Figure 1. Activation of PAR2 induces β-catenin stabilization and Lef/Tcf transcriptional activity.
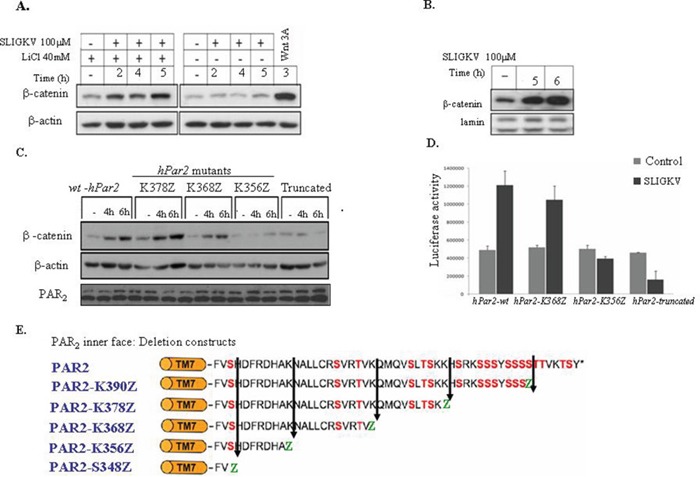
(A) PAR2 activation induces β-catenin stabilization in RKO cells. RKO cells were pretreated with LiCl (40 mM, 2h) followed by 2, 4, and 5 hours of PAR2 SLIGKV activation (100 μM). Whereas low levels of β-catenin were observed prior to PAR2 activation, a marked increase in β-catenin levels was seen following activation. RKO cells that were not pretreated with LiCl showed basal levels of β-catenin regardless of PAR2 activation. RKO cells pre-treated with Wnt3A conditioned medium for 3 hours were used as a positive control. (B) PAR2 activation induces nuclear β-catenin in HEK-293T cells. HEK-293T cells were transiently transfected with both flg-β-catenin and hPar2-wt plasmids. After PAR2 activation, the nuclear fraction was extracted and immunoblots were analyzed using anti-flg (for flg-β-catenin) and anti-lamin (used as a nuclearprotein loading control) antibodies. The amount of nuclear β-catenin was markedly increased in response to PAR2 activation. (C) PAR2-induced β-catenin stabilization depends on an intact K356 - K368 region. HEK-293T cells transiently transfected with hPar2 deletion constructs lacking the K356-K368 (i.e., hPar2-K356Z or hPar2-truncated) displayed a markedly reduced β-catenin stabilization level. In contrast, cells transfected with hPar2 constructs that possess the above-mentioned region (e.g., hPar2-K368Z/K378Z or wt) displayed increased β-catenin accumulation. (D) An intact K356 - K368 region is essential for PAR2-induced Lef/Tcf transcriptional activity. TOPflash luciferase transcription activity was analyzed in RKO cells following PAR2 activation in the presence of hPar2 wt or deletion constructs. While in hPar2-wt and hPar2-K368Z transfected cells, PAR2 activation elicits markedly elevated luciferase Lef/Tcf activity, in hPar2-K356Z and hPar2-truncated cells PAR2 activation failed to increase luciferase Lef/Tcf activity above the basal level. The results were evaluated using GraphPad InStat software and found to be statistically significant (p<0.01). (E) Scheme of PAR2 C-tail. Schematic representation of PAR2 C-tail and its various deleted constructs.
