Abstract
Acute myeloid leukemia is a hematologic malignancy with significant molecular heterogeneity. MicroRNAs have important biological functions and play critical roles in pathogenesis and prognosis in a variety of cancers including acute myeloid leukemia. Some reports have constructed risk stratification systems for adult acute myeloid leukemia patients using microRNAs to predict an optimal outcome of patients. However, little has been done in pediatric and adolescent patients. The purpose of this study is to identify a panel of microRNA signature that could predict prognosis in younger cytogenetically normal acute myeloid leukemia patients by analyzing the data from The Cancer Genome Atlas. A total of 59 cytogenetically normal acute myeloid leukemia patients under 21 years with corresponding clinical data were enrolled in our study. Using univariate Cox's model, we found 17 miRNAs were significantly related with overall survival in pediatric and adolescent cytogenetically normal acute myeloid leukemia patients but no clinical parameter was found significant related with overall survival. The multivariate Cox regression identified high expression of hsa-miR-146b was independent poor prognostic factor and high expression of hsa-miR-181c and hsa-miR-4786 appeared to be favorable factors. A model was proposed based on these three miRNAs. Leave-one-out Cross Validation method and Permutation Test was further used to evaluate this model. The function role of has-mir-181c was further studied by carrying out flow cytometry and cell counting kit-8 (CCK-8) in U937 cell line. The results indicate that the 3-microRNA-based signature is a reliable prognostic biomarker for pediatric and adolescent cytogenetically normal acute myeloid leukemia patients.
Keywords: pediatric and adolescent, CN-AML, miRNA, signature, prognosis
INTRODUCTION
Pediatric acute myeloid leukemia (AML) is a rare and heterogeneous childhood cancer, with an incidence of approximately 7 cases per million children annually [1]. Although board overlap lies between AML for pediatric patients, adolescents and adults in diagnosis, treatment and prognosis, differences still exist. Cytogenetically normal acute myeloid leukemia (CN-AML) is a kind of AML that no cytogenetic abnormality is detectable in leukemic cells. In pediatric CN-AML patients, outcome of the patients is associated with other co-occurring mutations. CN-AML without any molecular mutation is considered to be at intermediate risk. When harboring NPM1 or CEBPA double mutation, pediatric CN-AML patients are considered favorable prognosis. In contrast, a FLT3-ITD mutation to wild-type ratio (ITD allelic ratio) of > 0.4 is associated with adverse outcome [2]. MicroRNAs (miRNAs) are small non-coding RNA molecules that can regulate gene expression at the post-transcriptional level. MiRNAs are enrolled in tumorigenesis of acute myeloid leukemia and impact hematopoietic cell differentiation, proliferation and treatment response. Emerging evidence suggest miRNAs expression signatures can also predict outcomes of various human cancers [3–6], including hematological malignancies such as adult AML[7] and primary plasma cell leukemia [8]. However, there were few studies investigated the prognostic value of miRNAs in pediatric and adolescent AML patients. Whether miRNA-expression profiling can identify different outcomes of various molecular mutation remains unclear. Here we conducted a study using the dataset extracted from The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) and constructed a 3-miRNA signature which may be used to predict the outcome of pediatric and adolescent CN-AML patients.
RESULTS
Identification of miRNAs associated with outcome
A total of 301 patients with microRNA information were selected, 59 patients who are cytogenetically normal were enrolled in our study. MicroRNAs were transformed from continuous variable into binary variable—high miRNA expression (expression level greater than the median) status equals 1 and low status equals 0. A total of 549 miRNAs were selected for further analysis from 1539 miRNAs according to the exclusion criteria. After univariate cox's model filtering, 17 miRNAs were suggested to be correlated with prognosis (Table 1).
Table 1. Univariate Cox analysis of 549 miRNAs.
| MicroRNA | P-value | FDR | Type |
|---|---|---|---|
| hsa.mir.181c | 0.000 | 0.001 | Risky |
| hsa.mir.146b | 0.000 | 0.001 | Protective |
| hsa.mir.153.1 | 0.003 | 0.006 | Risky |
| hsa.mir.500a | 0.007 | 0.011 | Protective |
| hsa.mir.501 | 0.012 | 0.017 | Protective |
| hsa.mir.181d | 0.014 | 0.020 | Risky |
| hsa.mir.3174 | 0.018 | 0.024 | Risky |
| hsa.mir.30b | 0.018 | 0.024 | Risky |
| hsa.mir.3176 | 0.019 | 0.025 | Risky |
| hsa.mir.570 | 0.022 | 0.028 | Risky |
| hsa.mir.4484 | 0.023 | 0.029 | Risky |
| hsa.mir.378d.2 | 0.026 | 0.032 | Risky |
| hsa.mir.125b.1 | 0.027 | 0.034 | Risky |
| hsa.mir.4786 | 0.028 | 0.034 | Risky |
| hsa.mir.500b | 0.031 | 0.038 | Protective |
| hsa.mir.30d | 0.032 | 0.038 | Risky |
| hsa.mir.548s | 0.032 | 0.039 | Risky |
Construction of miRNA signature
By carrying out multivariate cox regression formula, we identified 3 miRNAs (mir-181c, mir-146b and mir-4786) that were independently associated with prognosis. High expression of hsa-miR-146b was independent poor prognostic factor and high expression of hsa-miR-181c and hsa-miR-4786 appeared to be favorable factors. Then the risk score was calculated through the three miRNA status and their weight on OS, which is represented by the β coefficient in multivariate cox model. The risk score = (1.652* status of has-mir-146b)–(1.838* status of hsa-mir-181c)–(1.455* status of has-mir-4786). Next, risk score was calculated in each of the 59 patients and the ones whose risk scores greater than the median were assigned to high risk group and the others belong to low risk group.
Clinical and molecular characteristics of patients associated with 3 miRNAs and the risk score
Clinical characteristics and molecular alteration information of the patients with the risk score were displayed in Tables 2, 3, 4. Clinical characteristics were utilized to fit the Univariate Cox's model and the results were shown in Table 5. In our study, age at diagnosis, gender, WBC at diagnosis, bone marrow blasts and peripheral blasts did not show significance with prognosis. NPM1 and CEBPA mutations are correlated with favorable prognosis, with P-value of 0.031 and 0.05, respectively.
Table 2. Correlation between miRNA score and clinical and laboratory features in pediatric and adolescent CN-AML patients (n = 59).
| Characteristics | Total | microRNA score | P-value | |
|---|---|---|---|---|
| Low (n = 30) | High (n = 29) | |||
| Age (Mean ± SD, years) | 11.5 ± 5.2 | 12.0 ± 4.7 | 10.9 ± 5.7 | 0.449 |
| Male sex-no. (%) | 36 (61.0%) | 17 (56.7%) | 19 (65.5%) | |
| WBC at diagnosis(10−9/liter)* | 69.9 (0.9–446) | 67.3 (5.1–210) | 72.5 (0.9–446) | 0.807 |
| Bone marrow blasts (%) | 71.3 (21-98) | 74.8 (21-98) | 67.5 (25–96) | 0.168 |
| Peripheral blasts (%) | 55.2 (0–97) | 67.1 (18.5–97) | 42.3 (0–95) | 0.002 |
| FAB Category | ||||
| M0 | 1 (1.7%) | 0 | 1 (3.4%) | NA |
| M1 | 16 (27.1%) | 12 (40.0%) | 4 (13.8%) | 0.049 |
| M2 | 14 (23.7%) | 8 (26.7%) | 6 (20.7%) | 0.815 |
| M4 | 13 (22.0%) | 5 (16.7%) | 8 (27.6%) | 0.486 |
| M5 | 4 (6.8%) | 2 (6.7%) | 2 (6.9%) | > 0.999 |
| NOS | 7 (11.9%) | 2 (6.7%) | 5 (17.2%) | 0.394 |
| Unknown | 4 (6.8%) | 1 (3.3%) | 3 (10.3%) | 0.580 |
*WBC at diagnosis: White Blood Cells at dignosis.
Table 3. Correlation between miRNA score and other gene alterations.
| Mutation | Total | microRNA score | P-value | |
|---|---|---|---|---|
| Low (n = 30) | High (n = 29) | |||
| FLT3-ITD allelic ratio > 0.4 | 14 (23.7%) | 5 (13.8%) | 9 (33.3%) | 0.322 |
| NPM1 | 15 (25.4%) | 12 (37.9%) | 3 (13.3%) | 0.021 |
| CEBPA | 14 (23.7%) | 12 (41.3%) | 2 (6.7%) | 0.007 |
Table 4. Association between molecular mutations and FAB category with 3 miRNAs.
| Variants | hsa-miR-181c | hsa-miR-146b | hsa-miR-4786 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| High (29) | Low (29) | P-value | High (29) | Low (20) | P-value | High(29) | Low (30) | P-value | |
| NPM1 | 9 | 6 | 0.500 | 7 | 8 | > 0.999 | 9 | 6 | 0.500 |
| CEBPA | 11 | 3 | 0.027 | 1 | 13 | 0.001 | 9 | 5 | 0.322 |
| FLT3-ITD | 9 | 13 | 0.479 | 16 | 6 | 0.012 | 11 | 11 | > 0.999 |
| FAB | |||||||||
| M0 | 0 | 1 | > 0.999 | 1 | 0 | > 0.999 | 0 | 1 | > 0.999 |
| M1 | 13 | 3 | 0.007 | 7 | 9 | 0.831 | 7 | 9 | 0.839 |
| M2 | 10 | 4 | 0.109 | 6 | 8 | 0.815 | 8 | 6 | 0.705 |
| M4 | 3 | 10 | 0.069 | 6 | 7 | > 0.999 | 7 | 6 | 0.945 |
| M5 | 0 | 4 | 0.129 | 1 | 3 | 0.629 | 3 | 1 | 0.580 |
| NOS | 2 | 5 | 0.449 | 5 | 2 | 0.394 | 4 | 3 | 0.962 |
| Unknown | 1 | 3 | 0.629 | 3 | 1 | 0.580 | 0 | 4 | 0.129 |
Table 5. Univariate Cox analysis of clinical parameters with the prognosis.
| Variants | P-value |
|---|---|
| Age at diagnosis | 0.249 |
| Gender | 0.543 |
| WBC at diagnosis | 0.533 |
| Bone marrow blasts | 0.733 |
| Peripheral blasts | 0.406 |
| FLT3-ITD positive | 0.153 |
| NPM1 mutation | 0.031 |
| CEBPA mutation | 0.050 |
MiRNA signature was an independent prognosis predictor of pediatric and adolescent CN-AML
After adjusting for NPM1 and CEBPA mutations, the 3-miRNA also independently predicted OS (P = 0.003) and DFS (P = 0.000).
Correlation of microRNA prognostic signature with clinical or laboratory features and gene alterations
Higher risk score is associated with lower percentage of peripheral blasts in laboratory features. In contrast, no association was found between risk score and clinical features (Table 1). Genetic alterations were found different between high and low risk score groups: Patients with lower scores more often had NPM1 (P = 0.021) and CEBPA mutations (P = 0.007), which means patients with lower risk score more likely had favorable mutations (Table 2). High expression level of miR-181c (P = 0.027) and low expression level of miR-146b (P = 0.001) were more often had CEBPA mutations. High expression level of miR-146b (P = 0.012) more often had FLT3-ITD mutations. Association between miRNAs, the risk score and FAB category were also assessed. High expression level of miR-181c and lower risk score more often had M1 subtype of leukemia. In our study, no association was found between FLT-ITD allelic ratio > 0.4 and the microRNA signature.
Performance of miRNA signature
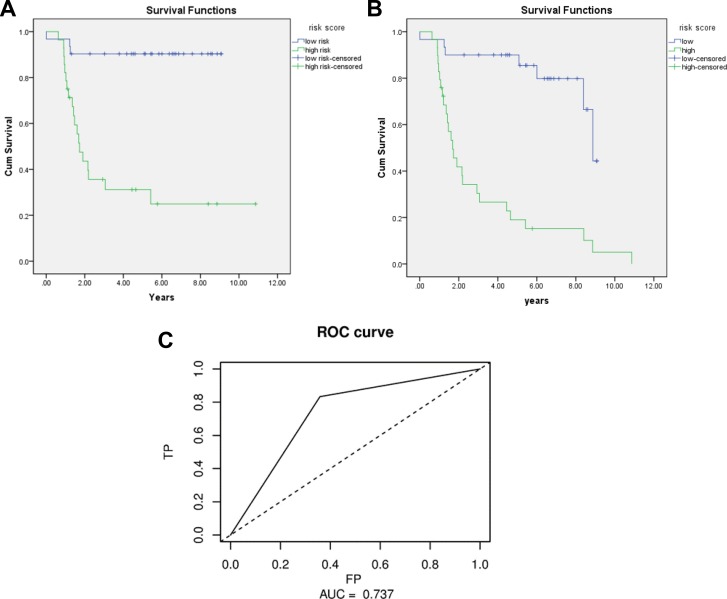
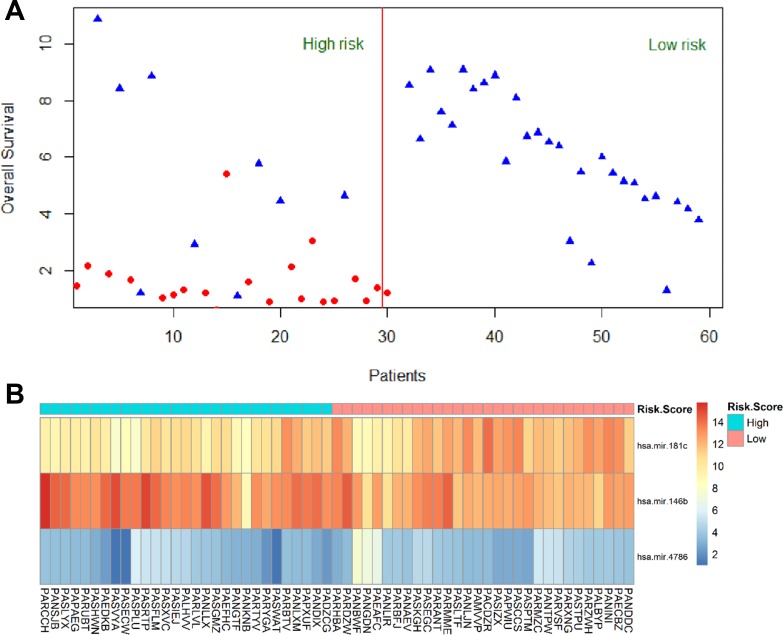
The Kaplan-Meier curve was applied to the 3-miRNA signature using the group separation of risk score. The results showed that patients in the high risk score group had significant worse OS/DFS than those in low risk score group (P = 0.000) (Figure 1A, 1C). The AUC of signature was 0.737 (Figure 1B). A figure and heatmap were created to evaluate the signature, and it was clearly shown that most dead patients were in high risk group and suffered worse OS (Figure 2). The results confirm that the 3-miRNA signature has the power to differentiate patients into high-risk and low-risk groups.
Figure 1. Kaplan-Meier for OS/DFS in low risk and high risk group and AUC curve for the risk score.
Figure 2. Heatmap for 3 miRNAs expression level and survival status in all 59 patients.
Permutation test and leave-one-out cross validation
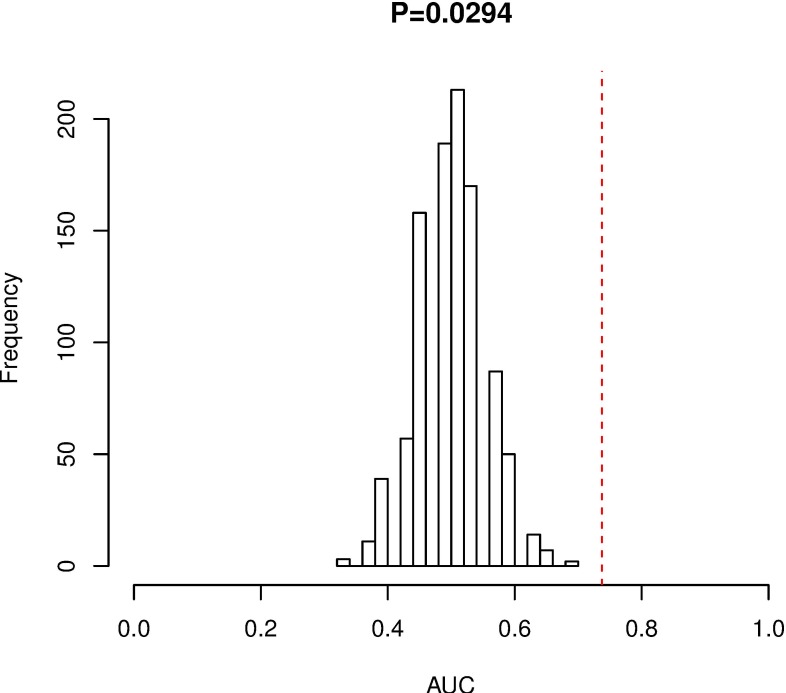
In order to validate whether the 3-miRNA signature is able to applied to other pediatric and adolescent CN-AML patients, we did permutation test and found that the AUC of random systems showed great significance with that of our studied cohort (P-value = 0.029) (Figure 3). These results indicate that our model could successfully predict the prognosis of pediatric and adolescent CN-AML patients. LOO-CV showed an AUC of 0.69 which also validates the 3-miRNA signature performs well.
Figure 3. Permutation test for 3-miRNA signature.
Bioinformatic analysis of target genes and pathways
Target genes of mir-181c, mir-146b and mir-4786 were predicted by three prediction tools—TargetScan, miRanda and miRTarBase. Considering false positive of prediction tools, TargetScan and miRanda were used to predict target genes of miR-181c and miR-146b, while TargetScan and miRTarBase were applied for miR-4786. MiRanda did not provide data of miR-4786. 327 target genes predicted by both tools were extracted (Supplementary Table 1). Part of the results were shown in Table 6. KEGG and part of GO results were displayed in Table 7 and Table 8. More GO results were shown in Supplementary Table 2.
Table 6. Part of target genes of three miRNAs.
| Target | Target | Target | |||
|---|---|---|---|---|---|
| ZNF667 | NOVA1 | YWHAE | |||
| RLF | TRAF6 | MORN4 | |||
| C16orf87 | CD80 | TTPAL | |||
| C7orf41 | NRAS | APP | |||
| BEND3 | FAM26E | APP | |||
| E | |||||
| LRRC8D | SCN3B | RNF115 | |||
| PLCL2 | GOSR1 | CA6 | |||
| hsa-mir-181c | SPP1 | hsa-mir-146b | ZNF148 | hsa-mir-4786 | IL17REL |
| HMGB2 | SIAH2 | TBC1D15 | |||
| LIN28A | MMP16 | COX6A1 | |||
| FLT1 | ZNRF3 | UBE2D3 | |||
| BAG4 | POU3F2 | COX6A1P2 | |||
| 2 | |||||
| IL1A | ROBO1 | LRTOMT | |||
| RABGEF1 | WASF3 | TCEB3 | |||
| 1 | |||||
| FIGN | FZD1 | TRIM17 |
Table 7. GO analysis results of 352 targets.
| Category | ID | Term | Counts | P-Value |
|---|---|---|---|---|
| Biological process | 0045449 | Regulation of transcription | 64 | 0.002 |
| 0006355 | Regulation of transcription, DNA-dependent | 47 | 0.003 | |
| 0051252 | Regulation of RNA metabolic process | 47 | 0.004 | |
| 0045664 | Regulation of neuron differentiation | 8 | 0.008 | |
| Cellular components | 0031974 | Membrane-enclosed lumen | 49 | 0.000 |
| 0043233 | Organelle lumen | 48 | 0.000 | |
| 0031981 | Nuclear lumen | 40 | 0.000 | |
| 0070013 | Intracellular organelle lumen | 45 | 0.000 | |
| 0005654 | Nucleoplasm | 26 | 0.002 | |
| 0044451 | Nucleoplasm part | 18 | 0.004 | |
| Molecular function | 0008270 | Zinc ion binding | 61 | 0.002 |
(P = 0.01).
Table 8. KEGG pathway results.
| Term | ID | P-Value | Gene |
|---|---|---|---|
| Alanine, aspartate and glutamate metabolism | 00250 | 0.009796 | GFPT1, AGXT, AGXT2, RIMKLB |
| Vitamin B6 metabolism | 00750 | 0.012 | PDXK, PNPO |
| Hippo signaling pathway -multiple species | 04392 | 0.0314 | RASSF6, MOB1B, RASSF1 |
| Acute myeloid leukemia | 05221 | 0.043011 | NRAS, RUNX1, MAPK1 |
| Prion diseases | 05020 | 0.048606 | IL1A, MAPK1, EGR1 |
(P = 0.05).
MiR-181c mimics treatment in U937 cells
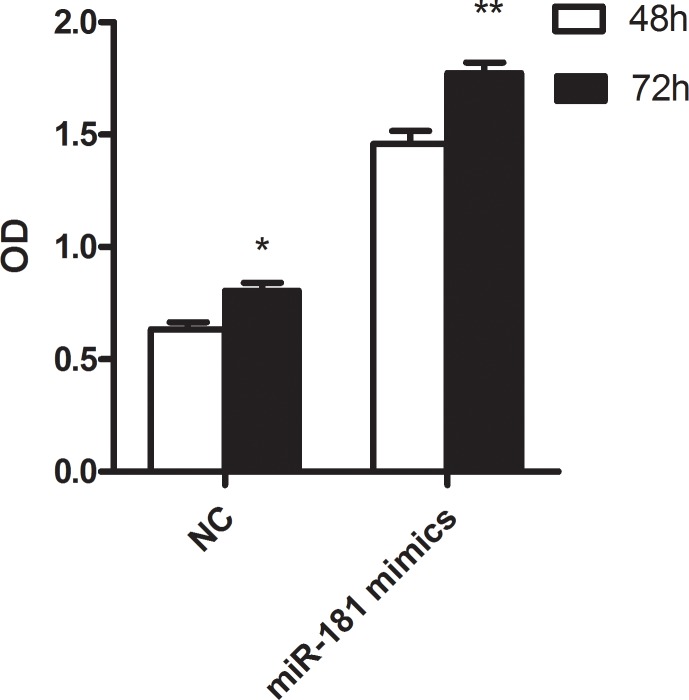
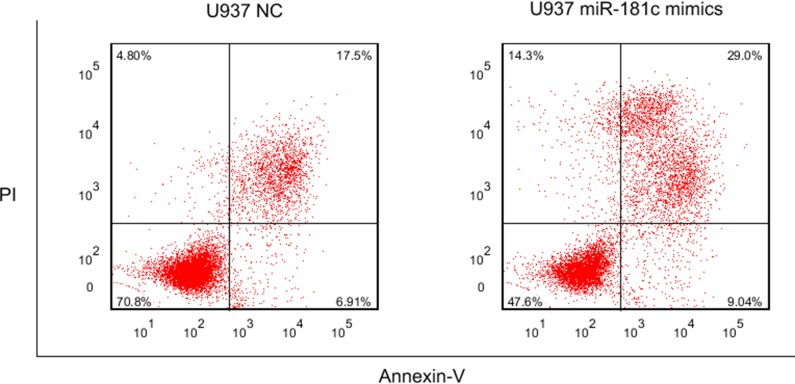
The functional roles of these miRNAs needs to be further studied. We focused on miR-181c, whose family members were broadly reported in AML including cytogenetically normal subtype. U937 was commonly regarded as cytogenetically normal AML cells and they were used in our study. After 48 and 72 hours transfection with miR-181c mimics and negative control, the proliferation activity was significantly lower in group treated with miR-181c mimics (P = 0.018 and 0.004, respectively.) (Figure 4). Flow cytometry was further carried out to observe apoptosis between miR-181c treatment and NC group. The result shows that miR-181c also exhibits ability of inducing apoptosis significantly in U937 cells (Figure 5).
Figure 4. The result of cell proliferation in U937 cell line treated with miR-181c mimics and NC.
Figure 5. The result of apoptosis in U937 cell line treated with miR-181c mimics and NC.
DISCUSSION
Ample evidence suggests microRNAs play important roles in progress and prognosis in pediatric and adolescent AML. Pediatric AML patients always carry cytogenetically abnormalities and the abnormal karyotypes rate reaches 70–80% compared to 55% in adult AML patients [2]. Little has been done to construct a miRNA signature scoring system to accurately predict the outcome of pediatric and adolescent CN-AML patients.
Here we used pediatric and adolescent TCGA AML dataset to construct a 3-miRNA signature (mir-181c, mir-146b and mir-4786) which underwent rounds of statistical analysis to ensure the strong and independent influence of outcome built for predicting and further exploring pediatric CN-AML molecular mechanisms .
CN-AML patients are considered to be an intermediate-risk prognosis category, and prognosis can be further divided into subgroups based on favorable (NPM1, CEBPA) or unfavorable (FLT3-ITD) genetic mutations. In our study, CEBPA and NPM1 mutations are not only predict favorable prognosis of pediatric CN-AML patients (CEBPA, P = 0.05, NPM1, P = 0.031), but also correlate with the risk score—lower risk score patients are more likely to have longer OS and DFS, and more often had these favorable mutations correlated with favorable outcomes. Lower expression level of aggressive miR-146b was more often had favorable CEBPA mutations and less often aggressive FLT3-ITD mutations. The condition was contrary in miR-181c. MiR-181 family (including miR-181c) has also been reported to be upregulated in patients with CEBPA mutations [9]. Our findings are consistent with the previous study.
However, FLT3-ITD neither shows relationship with the prognosis nor the 3-miRNA signature. One reason may be the small sample in our study cannot draw a significant conclusion statistically. Another reason may be 10 patients with FLT3-ITD mutations in our study also had NPM1 (8 patients) or CEBPA (2 patients) mutations and 8 of these patients are alive during the follow-up period. NPM1 and CEBPA are reported a positive effect on survival irrespective of FLT3-ITD mutations in some studies [9–11]. Therefore, FLT3-ITD did not present its predictive role in our study.
Over expression of miR-181 family was also reported associated with erythroid differentiation and monocytic differentiation, which means they potentially be involved in the differentiation block of M1 blasts [9, 12]. This may explain why higher expression level of miR-181c more often had M1 subtype of leukemia in our study. Lower risk score also more often possessed M1 subtype, which was mostly ascribed to miR-181c. MiR-181 family members were found downregulated in unfavorable adult hematologic malignancies [9, 13–16]. It has been reported that they increase expression of a series of homeobox superfamily genes (i.e., HOXA7, HOXA9, HOXA11 and PBX3) [15]. Homeobox superfamily genes are highly conserved and play important roles in a variety kinds of biological processes including apoptosis, differentiation and tumorigenesis, acting as tumor suppressors [17]. MicroRNA 181 family (including miR-181c) may act as a protective factor through HOX in pediatric and adolescent leukemia patients, our functional study in U937 cells consists with these findings. Su et al. suggest MiR-181 family (including miR-181c) expression level is elevated in adult acute myeloid leukemia patients compared with normal population and miR-181 family induces acute leukemogenesis through hindering granulocytic and macrophage-like differentiation in acute myeloid leukemia cell lines. However, the relationship between prognosis and miR-181 family expression level was not executed in this study [18]. More studies should be done to clarify relations between miR-181 family and pediatric acute myeloid leukemia progress and prognosis. MiR-146b was also reported to be a differential expression miRNA between M1 and M5 FAB subtypes, and was speculated participating in the block of M1 development and maturation, although its further function was not studied in AML [12]. It was found to play an oncogenic role in hematologic malignancies including T-ALL and lymphoma [19, 20], and to promote leukemic transformation of hematopoietic cells within BCR-ABL1-positive microvesicles [21]. MiR-146b is a key regulator accelerated the transformation by targeting NUMB and other genes, causing genome instability [20]. MiR-146b has been reported in a variety of kinds of cancer, acting as tumor promotor [22, 23] or suppresser [24–26] depending on different cancer types. In contrast, little was reported about mir-4786.
To gain a further insight into the functional role of the 3-miRNA signature, we extracted their target genes and carried out GO and KEGG pathway analysis. Interestingly, several pathways have compact relation with progression and prognosis of acute myeloid leukemia. For example, glutamate metabolism has critical role in AML progression. AML cells require glutamine to adapt to increased biosynthetic activity, and glutaminolysis inhibition activated mitochondrial apoptosis and provides a novel therapeutic strategy for AML [27]. In addition, vitamin B6 participates in many biological processes and is suggested to be related to carcinogenesis and tumor growth [28, 29]. B6 deficiency leads to uracil incorporation and chromosome breaks which makes organism susceptible to cancer [30]. Lower vitamin B6 status is also associated with pediatric AML [31]. Combining our KEGG results, we infer that vitamin B6 supplement may have favorable effect on prognosis of pediatric AML patients. Another important pathway is Hippo signaling pathway, which shows dual functions in tumorigenesis according to previous studies [32–34] and its role in pediatric AML remains to be detected. Three miRNA target genes also include NRAS, RUNX1, MAPK1, which are precisely enriched in acute myeloid leukemia pathway. Moreover, prion diseases pathway is also find significant in our analysis. Previous data suggest that prion protein participates in human leukocyte maturation [35] and the expression level of prion protein mRNA can be down-regulated in all-trans retinoic acid induced maturation in NB4 cells [36].
However, the limitations of our study should be considered for future applications. First, the number of pediatric and adolescent CN-AML patients enrolled in our study was only 59, which indicates our sample size is not large enough to draw a reliable conclusion, although we carried out permutation test and LOO-CV—validation techniques for small samples. Second, the number of pediatric and adolescent AML patients who were cytogenetically normal were relatively small and we did not find a suitable external dataset to validate our formula in previous study. External datasets are needed for evaluation of the 3-miRNA signature.
In conclusion, we identified a 3-miRNA signature through genome-wide miRNA expression profiling from TCGA, which could be used as an independent indicator for prognosis of pediatric and adolescent CN-AML patients.
MATERIALS AND METHODS
TCGA AML dataset
Expression levels of miRNA and clinical information of patients with AML were downloaded from The Cancer Genome Atlas (TCGA). Level 3 data of miRNA expression level includes 1539 miRNAs and clinical dataset contained 491 patients. Patients were screened by the following criteria. First, patients without miRNA information were excluded. Second, patients who were not cytogenetically normal or cytogenetically information unknown were removed. Third, patients who were alive and the last contact days were unavailable were discarded. Finally, 59 CN-AML patients who were under 21 at diagnosis were included in our study.
Overall survival times (OS) and disease free survival times (DFS) are considered as endpoints respectively. As the data was extracted from TCGA, an ethics committee was not needed.
Cell culture and transfection
U937 cells were obtained from ATCC. Cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum (FBS, Gibco). Cells were transfected with hsa-miR-181c mimics and negative control (NC) using lipofectamine 3000 (Thermo). Hsa-miR-181c mimics was purchased from GenePharma.
Statistical analysis
The expression level of 1539 miRNAs was presented as reads per million (RPM) miRNA mapped data and the miRNAs whose expression level equal 0 RPM in more than 25% of all observations were eliminated using R (version 3.3.2). Each miRNA was put into univariate Cox's model (miRNA was transformed into binary variable), P value and FDR adjustment were used to select miRNAs which are significantly associated with OS or DFS. Significant parameters were filtered out using 0.05 as the cutoff in both P value and FDR adjustment. Seventeen miRNAs were identified as biomarker of outcome. Clinical information including gender, race, white blood cell counts, and molecular mutations reported to be associated with prognosis (FLT3-ITD, NPM1, CEBPA) were also used to build univariate Cox's model under the same standard. Each significant miRNA identified by univariate proportional hazards regression was further evaluated in multivariate Cox's model (backward stepwise). To determine whether the miRNA signature can independently predict the prognosis, multivariate analysis was carried out using molecular mutations associated with prognosis and the risk score constructed by miRNA signature.
Permutation test was used to estimate whether the 3-miRNA expression signature can precisely predict OS in population of pediatric and adolescent CN-AML patients by R Bioconductor. It is used to evaluate the performance and randomness of the model. In brief, we take the combination of overall survival time and vital status of patients in the research as a label, then each individual in our study has a label and a risk score which is calculated using the proposed 3-miRNA scoring system. A random system was constructed by assigned labels randomly to individuals while the risk score keeps consistent with each individual. The random system was tested for survival significance. If the model performs well, a random system cannot predict the prognosis of patients. The area under Receiver Operating Characteristic curve (AUC) was supposed to equal 0.5. A thousand random systems were created by R, after all iterations, significance between AUC of random systems and the right label system was measured by P-value with a cutoff 0.05. P-value calculated greater than 0.05 means the 3-miRNA signatures have no effect on the outcome.
Leave-one-out Cross Validation (LOO-CV) was also applied to evaluate the model proposed above using R Bioconductor. LOO-CV is powerful in estimating a model's performance. Briefly, each time a observation was left out and all the other observations were used to construct a model using 3 miRNAs above and then makes a prediction for the observation left. 59 tests were conducted and the average AUC was carried out.
Bioinformatic analysis
We use TargetScan, miRanda and miRTarBase to find target genes of three miRNAs. Gene ontology (GO) analysis was carried out by The Database for Annotation, Visualization and Integrated Discovery (DAVID) online and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was carried out by KOBAS online.
Flow cytometry
Cells were stained with monoclonal antibodies in Annexin V-FITC/PI Apoptosis Detection Kit (BD Bioscience) by protocol. Data were collected using FACSCanto II (BD Bioscience) and were analyzed with FlowJo software (TreeStar, version 7.6.1).
Cell proliferation assay
Cell proliferation was carried out using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) according to the manufacturer's instruction. U937 cells were seeded into 96-well plates at a concentration of 5000–8000 cells/well. Next, 10 ul CCK-8 was added to the cells and the cells were incubated for 2.5 hours at 37°C at each time point. The optical density was read at 450nm with a microplate spectrophotometer.
SUPPLEMENTARY TABLES
Abbreviations
- AML
Pediatric acute myeloid leukemia
- CN-AML
Cytogenetically normal acute myeloid leukemia
- ITD allelic ratio
FLT3-ITD mutation to wild-type ratio
- TCGA
The Cancer Genome Atlas
- OS
Overall Survival Times
- DFS
Disease Free Survival Times
- AUC
The area under Receiver Operating Characteristic curve
- LOO-CV
Leave-one-out Cross Validation
- GO
Gene ontology
- DAVID
The Database for Annotation, Visualization and Integrated Discovery
- KEGG
Kyoto Encyclopedia of Genes and Genomes
Footnotes
Authors' contributions
Study design: Ruiqi Zhu; Statistical analysis: Ruiqi Zhu, Weiwei Zhao; Literature research: Jingdi Liu; Experimental studies: Ruiqi Zhu, Ting Luo, Fengjuan Fan, Jun Deng; Manuscript preparation: Ruiqi Zhu; Manuscript revision: Yu Hu, Liang Tang; Manuscript editing: Fengjuan Fan; Funding: Yu Hu, Liang Tang.
Ethical Approval
No human or animal experiment involved.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
FUNDING
Yu Hu: Integrated Innovative Team for Major Human Diseases Program of Tongji Medical College, HUST; Yu Hu: Clinical Research Physician Program of Tongji Medical College, HUST; Yu Hu: The National Natural Science Foundation of China: No.31620103909; Liang Tang: The Nationa Natural Science Foundation of China: No.81400099; Liang Tang: Novo Nordisk Haemophilia Research Fund.
REFERENCES
- 1.Zwaan CM, Kolb EA, Reinhardt D, Abrahamsson J, Adachi S, Aplenc R, De Bont ES, De Moerloose B, Dworzak M, Gibson BE, Hasle H, Leverger G, Locatelli F, et al. Collaborative Efforts Driving Progress in Pediatric Acute Myeloid Leukemia. Journal of clinical oncology. 2015;33:2949–2962. doi: 10.1200/JCO.2015.62.8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Creutzig U, van den Heuvel-Eibrink MM, Gibson B, Dworzak MN, Adachi S, de Bont E, Harbott J, Hasle H, Johnston D, Kinoshita A, Lehrnbecher T, Leverger G, Mejstrikova E, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. 2012;120:3187–3205. doi: 10.1182/blood-2012-03-362608. [DOI] [PubMed] [Google Scholar]
- 3.Liu B, Ding JF, Luo J, Lu L, Yang F, Tan XD. Seven protective miRNA signatures for prognosis of cervical cancer. Oncotarget. 2016;7:56690–56698. doi: 10.18632/oncotarget.10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vychytilova-Faltejskova P, Radova L, Sachlova M, Kosarova Z, Slaba K, Fabian P, Grolich T, Prochazka V, Kala Z, Svoboda M, Kiss I, Vyzula R, Slaby O. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis. 2016;37:941–950. doi: 10.1093/carcin/bgw078. [DOI] [PubMed] [Google Scholar]
- 5.Du F, Yuan P, Zhao ZT, Yang Z, Wang T, Zhao JD, Luo Y, Ma F, Wang JY, Fan Y, Cai RG, Zhang P, Li Q, et al. A miRNA-based signature predicts development of disease recurrence in HER2 positive breast cancer after adjuvant trastuzumab-based treatment. Scientific reports. 2016;6:33825. doi: 10.1038/srep33825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niyazi M, Pitea A, Mittelbronn M, Steinbach J, Sticht C, Zehentmayr F, Piehlmaier D, Zitzelsberger H, Ganswindt U, Rodel C, Lauber K, Belka C, Unger K. A 4-miRNA signature predicts the therapeutic outcome of glioblastoma. Oncotarget. 2016;7:45764–45775. doi: 10.18632/oncotarget.9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang MK, Chiu YC, Chou WC, Hou HA, Chuang EY, Tien HF. A 3-microRNA scoring system for prognostication in de novo acute myeloid leukemia patients. Leukemia. 2015;29:1051–1059. doi: 10.1038/leu.2014.333. [DOI] [PubMed] [Google Scholar]
- 8.Lionetti M, Musto P, Di Martino MT, Fabris S, Agnelli L, Todoerti K, Tuana G, Mosca L, Gallo Cantafio ME, Grieco V, Bianchino G, D'Auria F, Statuto T, et al. Biological and clinical relevance of miRNA expression signatures in primary plasma cell leukemia. Clinical cancer research. 2013;19:3130–3142. doi: 10.1158/1078-0432.CCR-12-2043. [DOI] [PubMed] [Google Scholar]
- 9.Marcucci G, Maharry K, Radmacher MD, Mrozek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus CD, Liu CG, Ruppert AS, Powell BL, Carroll AJ, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. Journal of clinical oncology. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollink IH, Zwaan CM, Zimmermann M, Arentsen-Peters TC, Pieters R, Cloos J, Kaspers GJ, de Graaf SS, Harbott J, Creutzig U, Reinhardt D, van den Heuvel-Eibrink MM, Thiede C. Favorable prognostic impact of NPM1 gene mutations in childhood acute myeloid leukemia, with emphasis on cytogenetically normal AML. Leukemia. 2009;23:262–270. doi: 10.1038/leu.2008.313. [DOI] [PubMed] [Google Scholar]
- 11.Gale RE, Green C, Allen C, Mead AJ, Burnett AK, Hills RK, Linch DC. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111:2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 12.Lutherborrow M, Bryant A, Jayaswal V, Agapiou D, Palma C, Yang YH, Ma DD. Expression profiling of cytogenetically normal acute myeloid leukemia identifies microRNAs that target genes involved in monocytic differentiation. American journal of hematology. 2011;86:2–11. doi: 10.1002/ajh.21864. [DOI] [PubMed] [Google Scholar]
- 13.Zhao L, Li Y, Song X, Zhou H, Li N, Miao Y, Jia L. Upregulation of miR-181c inhibits chemoresistance by targeting ST8SIA4 in chronic myelocytic leukemia. Oncotarget. 2016;7:60074–60086. doi: 10.18632/oncotarget.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. The New England journal of medicine. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, Radmacher MD, Maharry K, Elkahloun A, Yang X, He C, He M, Zhang Z, et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–2324. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwind S, Maharry K, Radmacher MD, Mrozek K, Holland KB, Margeson D, Whitman SP, Hickey C, Becker H, Metzeler KH, Paschka P, Baldus CD, Liu S, et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Journal of clinical oncology. 2010;28:5257–5264. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nature reviews Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 18.Su R, Lin HS, Zhang XH, Yin XL, Ning HM, Liu B, Zhai PF, Gong JN, Shen C, Song L, Chen J, Wang F, Zhao HL, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene. 2015;34:3226–3239. doi: 10.1038/onc.2014.274. [DOI] [PubMed] [Google Scholar]
- 19.Correia NC, Fragoso R, Carvalho T, Enguita FJ, Barata JT. MiR-146b negatively regulates migration and delays progression of T-cell acute lymphoblastic leukemia. Scientific reports. 2016;6:31894. doi: 10.1038/srep31894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burger ML, Xue L, Sun Y, Kang C, Winoto A. Premalignant PTEN-deficient thymocytes activate microRNAs miR-146a and miR-146b as a cellular defense against malignant transformation. Blood. 2014;123:4089–4100. doi: 10.1182/blood-2013-11-539411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang HM, Li Q, Zhu X, Liu W, Hu H, Liu T, Cheng F, You Y, Zhong Z, Zou P, Li Q, Chen Z, Guo AY. miR-146b-5p within BCR-ABL1-Positive Microvesicles Promotes Leukemic Transformation of Hematopoietic Cells. Cancer research. 2016;76:2901–2911. doi: 10.1158/0008-5472.CAN-15-2120. [DOI] [PubMed] [Google Scholar]
- 22.Geraldo MV, Yamashita AS, Kimura ET. MicroRNA miR-146b-5p regulates signal transduction of TGF-beta by repressing SMAD4 in thyroid cancer. Oncogene. 2012;31:1910–1922. doi: 10.1038/onc.2011.381. [DOI] [PubMed] [Google Scholar]
- 23.Fetahu IS, Tennakoon S, Lines KE, Groschel C, Aggarwal A, Mesteri I, Baumgartner-Parzer S, Mader RM, Thakker RV, Kallay E. miR-135b- and miR-146b-dependent silencing of calcium-sensing receptor expression in colorectal tumors. International journal of cancer. 2016;138:137–145. doi: 10.1002/ijc.29681. [DOI] [PubMed] [Google Scholar]
- 24.Yang W, Yu H, Shen Y, Liu Y, Yang Z, Sun T. MiR-146b-5p overexpression attenuates stemness and radioresistance of glioma stem cells by targeting HuR/lincRNA-p21/beta-catenin pathway. Oncotarget. 2016;7:41505–41526. doi: 10.18632/oncotarget.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Xu J, Li H, Sun C, Yu L, Li Y, Shi C, Zhou X, Bian X, Ping Y, Wen Y, Zhao S, Xu H, et al. miR-146b-5p functions as a tumor suppressor by targeting TRAF6 and predicts the prognosis of human gliomas. Oncotarget. 2015;6:29129–29142. doi: 10.18632/oncotarget.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu R, Liu C, Chen D, Yang WH, Liu X, Liu CG, Dugas CM, Tang F, Zheng P, Liu Y, Wang L. FOXP3 Controls an miR-146/NF-kappaB Negative Feedback Loop That Inhibits Apoptosis in Breast Cancer Cells. Cancer research. 2015;75:1703–1713. doi: 10.1158/0008-5472.CAN-14-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L, Saland E, Decroocq J, Maciel TT, Lambert M, Poulain L, Hospital MA, Sujobert P, et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126:1346–1356. doi: 10.1182/blood-2015-01-621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galluzzi L, Vacchelli E, Michels J, Garcia P, Kepp O, Senovilla L, Vitale I, Kroemer G. Effects of vitamin B6 metabolism on oncogenesis, tumor progression and therapeutic responses. Oncogene. 2013;32:4995–5004. doi: 10.1038/onc.2012.623. [DOI] [PubMed] [Google Scholar]
- 29.Zuo H, Ueland PM, Eussen SJ, Tell GS, Vollset SE, Nygard O, Midttun O, Meyer K, Ulvik A. Markers of vitamin B6 status and metabolism as predictors of incident cancer: the Hordaland Health Study. International journal of cancer. 2015;136:2932–2939. doi: 10.1002/ijc.29345. [DOI] [PubMed] [Google Scholar]
- 30.Ames BN, Wakimoto P. Are vitamin and mineral deficiencies a major cancer risk? Nature reviews Cancer. 2002;2:694–704. doi: 10.1038/nrc886. [DOI] [PubMed] [Google Scholar]
- 31.Pais RC, Vanous E, Hollins B, Faraj BA, Davis R, Camp VM, Ragab AH. Abnormal vitamin B6 status in childhood leukemia. Cancer. 1990;66:2421–2428. doi: 10.1002/1097-0142(19901201)66:11<2421::aid-cncr2820661130>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 32.Moroishi T, Hayashi T, Pan WW, Fujita Y, Holt MV, Qin J, Carson DA, Guan KL. The Hippo Pathway Kinases LATS1/2 Suppress Cancer Immunity. Cell. 2016;167:1525–1539.e17. doi: 10.1016/j.cell.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nature reviews Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 34.Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodelet VC, Cashman NR. Prion protein expression in human leukocyte differentiation. Blood. 1998;91:1556–1561. [PubMed] [Google Scholar]
- 36.Rybner C, Hillion J, Sahraoui T, Lanotte M, Botti J. All-trans retinoic acid down-regulates prion protein expression independently of granulocyte maturation. Leukemia. 2002;16:940–948. doi: 10.1038/sj.leu.2402443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.