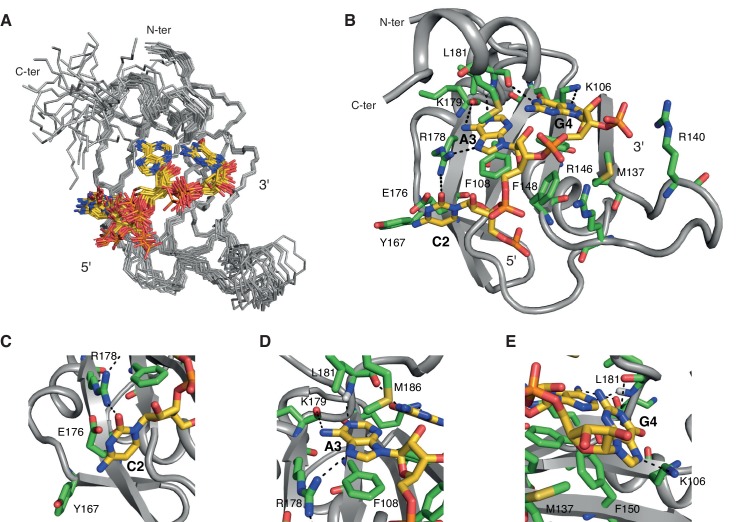
Figure 2. Overview of the solution structure of hnRNP A1 RRM2 bound to 5´-UCAGUU-3´ RNA.
(A) NMR ensemble. Overlay of the 20 final structures superimposed on the backbone of the structured part (103-112, 115-139, 144-187) and represented as a ribbon trace (N, Cα, C´). The not-well-defined N- and C-terminal residues as well as nucleotides U1, U5 and U6 are omitted for a better overview. The RNA is shown in stick representation with the carbon atoms in yellow, nitrogen in blue, phosphate in orange and oxygen in red. (B) Cartoon drawing of a representative structure of the NMR ensemble. Residues with important roles in RNA binding are shown as sticks, with the carbon atom in green. All other atoms have the same color code as in panel A. (C–E) Close-up views of each single nucleotide recognition by hnRNP A1 RRM2. Representation and colors are similar to panel B. Residues with important roles in RNA binding are labeled.