complete their lifecycle. However, the ISR may also influence the inflammatory and immuno-regulatory activities of dendritic cells and macrophages. Indeed, during microbial encounters by these cell types, a subset of inflammatory genes, including TNFα and IL-23 are synergistically upregulated by TLR signaling and the ISR-induced transcription factor CHOP (Goodall et al., 2010). Under these conditions, the signals that promote UPR pathway activation do not originate from a cellular stress per se, but rather via TLR signal transduction. This non-canonical mechanism of UPR pathway activation results in an atypical response, in that IRE1 activity, but not other ER stress sensors, is specifically activated. IRE1 then promotes the splicing of the xbp1 transcript, leading to the production of the transcription factor XBP1 (Martinon et al., 2010). Whereas XBP1 induces the expression of stress response genes during ER stress, TLR signaling prompts XBP1 to enhance transcription of CHOP, and inflammatory regulators such as IL-6, TNFα, and IFN.
While CHOP enhances inflammatory signaling, CHOP expression must be tightly regulated into order to prevent apoptosis. When TLR4 and the ISR are activated, downstream SMOCs serve a dual purpose. The Trif-containing triffosome activates inflammatory transcription factors to promote gene expression, and this complex renders cells resistant to phosphorylated eIF2α that occurs after PERK senses ER stress. A Trif-Src-PP2A signaling axis culminates in the dephosphorylation of eIF2Bε. This event stimulates GEF activity in the presence of the inhibitor, phosphorylated eIF2α (Woo et al., 2012). It is not known if phosphorylated eIF2α is the target of the Trif-induced GEF activity or if eIF2B targets a small subset of unphosphorylated eIF2α, but the activity is sufficient to resume host translation and prevent further translation of CHOP and thus CHOP-mediated apoptosis. Thus, cellular stress response systems that may be activated during infection may influence the cell fate decisions induced by PRRs within that same cell. These stress response systems can amplify the translation of some inflammatory mRNAs while preventing translation of viral transcripts, but long-term translational manipulation may ultimately prompt apoptotic cell death programs.
DAMPs from infected cells diversify the activation states of uninfected cells
Thus far, we have discussed several means by which infected cells can receive signals from diverse receptors to alter cell fate decisions in the innate immune system. In this section, we discuss recent work highlighting how uninfected cells can also achieve different activation states, depending on the PAMPs and DAMPs that they encounter. Like the aforementioned examples, this diversity of activation states of uninfected cells comes with immunological costs and benefits.
During virulent infections, pyroptotic programs not only limit intracellular pathogen replication by removing the infected living cell, these events also result in the release of a variety of DAMPs (de Vasconcelos et al., 2016). DAMPs are a heterogenous set of molecules, which include proteins (e.g. interleukin-1 (IL-1) family of cytokines), nucleic acids (DNA or RNA) and lipids (oxidized phosphorylcholines). Some DAMPs exhibit direct inflammatory activity, such as the IL-1 family of cytokines and nucleic acids (Netea et al., 2015). These molecules bind specific receptors whose signaling pathways promote an inflammatory response. Other DAMPs, in contrast, exhibit indirect inflammatory activity, such as the HMGB family of proteins (Pawaria et al., 2015). This latter set of molecules captures extracellular nucleic acids and facilitate their delivery to DNA or RNA sensing TLRs, thus helping to amplify the inflammatory response.
In recent years, there has become an increasing appreciation of the importance of PRR interactions with DAMPs. Like their microbial counterparts, DAMPs can engage PRRs directly or indirectly. An example of indirect PRR activation derives from studies of the inflammatory activities of extracellular ATP. High concentrations of extracellular ATP are rare under homeostatic conditions, as this molecule is present in the intracellular environment of living cells. During infections, the local stresses on the host can lead to regulated or spontaneous cellular damage, resulting in the release of cytosolic ATP. Under these conditions, released ATP can gate the P2X7 receptor and promote an ionic imbalance that results in potassium efflux from the cell (Munoz-Planillo et al., 2013). This efflux is coupled to a rush of calcium into the cell (Murakami et al., 2012). Through mechanisms that are unclear, this ionic imbalance promotes the activation of NLRP3, which induces the assembly of an inflammasome consisting of the adaptor ASC and the protease caspase-1 (Elliott and Sutterwala, 2015). Caspase-1 is activated within the NLRP3 inflammasome and consequently cleaves pro-IL-1 family members and GSDMD (Kayagaki et al., 2015; Shi et al., 2015). The N-terminal fragment then forms pores in the plasma membrane that leads to pyroptosis and the passive release of bioactive IL-1 and other cytosolic DAMPs (Ding et al., 2016; Liu et al., 2016). Among the DAMPs released is ATP, which may then act on another cell to trigger a pyroptotic feed-forward loop, resulting in a high degree of inflammation. As ATP binding to the P2X7 receptor leads to NLRP3 activation (Mariathasan et al., 2006), we consider this mechanism of PRR activation to be indirect. Moreover, because infected cells can be the initial source of the ATP released into the extracellular space, uninfected neighboring cells can be induced to undergo pyroptosis after they are exposed to ATP. Importantly, the pyroptosis-inducing activities of ATP is limited to cells that have been previously exposed to TLR ligands (Sutterwala et al., 2014). One possible explanation for the need to “prime” cells with TLR ligands in order for ATP to induce inflammasome assembly is based on the low basal expression of NLRP3 in resting cells. LPS treatment, for example, increases NLRP3 expression (and the expression of IL-1 family members), which may facilitate inflammasome assembly.
An example of direct engagement of PRRs by DAMPs comes from recent studies of a set of inflammatory lipids called oxPAPC (Zanoni et al., 2016). oxPAPC is a heterogenous population of lipids that are derived from non-oxidized phosphorylcholines that are present in the plasma membrane of all mammalian cells (Berliner et al., 2009). At sites of tissue injury, the reactive oxygen species released from dying cells results in the spontaneous (non-enzymatic) oxidation of these lipids to generate oxPAPC (Fu and Birukov, 2009; Imai et al., 2008). oxPAPC, like extracellular ATP, does not exhibit intrinsic inflammatory activities. In fact, oxPAPC was initially characterized as an anti-inflammatory factor because cells and mice pretreated with oxPAPC are poorly responsive to subsequent exposures to TLR4 ligands (Bochkov et al., 2002). Recent work has also highlighted the inflammatory activities of oxPAPC. When dendritic cells have been primed with TLR ligands, oxPAPC promotes the assembly of the NLRP3 inflammasome (Zanoni et al., 2016). The inflammasome assembled upon oxPAPC exposure differs from that assembled by ATP in several ways. First, the former is regulated by the cytosolic PRR caspase-11, whereas ATP-mediated NLRP3 inflammasome assembly does not require caspase-11 (Kayagaki et al., 2011). oxPAPC and ATP also differ in their dependence on potassium efflux for NLRP3 inflammasome assembly (Zanoni et al., 2016). Finally, and most notably, the inflammasome assembled by oxPAPC induces the processing and release of IL-1 family members from living cells, whereas ATP induces pyroptosis and IL-1 release (Zanoni et al., 2016). The ability of oxPAPC to add IL-1 family cytokines to the inflammatory repertoire normally released upon TLR ligand stimulation suggests that a heightened state of cell activation has been achieved. Dendritic cells exposed to TLR ligands and oxPAPC are therefore considered “hyperactive”, and are capable of eliciting a heightened degree of antigen-specific T-cell activation than dendritic cells treated solely with TLR ligands (Zanoni et al., 2016).
The ability of oxPAPC to promote dendritic cell hyperactivation depends on caspase-11, and oxPAPC binds directly to this PRR, thereby promoting its oligomerization (Zanoni et al., 2016). Interestingly, LPS also interacts with and oligomerizes caspase-11, yet this PRR interacts with PAMPs and DAMPs through different domains. The N-terminal CARD domain binds directly to LPS, whereas the C-terminal catalytic domain binds oxPAPC. oxPAPC also has the ability to form a complex with caspase-1, whereas LPS does not. Much remains to be learned about the mechanisms by which caspase-11 binding by LPS and oxPAPC influence inflammasome activities, but it is clear that the DAMPs oxPAPC and ATP differ in their abilities to influence cell fate decisions. ATP activates NLRP3 inflammasome assembly indirectly, by inducing potassium efflux from TLR ligand primed cells. These events lead to pyroptosis, IL-1 family cytokine release and local inflammation. oxPAPC, in contrast, activates NLRP3 inflammasome assembly directly, through interactions with the PRR caspase-11 (and perhaps caspase-1). These events do not result in pyroptosis, but rather dendritic cell hyperactivation, which leads to a heightened adaptive immune response.
Interestingly, dendritic cells are not the only cells that can achieve a hyperactive state, and PAMPs can also induce this cell fate choice. When cell wall peptidoglycans from gram-positive bacteria are degraded, the N-acetylglucosamine degradation product can somehow reach the cytosol of macrophages (Wolf et al., 2016). This bacterial product binds the glycolytic enzyme hexokinase, which results in its release from mitochondria (Wolf et al., 2016). Cytosolic hexokinase then leads to NLRP3 inflammasome assembly and the release of IL-1 from living macrophages. Similarly, in human and pig monocytes (but not murine cells), LPS has the ability to promote IL-1 release in the absence of pyroptosis (Gaidt et al., 2016). This process depends on TLR4 and a genetically-defined pathway that includes the adaptor protein Trif and caspase-8. CD14 has also been implicated in LPS-induced IL-1 release from human monocytes, as has caspase-5 (one of the human orthologs of murine caspase-11) (Vigano et al., 2015). While oxPAPC, N-acetylglucosamine and LPS are chemically distinct entities that are derived from different organisms, they all possess the ability to promote NLRP3 inflammasome assembly independent of potassium efflux. These unusual inflammasomes share the ability to promote IL-1 release from living uninfected cells, and can therefore all be considered capable of immune cell hyperactivation.
The findings that select DAMPs and PAMPs can hyperactivate phagocytes diversifies the activities of inflammasomes, as these SMOCs were previously thought to function to induce pyroptosis of cells that were infected directly. In an infected tissue, uninfected cells may also be an important source of inflammasome activities, as these cells should be in a position to detect PAMPs and DAMPs released from infected and/or damaged cells.
A third class of DAMPs also exists, which include molecules that may best be defined as accidental activators of PRRs. Unlike ATP and oxPAPC, which activate PRRs by mechanisms distinct from PAMPs, this third class of DAMPs engages PRRs by a mechanism that appears indistinguishable from microbial products. These molecules include self-nucleic acids that are released from dead or dying cells. In particular, DNA containing unmethylated CpG motifs, dsDNA and dsRNA are notable, as these features are identical to those detected within microbial nucleic acids. As such, TLR9 (CpG DNA), cGAS and AIM2 (dsDNA) and the RLRs (dsRNA) can detect self or microbial nucleic acids as DAMP or PAMP, and induce identical downstream effector responses. This finding is particularly notable when considering the central role of PRRs in self-non-self discrimination, as nucleic acids blur the line between the two. Consequently, nucleic acid sensing PRRs have been most commonly linked to autoimmune diseases (Gray et al., 2015; Pisitkun et al., 2006). This finding suggests that the problems associated with self-non-self discrimination by the innate immune system is not simply an academic curiosity, but one with clinical implications. Because of the similarity between nucleic acid DAMPs and their PAMP counterparts, it is possible that this class of DAMPs influences cell fate decisions by accident—meaning that the host did not evolve to recognize self-nucleic acids.
Perspectives
In this Review, we discussed several means by which host cell fate decisions are made during microbial encounters. Our major goal was to highlight the costs and benefits of any individual cell fate choice to the host. We now know of multiple examples of PRRs that recognize the same PAMP (or DAMP), and induce competing signaling pathways and responses. Understanding the high degree of dynamic regulation that likely underlies the activities of these seemingly competitive receptors represents a new challenge to the community.
Many of the studies that served as the foundation for this discussion have taken experimental approaches that focus on one ligand activating one pathway at a time. This approach has been hugely successful in defining the general principles of a pathway and how it may operate. However, it is also important to consider the complex environments that develop during natural infections. Host damage and the amount and variety of PAMPs will change dependent on many variables including the virulence of a pathogen and the stage of replication. For PRRs that detect the same ligand, we do not know how the timing of activation of one receptor compares with another, and how downstream cellular responses influence signaling from either PRR. When multiple PAMPs are present at the same time, is the activation state additively heightened or does one signal dominate other the other? It is also possible that the process of pathogen replication within a mammalian host will induce some amount of stress, damage or metabolic load. Thus, additional attention to how stress response pathways are activated and contribute to overall PRR-dependent cell fate decisions is warranted. In considering the ideas and questions presented in this Review, it is worth noting that our focus has been on gaining a better understanding of interactions between individual microbes and individual host cells. This focus is critical, as the cell fate decisions that occur upon these initial encounters will influence many downstream physiological responses. In animal models of infection, and in a clinical setting, these initial cell fate decisions may therefore have profound consequences, and efforts to gain a better understanding of these events will likely continue to keep the field of host-microbe interactions fresh and exciting for years to come.
Acknowledgments
We would like to thank members of the Kagan lab for helpful discussions. This work was supported by NIH grants AI093589, AI116550 and P30 DK34854 to J.C.K. and an unrestricted gift from Mead Johnson & Company. J.C.K. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. K.M.F. is supported by the Harvard Herchel Smith Graduate Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ades SE. Regulation by destruction: design of the sigmaE envelope stress response. Curr Opin Microbiol. 2008;11:535–540. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- Berliner JA, Leitinger N, Tsimikas S. The role of oxidized phospholipids in atherosclerosis. J Lipid Res. 2009;50(Suppl):S207–212. doi: 10.1194/jlr.R800074-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander JM, Medzhitov R. Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature. 2006;440:808–812. doi: 10.1038/nature04596. [DOI] [PubMed] [Google Scholar]
- Blander JM, Sander LE. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol. 2012;12:215–225. doi: 10.1038/nri3167. [DOI] [PubMed] [Google Scholar]
- Bochkov VN, Kadl A, Huber J, Gruber F, Binder BR, Leitinger N. Protective role of phospholipid oxidation products in endotoxin-induced tissue damage. Nature. 2002;419:77–81. doi: 10.1038/nature01023. [DOI] [PubMed] [Google Scholar]
- Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne ED, Roy CR. Recognition and delivery of effector proteins into eukaryotic cells by bacterial secretion systems. Traffic. 2006;7:929–939. doi: 10.1111/j.1600-0854.2006.00446.x. [DOI] [PubMed] [Google Scholar]
- Chen JJ, London IM. Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem Sci. 1995;20:105–108. doi: 10.1016/s0968-0004(00)88975-6. [DOI] [PubMed] [Google Scholar]
- Chow A, Toomre D, Garrett W, Mellman I. Dendritic cell maturation triggers retrograde MHC class II transport from lysosomes to the plasma membrane. Nature. 2002;418:988–994. doi: 10.1038/nature01006. [DOI] [PubMed] [Google Scholar]
- Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuenchor W, Jin T, Ravilious G, Xiao TS. Structures of pattern recognition receptors reveal molecular mechanisms of autoinhibition, ligand recognition and oligomerization. Curr Opin Immunol. 2014;26:14–20. doi: 10.1016/j.coi.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Woo SR, Williams JB, McWhirter SM, Dubensky TW, Jr, Gajewski TF. Antagonism of the STING Pathway via Activation of the AIM2 Inflammasome by Intracellular DNA. J Immunol. 2016;196:3191–3198. doi: 10.4049/jimmunol.1502538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P, Roy CR. Manipulation of host membrane machinery by bacterial pathogens. Curr Opin Cell Biol. 2010;22:547–554. doi: 10.1016/j.ceb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- de Vasconcelos NM, Van Opdenbosch N, Lamkanfi M. Inflammasomes as polyvalent cell death platforms. Cell Mol Life Sci. 2016;73:2335–2347. doi: 10.1007/s00018-016-2204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Pierson TC. Molecular Insight into Dengue Virus Pathogenesis and Its Implications for Disease Control. Cell. 2015;162:488–492. doi: 10.1016/j.cell.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- Elliott EI, Sutterwala FS. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol Rev. 2015;265:35–52. doi: 10.1111/imr.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber F, Tran L, Byndloss MX, Lopez CA, Velazquez EM, Kerrinnes T, Nuccio SP, Wangdi T, Fiehn O, Tsolis RM, et al. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature. 2016;534:697–699. doi: 10.1038/nature18597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu P, Birukov KG. Oxidized phospholipids in control of inflammation and endothelial barrier. Transl Res. 2009;153:166–176. doi: 10.1016/j.trsl.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt MM, Ebert TS, Chauhan D, Schmidt T, Schmid-Burgk JL, Rapino F, Robertson AA, Cooper MA, Graf T, Hornung V. Human Monocytes Engage an Alternative Inflammasome Pathway. Immunity. 2016;44:833–846. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Galan JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay NJ, Symmons MF, Gangloff M, Bryant CE. Assembly and localization of Toll-like receptor signalling complexes. Nat Rev Immunol. 2014;14:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- Gioannini TL, Teghanemt A, Zhang D, Coussens NP, Dockstader W, Ramaswamy S, Weiss JP. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc Natl Acad Sci U S A. 2004;101:4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioannini TL, Weiss JP. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol Res. 2007;39:249–260. doi: 10.1007/s12026-007-0069-0. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall JC, Wu C, Zhang Y, McNeill L, Ellis L, Saudek V, Gaston JS. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proc Natl Acad Sci U S A. 2010;107:17698–17703. doi: 10.1073/pnas.1011736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutieres Syndrome. J Immunol. 2015;195:1939–1943. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, Peschel C, Forster I, Ruland J. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YH, Wang H, et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, Zhang Y, Krantz BA, Miao EA. Pyroptosis triggers pore-induced intracellular traps (PITs) that capture bacteria and lead to their clearance by efferocytosis. J Exp Med. 2016;213:2113–2128. doi: 10.1084/jem.20151613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC. Signaling organelles of the innate immune system. Cell. 2012;151:1168–1178. doi: 10.1016/j.cell.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Barton GM. Emerging principles governing signal transduction by pattern-recognition receptors. Cold Spring Harb Perspect Biol. 2014;7:a016253. doi: 10.1101/cshperspect.a016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Magupalli VG, Wu H. SMOCs: supramolecular organizing centres that control innate immunity. Nat Rev Immunol. 2014;14:821–826. doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DC, Gopalkrishnan RV, Lin L, Randolph A, Valerie K, Pestka S, Fisher PB. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene. 2004;23:1789–1800. doi: 10.1038/sj.onc.1207300. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, Hinnebusch AG. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol Cell Biol. 2001;21:5018–5030. doi: 10.1128/MCB.21.15.5018-5030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, Portnoy DA. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Iwasaki A. Innate control of adaptive immunity: dendritic cells and beyond. Semin Immunol. 2007;19:48–55. doi: 10.1016/j.smim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, Schweighoffer E, Tybulewicz V, Brown GD, Ruland J, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- Levin D, London IM. Regulation of protein synthesis: activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proc Natl Acad Sci U S A. 1978;75:1121–1125. doi: 10.1073/pnas.75.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer O, Liu B, Barton GM. Nucleic acid-sensing TLRs: trafficking and regulation. Curr Opin Immunol. 2016;44:26–33. doi: 10.1016/j.coi.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30:766–775. doi: 10.1016/j.immuni.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427:154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- Meng J, Gong M, Bjorkbacka H, Golenbock DT. Genome-wide expression profiling and mutagenesis studies reveal that lipopolysaccharide responsiveness appears to be absolutely dependent on TLR4 and MD-2 expression and is dependent upon intermolecular ionic interactions. J Immunol. 2011;187:3683–3693. doi: 10.4049/jimmunol.1101397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, Sandercock AM, Robinson CV, Latz E, Gay NJ. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, Poppers J, Sternberg D, Mohr I. Regulation of eIF2alpha phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J Virol. 2003;77:10917–10928. doi: 10.1128/JVI.77.20.10917-10928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM, Nunez G. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Ockinger J, Yu J, Byles V, McColl A, Hofer AM, Horng T. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc Natl Acad Sci U S A. 2012;109:11282–11287. doi: 10.1073/pnas.1117765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, van de Veerdonk FL, van der Meer JW, Dinarello CA, Joosten LA. Inflammasome-independent regulation of IL-1-family cytokines. Annu Rev Immunol. 2015;33:49–77. doi: 10.1146/annurev-immunol-032414-112306. [DOI] [PubMed] [Google Scholar]
- Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, Kagan JC. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Medzhitov R. Tissue biology perspective on macrophages. Nat Immunol. 2016;17:9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- Orzalli MH, Knipe DM. Cellular sensing of viral DNA and viral evasion mechanisms. Annu Rev Microbiol. 2014;68:477–492. doi: 10.1146/annurev-micro-091313-103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Kawai T, Akira S. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb Perspect Biol. 2014;7:a016246. doi: 10.1101/cshperspect.a016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- Pawaria S, Moody K, Busto P, Nundel K, Choi CH, Ghayur T, Marshak-Rothstein A. Cutting Edge: DNase II deficiency prevents activation of autoreactive B cells by double-stranded DNA endogenous ligands. J Immunol. 2015;194:1403–1407. doi: 10.4049/jimmunol.1402893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Rauch I, Tenthorey JL, Nichols RD, Al Moussawi K, Kang JJ, Kang C, Kazmierczak BI, Vance RE. NAIP proteins are required for cytosolic detection of specific bacterial ligands in vivo. J Exp Med. 2016;213:657–665. doi: 10.1084/jem.20151809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare M, Barton GM, Holt AC, Takeda K, Akira S, Medzhitov R. Toll-like receptors control activation of adaptive immune responses. Nat Immunol. 2001;2:947–950. doi: 10.1038/ni712. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Schulz O, Pichlmair A, Rehwinkel J, Rogers NC, Scheuner D, Kato H, Takeuchi O, Akira S, Kaufman RJ, Reis e Sousa C. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe. 2010;7:354–361. doi: 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Stojdl DF, Abraham N, Knowles S, Marius R, Brasey A, Lichty BD, Brown EG, Sonenberg N, Bell JC. The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J Virol. 2000;74:9580–9585. doi: 10.1128/jvi.74.20.9580-9585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterwala FS, Haasken S, Cassel SL. Mechanism of NLRP3 inflammasome activation. Ann N Y Acad Sci. 2014;1319:82–95. doi: 10.1111/nyas.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigano E, Diamond CE, Spreafico R, Balachander A, Sobota RM, Mortellaro A. Human caspase-4 and caspase-5 regulate the one-step non-canonical inflammasome activation in monocytes. Nat Commun. 2015;6:8761. doi: 10.1038/ncomms9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AJ, Reyes CN, Liang W, Becker C, Shimada K, Wheeler ML, Cho HC, Popescu NI, Coggeshall KM, Arditi M, et al. Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan. Cell. 2016;166:624–636. doi: 10.1016/j.cell.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CW, Kutzler L, Kimball SR, Tabas I. Toll-like receptor activation suppresses ER stress factor CHOP and translation inhibition through activation of eIF2B. Nat Cell Biol. 2012;14:192–200. doi: 10.1038/ncb2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Dittmann M, Peisley A, Hoffmann HH, Gilmore RH, Schmidt T, Schmid-Burgk JL, Hornung V, Rice CM, Hur S. ATP-dependent effector-like functions of RIG-I-like receptors. Mol Cell. 2015;58:541–548. doi: 10.1016/j.molcel.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zanoni I, Tan Y, Di Gioia M, Broggi A, Ruan J, Shi J, Donado CA, Shao F, Wu H, Springstead JR, et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science. 2016;352:1232–1236. doi: 10.1126/science.aaf3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Shi J, Shi X, Wang Y, Wang F, Shao F. Genetic functions of the NAIP family of inflammasome receptors for bacterial ligands in mice. J Exp Med. 2016;213:647–656. doi: 10.1084/jem.20160006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]