Abstract
To start DNA replication, the Origin Recognition Complex (ORC) and Cdc6 load a Mcm2-7 double hexamer onto DNA. Without ATP hydrolysis, ORC-Cdc6 recruits one Cdt1-bound Mcm2-7 hexamer, forming an ORC-Cdc6-Cdt1-Mcm2-7 (OCCM) helicase loading intermediate. Here we report a 3.9Å structure of the OCCM on DNA. Flexible Mcm2-7 winged-helix domains (WHD) engage ORC-Cdc6. A three-domain Cdt1 configuration embraces Mcm2, Mcm4, and Mcm6, nearly half of the hexamer. The Cdt1 C-terminal domain extends to the Mcm6 WHD, which binds Orc4 WHD. DNA passes through the ORC-Cdc6 and Mcm2-7 rings. Origin DNA interaction is mediated by an α-helix in Orc4 and positively charged loops in Orc2 and Cdc6. The Mcm2-7 C-tier AAA+ ring is topologically closed by a Mcm5 loop that embraces Mcm2, but the N-tier ring Mcm2-Mcm5 interface remains open. This structure suggests loading mechanics of the first Cdt1-bound Mcm2-7 hexamer by ORC-Cdc6.
INTRODUCTION
The S. cerevisiae Origin Recognition Complex (ORC) is an ATPase complex composed of Orc1-61–3. The composition and architecture of ORC is conserved in all eukaryotes4. Low resolution electron microscopy (EM) showed that the six subunits are arranged into a crescent in the order of Orc1-Orc4-Orc5-Orc3-Orc2, with Orc6 binding to Orc2/Orc35,6. This architecture is confirmed by a recent crystal structure of an inactive Drosophila ORC (DmORC)7. This DmORC core is a notched two-tiered ring composed of the N-tier ring of five AAA+ domains and the C-tier ring of five winged helix (WH) domains of Orc1-5. Because the DmOrc1 AAA+ domain blocks the putative central DNA binding channel in the auto-inhibited conformation7, the configuration of an active ORC has been unknown. The budding yeast ORC binds the replication origins throughout the cell division cycle1, but they are “licensed” during the G1 phase8. An early step is the binding of initiation factor Cdc6 to DNA-bound ORC to form the ORC-Cdc6-DNA complex9. EM has shown that Cdc6 closes a gap in the crescent-shaped ORC to form a ring9,10 and apparently activates a molecular switch in ORC, converting it from an origin DNA binder to an active Mcm2-7 loader6,9. However, the physical nature of the molecular switch is currently unknown due to the lack of a high-resolution structure of ORC-Cdc6 on DNA.
The next steps involve the sequential recruitment onto the origin DNA of two Cdt1-bound hexamers of Mcm2-7 by ORC-Cdc6 to form a Mcm2-7 double-hexamer (D-H) that forms part of the pre-Replicative Complex (pre-RC)8,11. In vitro reactions using purified components have demonstrated that a high salt-stable Mcm2-7 D-H is loaded on DNA in an ATP dependent manner12,13. Each hexamer within the D-H is assembled in a way that their respective Mcm3 and Mcm6 subunits face each other, as revealed by EM of Maltose Binding Protein (MBP) tagged D-H as well as a 3.8 Å resolution cryo-EM structure14,15. The Mcm2-7 double-hexamer has a central channel that is wide enough for passage of double stranded DNA, in agreement with biochemical findings12,13. Because the two Mcm2-7 hexamers are twisted relative to each other, it was speculated that within the interface between the two hexamers an inflection point of the DNA path is created, possibly promoting melting of the double stranded DNA when the helicase becomes activated in S phase14. At the G1-S transition, the inactive D-H is converted into an active replicative helicase that consists of a Mcm2-7 hexamer bound to Cdc45 and the four-subunit GINS complex, called the CMG16,17. Disruption of the Mcm2-7 D-H and assembly of CMG require activation by the Dbf4-Cdc7 protein kinase (DDK) and Cyclin-Dependent Kinase (CDK; Clb5-Cdc28), which phosphorylate some of the pre-RC components, such as Mcm2-7 subunits and accessory loading proteins Sld2 and Sld31,8,18–26. Subsequently, primase and DNA polymerases load, along with many other replication factors, to form the replisome that executes DNA synthesis27,28.
The two Mcm2-7 hexamers are loaded on DNA sequentially15,29,30. In the presence of ATPγS, ORC-Cdc6 loads the first Mcm2-7 hexamer on DNA, forming an ORC-Cdc6-Cdt1-Mcm2-7 intermediate (OCCM)15,31. Then ATP hydrolysis is triggered and Cdc6 and Cdt1 are released30,32–34. Single-molecule analysis suggests that a second Cdc6 protein is recruited to ORC, which functions to load a second Cdt1-bound Mcm2-7 to form an ORC-Cdc6-Mcm2-7-Mcm2-7 (OCMM) complex prior to Mcm2-7 D-H formation15,30,35. In vivo evidence suggests that ATP hydrolysis by Cdc6 causes the separation of the D-H from ORC-Cdc636. In this work, we describe a 3.9 Å resolution cryo-EM structure of the 1.1-MDa 14-protein OCCM complex on DNA and interactions between individual proteins using mass spectrometry. The structure revealed how ORC-Cdc6 recognizes origin DNA and how this complex recruits the first Cdt1-bound Mcm2-7 hexamer, thereby illuminating a crucial step in eukaryotic DNA replication initiation.
RESULTS
Overall structure of the OCCM-DNA complex
We prepared the OCCM complexes in the presence of ATPγS from purified proteins on a replication origin containing plasmid attached to magnetic beads. Upon DNaseI treatment the OCCM samples were released from the beads and directly processed for cryo-EM grid preparation. We derived a 3.9 Å resolution cryo-EM 3D map of the OCCM from 304,288 particles that were selected from 7500 raw electron micrographs and nearly 1,000,000 raw particles that were recorded on a K2 camera in a Titan Krios microscope operated at a high tension of 300 kV (Fig. 1a–c, Supplementary Figs. 1–3, Table 1, Supplementary Video 1, Online methods). The 3D map had well-defined densities for the double strand DNA and 13 subunits of the 14-protein complex (Fig. 1d and e). The only protein not well resolved was Orc6; its density was visible only at a lower display threshold. Nevertheless, a conserved C-terminal α-helix of Orc6 was resolved, which is important, as it is mutated in Meier Gorlin syndrome37,38. To better understand the architecture of the complex, and particularly of the flexible sections involving Orc6, cross-linking/mass spectrometry (CLMS) analysis was performed, which confirmed the general architecture and also identified several interactions between Orc6 and Orc2/Mcm2 (Fig. 2a–c, Supplementary Fig. 4, Supplementary Datasets 1–2). An atomic model of the OCCM was built into the EM densities guided by the published structures of the Mcm2-7 hexamer14, the Drosophila ORC7, an archaeal Cdc6 homolog39, a homolog of the N-terminal domain (NTD) of Cdt1, the middle helical domain (MHD) and the C-terminal domain (CTD) of the human Cdt140–42. The double stranded DNA was manually built into the EM density (Supplementary Fig. 5). Electron densities for eight nucleotides were observed at the interface between Mcm2-Mcm6, Mcm6-Mcm4, Mcm4-Mcm7, Mcm7-Mcm3, Cdc6-Orc1, Orc1-Orc4, Orc4-Orc5, and Orc5-Orc3 (Supplementary Fig. 6). No nucleotide densities were resolved at interfaces between Mcm3-Mcm5 and Mcm5-Mcm2 due to the reduced resolution and increased flexibility, as they were either at or near the DNA loading gate.
Figure 1. Cryo-EM and overall structure of the S. cerevisiae OCCM complex.
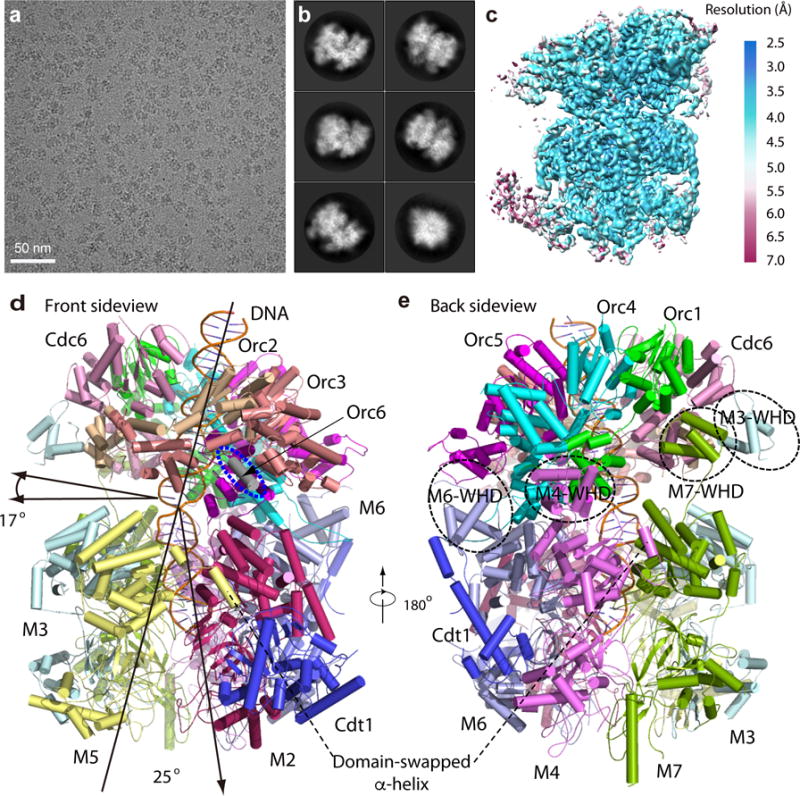
(a) A typical motion-corrected raw image of frozen OCCM particles recorded on a direct detector. (b) Selected six 2D averages representing the particles in different views. (c) 3D cryo-EM map of OCCM color coded by local resolution. Overall resolution is 3.9 Å. (d) Cartoon view of the atomic model of OCCM as viewed from Front side. The two black arrows in left indicates that the ORC-Cdc6 ring lays on the MCM ring tilted by an angle of ~17°. The two black arrows in middle shows the DNA in central channel is bent by ~25°. The blue oval marks the short helix of Orc6. (e) Cartoon view of the OCCM model as viewed from the backside. The black circles mark the WHDs of Mcm3, Mcm4, Mcm6 and Mcm7, respectively.
Table 1.
Cryo-EM data collection and refinement statistics
| Data Collection | |
|---|---|
| EM equipment | FEI Titan Krios |
| Voltage (kV) | 300 |
| Detector | Gatan K2 |
| Pixel size (Å) | 1.01 |
| Electron dose (e−/Å2) | 50 |
| Defocus range (μm) | 1.5~3.5 |
| Reconstruction | |
| Software | RELION 1.4 |
| Number of used Particles | 304,288 |
| Resolution (Å) | 3.9 |
| Map sharpening B-factor (Å2) | 123 |
| Model composition | |
| Peptide chains | 14 |
| Protein residues | 6907 |
| Nucleotides | 78 |
| R.m.s deviations | |
| Bonds length (A°) | 0.007 |
| Bonds Angle (°) | 1.292 |
| Ramachandran plot | |
| Preferred (%) | 89.00 |
| Allowed (%) | 9.52 |
| Outlier (%) | 1.48 |
| Validation | |
| Molprobity score | 2.56 (98%) |
| Good rotamer (%) | 93.58 |
| Clashscore, all atoms | 27.74 (86%) |
Figure 2. Cross-linking/mass spectrometry analysis of S. cerevisiae OCCM complex.
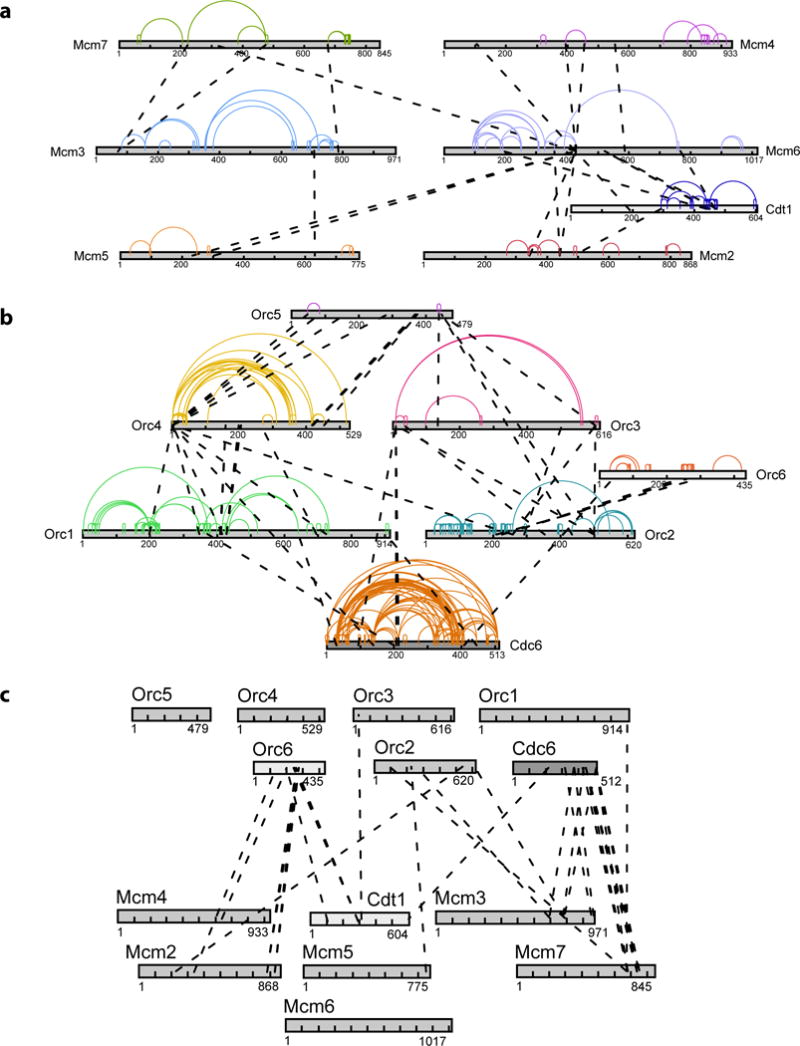
(a) Linkage map showing the observed cross-linked residue pairs within the MCM2-7/Cdt1 complex. Intra-molecular cross-links are color coded, while inter-molecular cross-links are shown in black. (b) Linkage map showing the observed cross-linked residue pairs within the ORC/Cdc6 complex. Intra-molecular cross-links are color coded, while inter-molecular cross-links are shown in black. (c) Linkage map showing the observed cross-linked residue pairs between ORC/Cdc6 complex and Mcm2-7/Cdt1 complex. Orc6, which was only partially resolved by cryo-EM, is in close proximity to Mcm2 and Cdt1. The Winged Helix Domain of Mcm5, that was only partially resolved by cryo-EM, is in close proximity to the N-terminal region of Orc2.
In the top ORC-Cdc6 tier of the OCCM, Orc1-5 and Cdc6 formed a six-membered ring structure with Cdc6 bridging the gap between Orc1 and Orc2, in agreement with a previous lower resolution EM study (Fig. 3)5. Interestingly, four WH domains of Mcm3, Mcm4, Mcm6, and Mcm7 spiraled upwards from the Mcm2-7 hexamer and engaged the ORC-Cdc6 ring, each interacting with ORC-Cdc6 subunits (Fig. 1d and e, Supplementary Fig. 7), while an interaction between the Mcm5 WH domain and Orc2 was seen by CLMS (Fig. 2c). In the bottom Cdt1-Mcm2-7 tier, the six Mcm subunits formed a ring structure in the order of 2-6-4-7-3-5, consistent with previous studies14,15,22,43,44. At the interface between the Mcm2 AAA+ domain and Mcm5 AAA+ domain, the domain swapped α-helix of Mcm5 bound to the Mcm2 AAA+ domain, thereby topologically closing the DNA-loading gate (Fig. 1c–d, Supplementary Fig. 8). However, the N-tier ring was still open at the Mcm2-5 interface. Hence the Mcm2-7 was in a half-open and half-closed state. This structural feature explains why the Mcm2-7 ring in the OCCM is partially salt stable on DNA, but not as salt stable as the Mcm2-7 double-hexamer15,31,44. The structure also explains why the DNA is intact in the C-tier AAA+ ring but invisible in the N-tier ring as DNA in this region may be less constrained or digested by the nuclease, which was used to release the DNA bound OCCM during sample preparation.
Figure 3. ORC-Cdc6 encircles the origin DNA with the Orc4 insertion helix binding to a major groove.
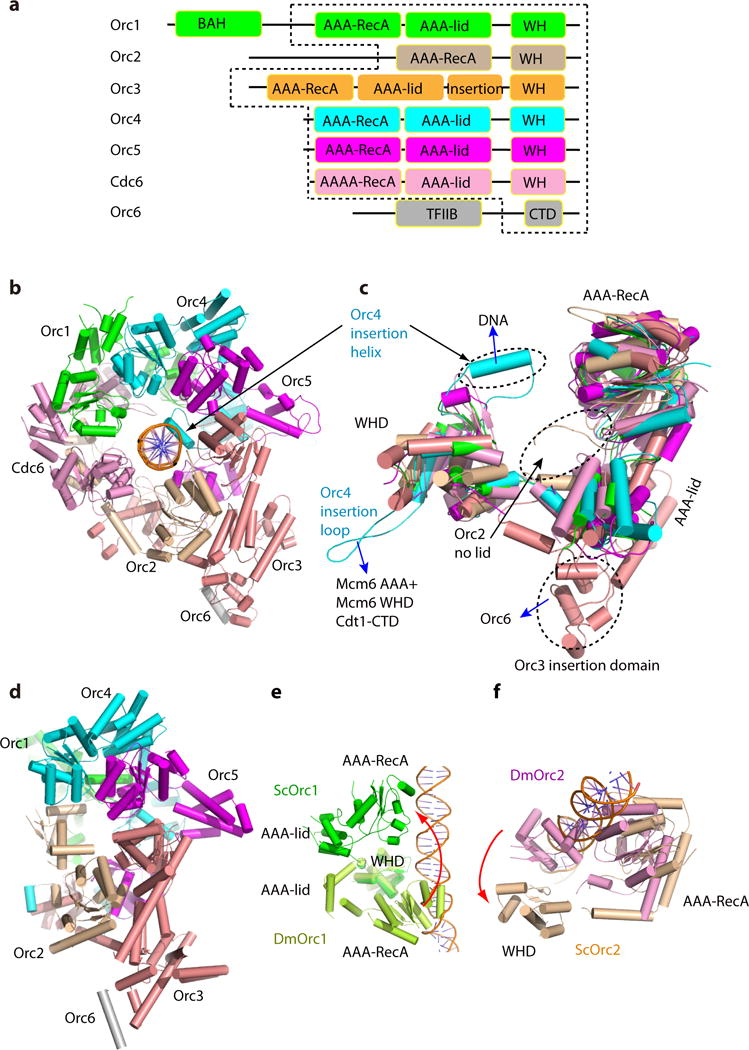
(a) Domain organization of S. cerevisiae Orc1-5 subunits and Cdc6. Dashed lines mark the ORC-Cdc6 core regions resolved in our model (TFIIB and CTD, transcription factor-II-like and C-terminal domains in Orc6; BAH, bromo-adjacent homology domain in Orc1). (b) The ORC-Cdc6 structure in our S. cerevisiae OCCM model in top view. (c) Superposition of Orc1-5 and Cdc6, highlighting their similar overall structures. Orc2 lacked the AAA-lid domain, resulting in a relatively open interface between Orc2-Cdc6. Orc3 had an insertion domain between the AAA-lid domain and the WHD domain that interacted with Orc6. The blue arrows point to structures with which these marked elements interact. The black arrow points to the missing lid domain in Orc2. (d) Crystal structure of Drosophila ORC complex in a similar subunit color scheme. (e, f) Alignment of DmORC with ScORC-Cdc6 using the most similar Orc3-5 region as a reference showed that the AAA-RecA-fold domain of DmOrc1 (e) and WHD of DmOrc2 (f) needed to move and rotate by 180° to assume their respective position in ScOCCM. See also Supplemental video 1.
ORC-Cdc6 forms a closed ring with a pseudo 6-fold symmetry
In the OCCM structure, ORC-Cdc6 assembled into a complete ring encircling DNA with a pseudo six-fold symmetry (Fig. 3a–b, Fig. 4). The N-terminal extension on Orc2 and an insertion in Orc3 give ORC a helical shape44. The four observed ATPγS molecules of ORC-Cdc6 define one circle on top and the four observed nucleotides in Mcm2-7 define another circle below. The top circle is larger, off center, and is tilted by 17° with respect to the lower circle (Fig. 4a–b). The six predicted AAA+ proteins, Orc1-5 and Cdc6, all had one AAA+ domain with an AAA-RecA-fold (the RecA fold), an α-helical-lid domain (the lid), and a C-terminal (CT), α-helical winged helix domain (WHD), and they were superimposable (Fig. 3c)45,46. There were variations to this general rule in that Orc2 lacked the α-helical lid similar to the DmOrc2, and Orc3 had an insertion consisting of a helical domain. Among the six initiator AAA+ subunits, Orc4 was unique as it had one α-helix insertion and one insertion loop in the WHD. The six AAA-RecA-folds and the six WHD formed a two-tiered ring structure that surrounded the DNA within the central channel. In contrast, the six AAA-lid domains that each bridged the AAA-RecA-like domain and the WH domain formed an outer brace that spiraled around the DNA interacting domains. The bottom tier ring of the WH domains was largely responsible for interacting with the Mcm CTD domains below, as suggested7. The peripheral brace of the AAA-lids also interacted with the Mcm subunit WH domains.
Figure 4. Nucleotide binding sites and configuration in OCCM.
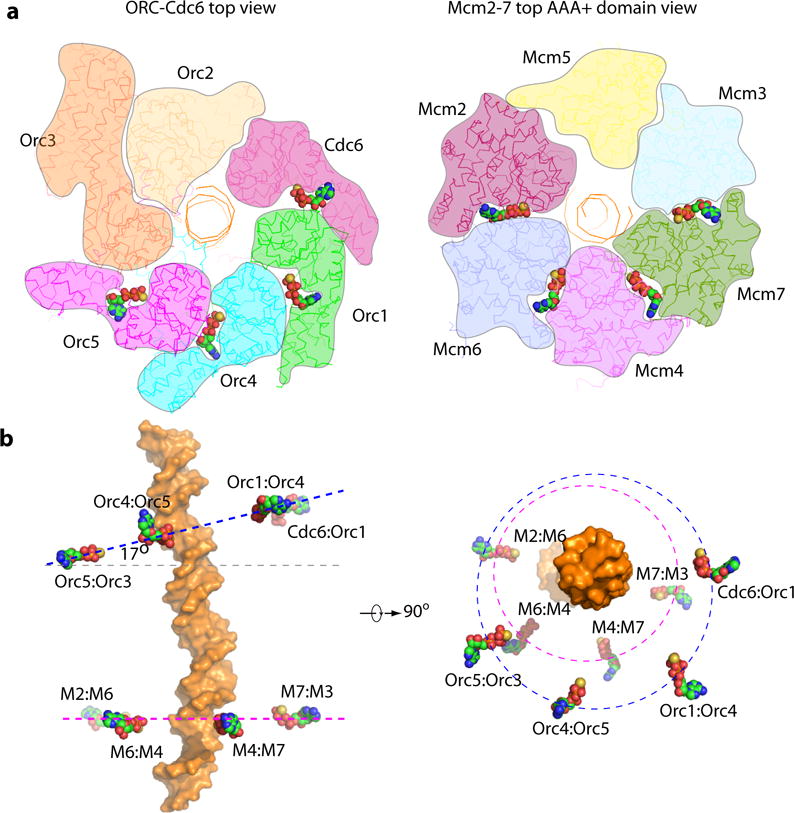
(a) Cut-open top view of ORC-Cdc6 and Mcm2-7 shown in surface view. The four ATPγS molecules identified in ORC-Cdc6 at the interface between Cdc6-Orc1, Orc1-Orc4, Orc4-Orc5, and Orc5-Orc3 (right), and four ATPγS molecules in Mcm2-7 at the interface between Mcm2-Mcm6, Mcm6-Mcm4, Mcm4-Mcm7, and Mcm7-Mcm3 are shown as spheres (carbon in green, oxygen in red, nitrogen in blue, sulfur in yellow). (b) The positons of the observed nucleotides in OCCM relative to the DNA, which is shown in orange surface. The left panel is a side view with Mcm4 in front and the right panel is a top view with ORC-Cdc6 on top, but proteins are not shown in order to highlight the nucleotides. The four ATPγS molecules in ORC-Cdc6 are co-planar, but the plane is tilted by ~17° with respect to the plane formed by the nucleotides in Mcm2-7. An imaginary circle defined by the nucleotide in ORC-Cdc6 is larger (75 Å) than the circle defined by nucleotides in Mcm2-7 (65 Å), and the two circles are acentric.
The crystal structure of the DmORC core revealed a conformation that is not compatible with DNA binding7 (Fig. 3d). DmORC structure is also incompatible with Cdc6 binding. However, we found that Orc3-4-5 were in a similar configuration in both ScORC and DmORC. By aligning the two ORC structures using the common Orc3-4-5 region as a reference, we found that the RecA-fold of DmOrc1 and the WH domain of DmOrc2 needed to move and flip by ~180° in order to match their respective yeast counterparts (Fig. 3e, f). These changes created a gap between Orc1-Orc2 for DNA passage as well as for Cdc6 insertion between Orc1-Orc2 following DNA binding by ORC (Fig. 3b, d, Supplementary Video 2). Since the Mcm2-7 hexamer has been loaded onto DNA by ORC-Cdc6, the conformation of ORC-Cdc6 in the OCCM structure is clearly in its active form, allowing DNA binding9 and both Cdc6-Orc1 and Orc1-Orc4 ATPase activities that are required for Mcm2-7 double hexamer assembly or subsequent regulated initiation of DNA replication once per cell division cycle9,29,30,32,47,48.
Cdt1 forms an extended three-domain structure
In the OCCM structure, Cdt1 exists in an unusually extended three-domain structure (Fig. 5a–c). The density of Cdt1 NTD was relatively weak, indicating a degree of flexibility. The Cdt1 NTD bound only to the Mcm2 CTD with an interface of ~ 600 Å2. Surprisingly, the Cdt1 CTD was linked to the MHD by a long loop and was 60 Å away from MHD, located between Mcm6 and Mcm4. In contrast, the Cdt1 MHD bound to both NTD and CTD of Mcm2 as well as NTD of Mcm6 with a larger interface of ~ 1000 Å2, which was also seen by CLMS (Fig. 2a). The Cdt1 CTD interacted extensively with all of the major domains of Mcm6, the Mcm4 NTD, and the Orc4 WH domain insertion loop. Consistent with this observation, a previous NMR study showed interaction between the Mcm6 WH domain and a short peptide in the CTD of Cdt140. Importantly, the Cdt1 CTD formed an arch toward the Mcm6 WH domain, which in turn interacted with the Orc4 WH domain and the Orc5 AAA-lid. The Mcm6 WH domain in the Mcm2-7 hexamer before encountering ORC-Cdc6 is likely located in the middle of the ring between Mcm2 and Mcm6, because this is where the domain is found in both active helicase CMG and in the inactive Mcm2-7 double-hexamer (Fig. 5d). Hence, the CT arch of Cdt1 is likely responsible for displacing the Mcm6 WHD domain by 40 Å to the periphery where the Mcm6 WHD is found in the OCCM structure. This conformational change likely explains the inhibitory role of the Mcm6 WHD, which blocks OCCM formation in the absence of Cdt132. Therefore, Cdt1 appeared to play a dual role in Mcm2-7 hexamer loading: it created the ORC-Cdc6-binding surface on the CT surface of Mcm2-7 hexamer by moving outward the obstructing Mcm6 WHD, and at the same time formed an extended 3-domain side-brace that stabilized the Mcm2-Mcm6-Mcm4 half ring, potentially allowing the other half ring Mcm5-Mcm3-Mcm7 to move (Fig. 1d, e). We suggest that these interactions underlie the essential roles of Cdt1 in Mcm2-7 loading on DNA13. Cdt1 bound in the OCCM, particularly its interaction with Mcm2 and the Mcm2-7 subunits that bind ATP (Mcm2,4,7 and 3) may keep the Mcm2-Mcm5 N-tier interface open and prevent Mcm2-7 ATP hydrolysis. ORC-Cdc6 ATPase activity, the next step after OCCM assembly, removes Cdt130 and may promote Mcm2-7 ATPase activity to close the first Mcm2-7 hexamer33,34.
Figure 5. Extensive interactions between Cdt1 and MCM hexamer.
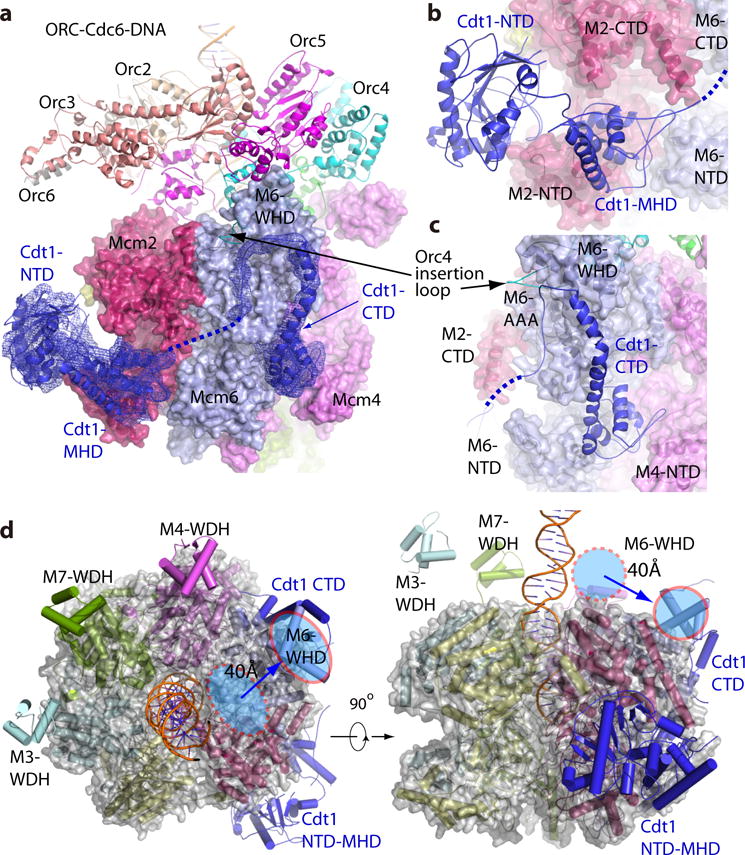
(a) OCCM structure with Cdt1 electron density shown in blue mesh. The CTD of Cdt1 locates between Mcm6 and Mcm4, over 60 Å away from the NTD and MHD of Cdt1. (b) Zoomed view of the Cdt1 NTD and MHD showing their interactions with Mcm2 and Mcm6. (c) Zoomed view showing the Cdt1 CTD interacting with Mcm6 WHD. The dotted blue line in (a-c) indicates a flexible loop connecting Cdt1 MHD and CTD. (d) The top view (left) and front side view (right) of Mcm2-7 structure in cartoon and semi-transparent surface view. The red oval marks Mcm6-WHD in OCCM, and the dashed red oval the position of Mcm6-WHD in CMG helicase. The blue arrow shows the displacement of Mcm6 WHD in OCCM due to interaction with Cdt1 CTD. Such displacement forms an unobstructed Mcm2-7 C-terminal face for binding with ORC-Cdc6.
To further investigate the interactions between Cdt1 and Mcm2-7, we expressed in baculoviruses each Mcm subunit and Cdt1 as a Strep-Strep-SUMO-Cdt1 (SSS-Cdt1). Each Mcm subunit alone or all six in combination were expressed and a pull down with purified SSS-Cdt1 was performed (Supplementary Fig. 8). Cdt1 interacted with all six Mcm proteins when they are expressed together. Individually, Mcm2, Mcm6 (most strongly) but also Mcm7 interact with Cdt1. The former two interactions were found in the OCCM model. We did not see an interaction with Mcm4, suggesting that the Cdt1-CTD interaction with Mcm4 seen in OCCM structure must depend on the prior binding to Mcm2-Mcm6 in the Mcm2-7 hexamer. The interaction with Mcm7 is not present at the stage of OCCM, but could be functional downstream, when OCM recruits the second Cdt1-bound Mcm2-7 hexamer to form the D-H35. Indeed, a long Mcm7 α-helix projects down toward the incoming second Mcm2-7-Cdt1 complex (Fig. 1e).
By comparing the Mcm2-7 in the OCCM with that in the Mcm2-7 D-H, we found that the Mcm2-7 needed to undergo large conformational changes during the OCCM – D-H transition, in particular within the entire Mcm2-7 NTD ring as well as the CTDs of Mcm2 and Mcm5 (Fig. 6, Supplementary Video 3). Specifically, the Mcm2-7 NTD ring needed to rotate by ~25° relative to the Mcm2-7 CTD to match the MCM ring in the D-H, and the CTDs of Mcm2 and Mcm5 had to rotate by ~5° and ~15°, respectively, to form the closed interface in the D-H (Fig. 6b,c). Because the ATPase activity of Orc1 and Cdc6 is required during the loading reaction29,30,32, and Orc1 and Cdc6 appear in a conformation poised to hydrolyze ATP, it is possible that the conformational changes outlined here are driven by ORC-Cdc6 ATP hydrolysis. Conceivably, the large conformational changes, which could be mediated by interactions between Orc4, Mcm6 and Cdt1 (Fig. 5a), would alter the Cdt1 binding surface, leading to its release from Mcm2-7. Since Cdt1 release is known to occur immediately before the recruitment of the second Cdt1-bound Mcm2-730,32,35, we suggest that ATP hydrolysis by ORC-Cdc6 could facilitate Cdt1 release, completely closing the first Mcm2-7 ring and establishing a condition for recruitment of the next Cdt1-bound Mcm2-7 hexamer. Alternatively, MCM ATP-hydrolysis could be involved33,34; however the Mcm2-7 ring is broken in the OCCM, thus ATPase activity of Mcm is likely blocked at this stage45.
Figure 6. Conformational changes between the Mcm2-7 in OCCM and double-hexamer (D-H).

(a) Comparison of the top CTD view (left) and the bottom NTD view (right) of the Mcm2-7 structure in the double-hexamer (gray cartoon) with the Mcm2-7 structure in the OCCM. The two structures were aligned using CTDs of Mcm4-6-7 as reference. Changes in the CTD ring are focused in Mcm2-5-3. The NTD ring rotated en bloc by about 25°. (b) Front Mcm2/5 side view of the Mcm2-7 hexamer in the OCCM structure (left) as compared to that in the D-H (right). Transitioning from OCCM to double hexamer, each CTD AAA+ domain and NTD of Mcm2 and Mcm5 undergoes a combination of rotation and translation, with the degree of rotation and translation shown as labeled. Mcm5 NTD needs to rotate by as much as 50° to close the DNA entry gate. (c) A sketch showing how the gate between Mcm2 and Mcm5 can be open for DNA insertion in OCCM (left) and how the gate is closed in the D-H (right).
Asymmetric interaction between ORC-Cdc6 and Cdt1-Mcm2-7 enables DNA insertion
Although both the Mcm2-7 hexamer and ORC-Cdc6 form ring-like structures with a pseudo 6-fold symmetry, the interaction between the two rings is asymmetric due to the ~17° tilt of the ORC-Cdc6 ring with respect to the Mcm2-7 ring (Fig. 1d, Fig. 4). As a consequence, the DNA is bent by ~20–25° at their interface. Furthermore, the tilt led to a tight interface between Orc1-Orc4-Orc5 with Mcm4-6-2, and an apparent “loose” interface between Orc3-Orc2-Cdc6 and Mcm5-3-7. At the tight interface, the WH domains of Orc1 and Orc4 insert into the gaps between the WH domains and the AAA-lid domains of Mcm4 and Mcm6; the Orc5 WH domain interacted only with Mcm2 AAA-RecA-fold because Mcm2 lacked a WH domain (Fig. 1d–e, Supplementary Fig. 7). At the “loose interface”, the WH domain of Mcm5 was not visible in the EM map, but the CLMS data identified it across-the-interface partner of Orc2. Interestingly, the WH domains of Mcm3 and Mcm7 reached upwards more than 30 Å via their long loops to interact with Orc2 and Cdc6, respectively (Fig. 1e, Supplementary Fig. 7). Accordingly, we propose that the asymmetric interaction leaves half of the Mcm2-7 ring (Mcm5-3-7) only loosely tethered, such that the Mcm5-3-7 half ring can move away from the tightly-tethered Mcm4-6-2 half ring to open up the Mcm2/Mcm5 gate for DNA insertion and then move back to close the gate. This conformation is most likely stabilized by Cdt1.
Protein-DNA interactions in the OCCM
We modeled 39 base pairs (bp) of DNA in the OCCM density map; 24 bp were encircled by the ORC-Cdc6 ring and the remaining 15 bp by the C-tier ring of Mcm2-7 (Fig. 7a, Supplementary Fig. 9). There was no apparent DNA density inside the N-tier ring of Mcm2-7. Because the N-tier ring was open at the Mcm2-Mcm5 interface, the dsDNA there might have been digested by the DNase I nuclease that was used to cleave the loading intermediate off of the plasmid DNA. In the top ORC-Cdc6 region, Orc1, Orc3 and Orc5 had little direct interaction with DNA. DNA was held in place by interactions with the initiator-specific motif (ISM) in the AAA+ domains of Orc2, a unique Orc4-specific insertion α-helix, and by the ISM and the WH domains of Cdc6 (Fig. 7a–e). These four binding components spiraled around the DNA just like RFC clamp loaders spiral around the DNA44,49,50, although the overall ORC-Cdc6 ring itself appears flat because the Orc2 and Orc3 subunits break the helical path. Archaeal AAA+ replication initiators also interact with DNA via their respective ISM39,51. The archaeal WH domain is known to bind DNA with both the helix-turn-helix (HTH) motif and the β-hairpin wing loop51. However, in the yeast ORC-Cdc6, we found that only the β-hairpin wing loops of the WH domains of Cdc6 and Orc4 bound to DNA, their respectively HTH motif did not bind to DNA but rather were engaged in subunit-subunit interactions. This different DNA binding mode was caused by a ~90° rotation of the Orc4 and Cdc6 WH domains away from the central DNA channel (Supplementary Fig. 10). We have previously demonstrated that ORC-Cdc6 causes a nuclease protected footprint on the origin DNA that extends to 70–78 base pairs, greater than the 44–50 base-pair footprint of ORC alone9. Even allowing for limited nuclease access to the DNA near edges of the ORC-Cdc6 complex, which accounts to 10 bp52, the amount of DNA found interacting with ORC-Cdc6 by nuclease footprinting is more than twice that found interacting with ORC-Cdc6 in the OCCM structure. There are positively charged patches in the front side (Orc2,3,6) and the bottom surface of ORC-Cdc6 that is proximal to Mcm2-7 (Supplementary Fig. 10). It is possible that DNA bends and binds to some of these surfaces in addition to the central channel.
Figure 7. Interactions between OCCM and DNA.
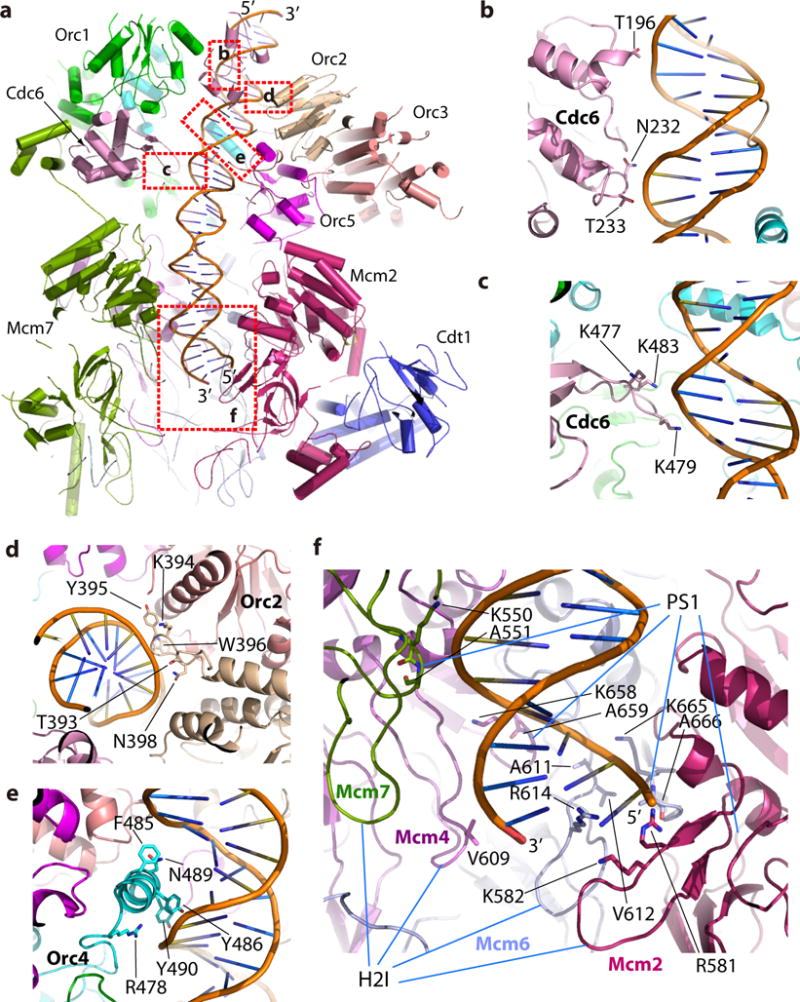
(a) An overview of OCCM-DNA structure in side view. Subunits in front of DNA including parts of Orc1 and Cdc6, and all of Mcm3 and Mcm5 are removed to show DNA. Five rectangle-marked areas are enlarged in panels (c-f). (b-c) Detailed view of the Cdc6 interaction with dsDNA. (d) Orc2 interaction with DNA. (e) Orc4 interaction with DNA. (f) Mcm2-6-4-7 and DNA interfaces. PS1: Pres-sensor 1 β-hairpin loop, H2I: Helix-2-insert β-hairpin loop.
In the lower Cdt1-Mcm2-7 region, Mcm3 and Mcm5 did not interact with DNA. Although the Mcm2-7 ring was nearly flat, the hairpin loops in the AAA+ domains of Mcm2-Mcm4-Mcm6-Mcm7 were arranged in a spiral to grip onto the DNA (Fig. 7a, f). We found that Mcm2, Mcm4 and Mcm6 interacted with DNA via their respective helix-2-insert (H2I) β-hairpin loop. Mcm4, Mcm6 and Mcm7 contacted the DNA with their respective Presensor 1 (PS1) β-hairpin loops, in particular the well-conserved KA motifs (Fig. 7f, Supplementary Fig. 9). In contrast, the same H2I and PS1 within the Mcm2-7 D-H are involved in inter-subunit interactions14. In the apo form of the active CMG helicase the PS1 loops face the central DNA channel53, and in a recent cryo-EM structure of CMG at a sub-nanometer resolution, the PS1 loops were found to interact with a 6-base ssDNA54. Interestingly, most of these hairpin loops, except for the Mcm4 H2I loop, interacted with the same strand of the duplex DNA (Fig. 7f). It is unclear if this strand functions as the leading or the lagging strand in the CMG helicase, because extensive conformation changes must occur in Mcm2-7 to form the active helicase, such that Mcm-DNA interaction in the CMG may be very different.
DISCUSSION
Since the discovery of ORC more than two decades ago2, it has been a key issue in the DNA replication field to identify how ORC recognizes the dsDNA and how ORC cooperates with Cdc6 to load the Mcm2-7 hexamers onto DNA. The current atomic model of the OCCM provides the first high-resolution structure of ORC-Cdc6 bound to origin DNA. Overall, the ORC-Cdc6 structure is flat, but within this structure the subunits that bind ATP, Orc1, Orc4, Orc5 and Cdc6, form a right-handed spiral around the double stranded DNA, similar to the spiral of AAA+ subunits around primer-template DNA in RFC clamp loaders49,50. The human ORC has an almost identical structure (A. Tocilj, K. On, C. Yuan, J. Sun, E. Elkayam, H. Li, B. Stillman and L. Joshua-Tor, submitted). Orc4 had an unusual insertion α-helix that appears to contact the major groove of the dsDNA. This helix is unique to budding yeast Orc4 subunits, being absent in the DmORC and HsORC4 structures7 (A. Tocilj, K. On, C. Yuan, J. Sun, E. Elkayam, H. Li, B. Stillman and L. Joshua-Tor, submitted), may help to explain the sequence specificity of ORC that is characteristic of origin binding in Saccharomyces sp. Cdc6 is particularly important for DNA binding with two principal DNA binding sites: the ISM and the WH domain, explaining why Cdc6 enhances ORC’s specificity for DNA9,10.
The most prominent feature of Mcm2-7 loading by ORC-Cdc6 is the extensive use of WH domains of these replication proteins. There were six resolved WH domains in ORC-Cdc6 that formed a larger WH ring in the upper tier of the OCCM, and 5 cryo-EM and CLMS resolved WH domains in Mcm2-7 that formed a second slightly smaller WH ring on the lower Mcm2-7 tier of the OCCM. This appears to be an evolutionary conserved interaction, as it was reported recently for archaeal MCM and Orc155. However, here we observed that the WHD mediated correct stacking of the two rings mediates much of the recruitment mechanism. The position of the WHD of the Mcm proteins varies widely, and can sit either right above the AAA+ domain, or move away from the main body of the protein, or even move to the side of the AAA+ domain in the case of Mcm5, as seen in the active helicase (Supplementary Fig. 7). The CTD of Cdt1 plays a special role in displacing the Mcm6 WH domain to create the ORC-Cdc6 binding surface; we showed previously that the WH domain of Mcm6 is inhibitory and blocks OCCM formation in the absence of Cdt132. The attachment of the winged helix domains by flexible linker provide these domains manifold possibility to interact with ORC-Cdc6, but after helicase activation, they could also be important interaction partners with other proteins at the DNA replication fork.
The Mcm2-7 double-hexamer structure, although obtained in the absence of DNA, showed that six H2I hairpin loops are arranged in an approximately helical trajectory that was suggested to facilitate DNA translocation and unwinding. However, in the OCCM, only three H2I hairpin loops of Mcm2-Mcm4-Mcm6 contact DNA. In addition, the three PS1 hairpin loops of Mcm4, Mcm6 and Mcm7 make contact with DNA. In the D-H, the PS1 loop is involved in inter-subunit interaction, not in DNA binding, but in the CMG helicase, the PS1 loop contacts DNA14,54. Another interesting feature of the OCCM is the partially open Mcm2-Mcm5 interface. Thus the data provide the first structural proof that the Mcm2-Mcm5 interface is the DNA entry gate in Mcm2-7 during pre-RC formation22,31,43. ATPγS prevents Cdt1 removal and the gate from closing30,32 and thus it is interesting that the N-tier ring is not yet closed given the dsDNA has been loaded into the Mcm2-7 hexamer channel. We suggest that the Mcm2-7 N-tier ring closure requires ATP hydrolysis by Orc1 and Cdc6 and removal of Cdt1 from the Mcm2-7 hexamer. The unusually extended three-domain structure of Cdt1 most likely keeps Mcm2-7 subunits vertically aligned, preventing the left-handed spiral of the Mcm2-7 subunits in the completely assembled Mcm2-7 double hexamer (Fig. 6c). Thus, like the RFC-PCNA clamp loader and clamp structures, the OCCM structure provides further insight into how a AAA+ complex loads a ring-shaped, multi-subunit complex of proteins around double stranded DNA to promote DNA replication. We suggest that ORC-Cdc6 ATPase promotes complete Mcm2-7 ring closure just as ATP hydrolysis by the clamp loader RFC locks the PCNA DNA polymerase clamp onto double stranded DNA50.
METHODS
Methods and any associated references are available in the ONLINE METHODS SECTION.
Online methods
Sample preparation and electron microscopy
The Saccharomyces cerevisiae loading intermediate OCCM was assembled in vitro with purified ORC, Cdc6, Cdt1 and Mcm2-7 on plasmid DNA containing the ARS1 sequence in the presence of ATPγS, and isolated by the magnetic beads pull-down approach as described previously44 with minor modifications. 24 pre-RC reactions containing 40 nM ORC, 80 nM Cdc6, 40 nM Cdt1, 40 nM MCM2-7 and 6 nM pUC19-ARS1 beads in 50 μl buffer A (50 mM HEPES-KOH (pH 7.5), 100 mM potassium glutamate, 10 mM magnesium acetate, 50 μM zinc acetate, 3 mM ATPγS, 5 mM DTT, 0.1% Triton X-100 and 5% glycerol) were incubated for 15 min at 24 °C. After three washes with buffer B (50 mM HEPES-KOH (pH 7.5), 100 mM K acetate, 3 mM ATPγS) the complex was eluted with 1 U of DNase I in buffer B and 1 mM CaCl2.
To prepare cryo-EM grids, we pooled all the elutions together and concentrated the sample to about 0.9 mg/ml in buffer B with a Microcon centrifugal filter unit (YM-100 membrane). Before EM grid preparation, we checked the sample for homogeneity by negative-stain electron microscopy. We then applied 3 μl of OCCM sample at a final concentration of 0.9 mg/ml to glow-discharged C-flat 1.2/1/3 holey carbon grids, incubated for 10 s at 6 °C and 95% humidity, blotted for 3 s then plunged into liquid ethane using an FEI Vitrobot IV. We loaded the grids into an FEI Titan Krios electron microscope operated at 300 kV high tension and collected images semi-automatically with SerialEM under low-dose mode at a magnification of ×29,000 and a pixel size of 1.01 Å per pixel. A Gatan K2 summit direct electron detector was used under super resolution mode for image recording with an under-focus range from 1.5 to 3.5 μm. The dose rate was 10 electrons per Å2 per second and total exposure time was 5 seconds. The total dose was divided into a 25-frame movie and each frame was exposed for 0.2 s.
Image processing and 3D reconstruction
Approximately 7500 raw movie micrographs were collected. The movie frames were first aligned and superimposed by the program Motioncorr56. Contrast transfer function parameters of each aligned micrograph were calculated using the program CTFFIND457. All the remaining steps, including particle auto selection, 2D classification, 3D classification, 3D refinement, and density map post-processing were performed using Relion-1.458. We manually picked ~10,000 particles from different views to generate 2D averages, which were used as templates for subsequent automatic particle selection. Automatic particle selection was then performed for the entire data set. 1,371,667 particles were initially selected. Then we carefully checked the particles obtained from automatically picking, removed the bad particles and re-picked the missing good ones. Particles were then sorted by similarity to the 2D references; the 10% of particles with the lowest z-scores were deleted from the particle pool. 2D classification of all remaining particles was performed and particles in unrecognizable classes by visual inspection were removed. A total of 601,095 particles were used for 3D classification. We derived six 3D models from the dataset, and found two models were similar to each other and their associated particles were combined for further refinement; the other four models were distorted and those particles were discarded, leading to a dataset size of 304,288 particles. This final dataset was used for further 3D refinement, resulting in the 3.91 Å 3D density map. The resolution of the map was estimated by the so-called gold-standard Fourier shell correlation, at the correlation cutoff value of 0.143. The 3D density maps were corrected for the detector modulation transfer function and sharpened by applying a negative B-factor of -112 Å2. The particles had some preference for end-on views but because of the large number of particles used virtually all of the angular space was sampled.
Structural modeling, refinement, and validation
The yeast Mcm2-7 models were directly extracted from the cryo-EM structure of the yeast Mcm2-7 double hexamer (PDB code 3JA8)14. For subsequent docking, each Mcm proteins was split into 2 parts: NTD and CTD. The initial models of the S. cerevisiae Orc1-5 subunits were generated from the crystal structure of the Drosophila ORC complex (PDB ID: 4XGC)7 and Cdc6 subunit from the homologue Archaeal structure (PDB ID: 2V1U)39 using the SWISS-MODEL server59. These models were first docked and fitted into the density map using COOT60 and Chimera61. We found Orc1 and Orc2 in the yeast OCCM had gone through dramatic conformational changes compared to the fly ORC structure. Thus, we split each of the two proteins into 3 parts: the AAA-RecA-fold and the AAA-lid domain (where present) and the C-terminal WH domain, and fitted these domains independently into the 3D density map. Based on the structural features, the extra density outside Mcm2 and Mcm6 clearly belonged to NTD and middle helical domain (MHD) of Cdt144. We used the SWISS-MODEL server to generate the atomic models of Cdt1 NTD and MHD from their respective homologous structures (PDB ID: 5C3P and 2ZXX)42,62. The Cdt1 CTD model was generated from human Cdt1 crystal structure (PDB ID: 2WVR)63, and this domain was found in the region between Mcm6 and Mcm4. Four bulky densities outside the core structure of Cdc6-ORC were identified to be the WH domains of Mcm3, Mcm4, Mcm6, and Mcm7, respectively. Their atomic models were generated from their corresponding homologue structures (PDB ID: 3NW0, 2M45, 2KLQ, and 2OD5)41,64,65 using the SWISS-MODEL server. The Orc6 density was very weak, indicating high flexibility of this protein. However, based on the homologous Drosophila ORC core complex structure (PDB ID: 4XGC)7, we identified a short α-helix at the C-terminus of Orc6 that bond to and was stabilized by Orc3. Finally, the double stranded DNA was built into the long helical density that ran through the OCCM structure in the program COOT. The entire OCCM atomic model was subsequently adjusted manually and rebuilt in COOT. Clearly resolved bulky residues such as Phe, Tyr, Trp, and Arg were used for sequence registration.
The manually built atomic model was then iteratively refined in real space by phenix.real_space_refine66 and rebuilt in COOT. We also performed the reciprocal space refinement procedure with the application of secondary structure and stereochemical constraints in the program Phenix67. The structure factors (including phases) were calculated by Fourier transform of the experimental density map with the program Phenix.map_to_structure_factors. The atomic model was validated using MolProbity68. Structural figures were prepared in Chimera and Pymol (https://www.pymol.org).
The final model was cross validated using a method described previously69. Briefly, we randomly added 0.1 A° noise to the coordinates of the final model using the PDB tools in Phenix, then refined the noise-added against the first half map (Half1) that was produced from one half of the particle dataset during refinement by RELION. We performed one round of coordinate refinement, followed by a B-factor refinement. The refined model was then correlated with the 3D maps of the two half maps (Half1, Half2) in Fourier space to produce two FSC curves: FSCwork (model versus Half1 map) and FSCfree (model versus Half2 map), respectively. A third FSC curve was calculated between the refined model and the final 3.9 A° resolution density map produced from all particles. The general agreement of these curves was taken as an indication that the model was not over-fitted.
Cross-linking/Mass Spectrometry analysis
240 nM pUC19-ARS1 beads were used to assemble the OCCM complex as previously described. The complex was cross-linked in the presence of the beads with BS3 1:8100 (molar protein:cross-linker ratio) for 2 hours at 4 °C. Then, the reaction was quenched with 50 ul of saturated ammonium bicarbonate for 45 minutes at 4 °C and it was transferred into digestion buffer (50 mM ammonium bicarbonate, 8 M Urea) followed by reduction with DTT and alkylation with iodoacetamide. 3 ug LysC (with estimated 1:50 enzyme to protein ratio) were added and incubated at room temperature for 4 hours. The digestion buffer was then diluted with 50 mM ammonium bicarbonate to a final Urea concentration of 2M. 3 ug of trypsin were added (at estimated 1:50 enzyme to protein ratio) and incubated for 16 hours at room temperature. After digestion, the supernatant was collected and acetified using 200 ul 10% Trifluoroacetic acid (TFA). The peptide mixture was then desalted using C18-Stage-Tips for mass spectrometric analysis70. LC-MS/MS analysis was performed using an Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer (Thermo Scientific) applying a “high-high” acquisition strategy71. 2 ug peptide mixture was injected for each mass spectrometric acquisition. Peptides were separated on a 50 centimeter EASY-Spray column (Thermo Scientific) assembled in an EASY-Spray source (Thermo Scientific), operated at 50 °C column temperature. Mobile phase A consisted of water, 0.1% v/v formic acid and 5% v/v DMSO. Mobile phase B consisted of 80% v/v acetonitrile, 0.1% v/v formic acid and 5% v/v DMSO. Peptides were loaded at a flow-rate of 0.3 μl/min and eluted at 0.2 μl/min using a linear gradient going from 2% mobile phase B to 40% mobile phase B over 139 minutes, followed by a linear increase from 45% to 95% mobile phase B in 12 minutes. The eluted peptides were directly introduced into the mass spectrometer. MS data was acquired in the data-dependent mode with the top-speed option. For each three-second acquisition cycle, the mass spectrum was recorded in the Orbitrap with a resolution of 120,000. The ions with a precursor charge state between 3+ and 8+ were isolated and fragmented. The fragmentation spectra were then recorded in the Orbitrap. Dynamic exclusion was enabled with single repeat count and 60-second exclusion duration. To improve the identification of cross-linked peptides, in total 9 acquisitions were carried out with variations on parameters mainly related to criteria for ion selection for fragmentation and fragmentation methods.
Identification of cross-linked peptides
The raw mass spectrometric data files were processed into peak lists using MaxQuant version 1.5.2.872 with default parameters, except for “FTMS top peaks per 100 Da” was set to 20 and “FTMS de-isotoping” was disabled. The peak lists were searched against the sequences as well as the reversed sequences (as decoy) of 14 OCCM subunits using Xi software (ERI, Edinburgh) for identification of cross-linked peptides. Search parameters were as follows: MS accuracy, 6 ppm; MS2 accuracy, 20 ppm; enzyme, trypsin; specificity, fully tryptic; allowed number of missed cleavages, four; cross-linker, BS3; fixed modifications, carbamidomethylation on cysteine; variable modifications, oxidation on methionine, modifications by BS3 that are hydrolyzed or amidated on the other end. The reaction specificity for BS3 was assumed to be for lysine, serine, threonine, tyrosine and protein N-termini. For acquisitions where CID and HCD fragmentations were applied (acquisition 1.3.4.6.7 and 9), only b- and y-ions were considered for the fragment ion matches; while for data acquired using a combined fragmentation of ETD and CID or HCD (acquisition 2,5 and 8), b-, y-, c-ions and z-ions were considered for fragment ion matches. Two independent quality control approaches have been applied for identified cross-linked peptide candidates. All cross-linked peptides with estimated 5% FDR at residue pair level were accepted for further structural interpretation. We also accepted cross-linked peptides identified with MS2 spectra that passed machine-learning based auto-validation.
In vitro pulldown of Cdt1 with Mcm2-7 proteins
Baculoviruses expressing Mcm2-7 subunits (the Mcm3 was tagged with HA at the N-terminus) and Strep-Strep-SUMO-Cdt1 were prepared in the Profold-C1 expression vector (AB Vector, San Diego). 2.5 × 107 Hi-Five insect cells were infected at a multiplicity of infection of 10 with each MCM subunit alone or in combination with SSS-Cdt1, and harvested at 55 hours post infection. After washing cells in cold PBS, they were placed for 10 minutes on ice in < 1 ml of hypotonic buffer (25 mM Hepes-KOH pH 7.5, 20 mM K glutamate, 1 mM Mg acetate, 1 mM DTT, 5 mM ATP, Protease Inhibitor (Roche, 1 tablet in 50 ml). Cell extracts were prepared by Dounce B homogenization and centrifugual clarification (10,000 rpm in Sorvall SS34 rotor), then 100 μl of supernatant was incubated of 2 hours on ice with 50 μl of Strep-Tactin sepharose (IBA) that had previously been washed in PBS and hypotonic buffer. Beads were washed (3 × 5 minutes) with IP buffer (25 mM Hepes-KOH pH 7.5, 300 mM KGlutamate, 10 mM MgAcetate, 0.04% NP40, 1 mM DTT, 5 mM ATP and protease inhibitor). Bead bound proteins were run on a SDS-PAGE gel and stained with silver.
Data Availability
The 3D cryo-EM map of OCCM at 3.9 Å resolution has been deposited at the EMDB database with accession code EMD-8540. The corresponding atomic model was deposited at RCSB PDB bank with accession code 5UDB. Source data files including both the intra-molecular and inter-molecular crosslinks for figure 2 and supplementary 4 are presented as Source Data files 1–2, available with the paper online.
Supplementary Material
Acknowledgments
Cryo-EM data was collected on a FEI Titan Krios at HHMI Janelia Farm. We also collected a cryo-EM dataset on an FEI Technai F20 equipped with a K2 detector at NRAMM in the Scripps Research Institute, which was supported by NIH grants P41 GM103310. We thank Zhiheng Yu, Chuan Hong, Rick Huang at HHMI, and Clint Porter and Bridget Carragher at Scripps for help with data collection. H.L dedicates this work to the loving memory of his son Paul J. Li. This work was funded by the US National Institutes of Health (GM111472 and OD12272 to H.L. and GM45436 to B.S.), the Biotechnology and Biological Sciences Research Council UK (P56061 to CS) and the Wellcome Trust (Investigator Award P56628 to CS, Senior Research Fellowship 103139 to JR, a Centre core grant 092076 to JR and an instrument grant 108504 to JR).
Footnotes
Author Contributions Y.Z., A.R., L.B., J.S., J.R., Z.A.C., B.S., C.S. and H.L. designed experiments. Z.Y., A.R., L.B., C.SPA., M.B. and J.S. performed experiments. Z.Y., A.R., L.B., J.S., Z.A.C., J.R., B.S., C.S. and H.L. analyzed the data. L.B., B.S., C.S. and H.L. wrote the manuscript.
COMPETING FINANCNCIAL INTNTERESTS
The authors declare no competing financial interests.
References
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–74. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–34. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 3.Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–84. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Stillman B. The origin recognition complex: a biochemical and structural view. Subcell Biochem. 2012;62:37–58. doi: 10.1007/978-94-007-4572-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, et al. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2008;105:10326–31. doi: 10.1073/pnas.0803829105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun J, et al. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure. 2012;20:534–44. doi: 10.1016/j.str.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleichert F, Botchan MR, Berger JM. Crystal structure of the eukaryotic origin recognition complex. Nature. 2015;519:321–6. doi: 10.1038/nature14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell SP, Labib K. Chromosome Duplication in Saccharomyces cerevisiae. Genetics. 2016;203:1027–67. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–71. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speck C, Stillman B. Cdc6 ATPase activity regulates ORC × Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J Biol Chem. 2007;282:11705–14. doi: 10.1074/jbc.M700399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–2. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 12.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–5. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–30. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li N, et al. Structure of the eukaryotic MCM complex at 3.8 A. Nature. 2015;524:186–91. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 15.Sun J, et al. Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function. Genes Dev. 2014;28:2291–303. doi: 10.1101/gad.242313.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–41. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–66. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 18.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21:771–7. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell. 2010;40:834–40. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botchan M, Berger J. DNA replication: making two forks from one prereplication complex. Molecular Cell. 2010;40:860–1. doi: 10.1016/j.molcel.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aparicio T, Guillou E, Coloma J, Montoya G, Mendez J. The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic Acids Res. 2009;37:2087–95. doi: 10.1093/nar/gkp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–7. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–5. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 24.Araki H. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol. 2010;22:766–71. doi: 10.1016/j.ceb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 25.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–7. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deegan TD, Yeeles JT, Diffley JF. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J. 2016;35:961–73. doi: 10.15252/embj.201593552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–5. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evrin C, et al. In the absence of ATPase activity, pre-RC formation is blocked prior to MCM2-7 hexamer dimerization. Nucleic Acids Res. 2013;41:3162–72. doi: 10.1093/nar/gkt043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ticau S, Friedman LJ, Ivica NA, Gelles J, Bell SP. Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell. 2015;161:513–25. doi: 10.1016/j.cell.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samel SA, et al. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 2014;28:1653–66. doi: 10.1101/gad.242404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez-Cid A, et al. An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol Cell. 2013;50:577–88. doi: 10.1016/j.molcel.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 33.Kang S, Warner MD, Bell SP. Multiple functions for Mcm2-7 ATPase motifs during replication initiation. Mol Cell. 2014;55:655–65. doi: 10.1016/j.molcel.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coster G, Frigola J, Beuron F, Morris EP, Diffley JF. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol Cell. 2014;55:666–77. doi: 10.1016/j.molcel.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evrin C, et al. The ORC/Cdc6/MCM2-7 complex facilitates MCM2-7 dimerization during prereplicative complex formation. Nucleic Acids Res. 2014;42:2257–69. doi: 10.1093/nar/gkt1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang F, et al. Cdc6 ATPase activity disengages Cdc6 from the pre-replicative complex to promote DNA replication. Elife. 2015;4 doi: 10.7554/eLife.05795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bleichert F, et al. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. Elife. 2013;2:e00882. doi: 10.7554/eLife.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balasov M, Akhmetova K, Chesnokov I. Drosophila model of Meier-Gorlin syndrome based on the mutation in a conserved C-Terminal domain of Orc6. Am J Med Genet A. 2015;167A:2533–40. doi: 10.1002/ajmg.a.37214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 40.Liu C, et al. Structural insights into the Cdt1-mediated MCM2–7 chromatin loading. Nucleic acids research. 2012;40:3208–3217. doi: 10.1093/nar/gkr1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei Z, et al. Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J Biol Chem. 2010;285:12469–73. doi: 10.1074/jbc.C109.094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee C, et al. Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature. 2004;430:913–917. doi: 10.1038/nature02813. [DOI] [PubMed] [Google Scholar]
- 43.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–93. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 44.Sun J, et al. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20:944–51. doi: 10.1038/nsmb.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–57. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 47.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–78. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 48.Frigola J, Remus D, Mehanna A, Diffley JF. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–43. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Donnell M, Kuriyan J. Clamp loaders and replication initiation. Curr Opin Struct Biol. 2006;16:35–41. doi: 10.1016/j.sbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Kelch BA, Makino DL, O’Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–80. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dueber ELC, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 52.Suck D, Oefner C. Structure of DNase I at 2.0 A resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986;321:620–5. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- 53.Yuan Z, et al. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016;23:217–24. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abid Ali F, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samson RY, Abeyrathne PD, Bell SD. Mechanism of Archaeal MCM Helicase Recruitment to DNA Replication Origins. Mol Cell. 2016;61:287–96. doi: 10.1016/j.molcel.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–21. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheres SH. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–22. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research. 2014:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 61.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Li W, Zhang T, Ding J. Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi. Nucleic acids research. 2015:10026–10038. doi: 10.1093/nar/gkv979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Marco V, et al. Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing. Proceedings of the National Academy of Sciences. 2009;106:19807–19812. doi: 10.1073/pnas.0905281106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Molecular cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiedemann C, et al. Structure and regulatory role of the C-terminal winged helix domain of the archaeal minichromosome maintenance complex. Nucleic acids research. 2015;43:2958–2967. doi: 10.1093/nar/gkv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–67. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–9. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 71.Chen ZA, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29:717–26. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 3D cryo-EM map of OCCM at 3.9 Å resolution has been deposited at the EMDB database with accession code EMD-8540. The corresponding atomic model was deposited at RCSB PDB bank with accession code 5UDB. Source data files including both the intra-molecular and inter-molecular crosslinks for figure 2 and supplementary 4 are presented as Source Data files 1–2, available with the paper online.


