Abstract
T-cell-originated protein kinase (TOPK) is highly and frequently expressed in various cancer tissues and plays an indispensable role in the mitosis of cancer cells, and therefore, it is an important target for drug treatment of tumor. Ilaprazole was identified to be a potent TOPK inhibitor. The data indicated that ilaprazole inhibited TOPK activities with high affinity and selectivity. In vitro studies showed that ilaprazole inhibited TOPK activities in HCT116, ES-2, A549, SW1990 cancer cells. Moreover, knockdown of TOPK in these cells decreased their sensitivities to ilaprazole. Results of an in vivo study demonstrated that gavage of ilaprazole in HCT116 colon tumor-bearing mice effectively suppressed cancer growth. The TOPK downstream signaling molecule phospho-histone H3 in tumor tissues was also decreased after ilaprazole treatment. Our results suggested that ilaprazole inhibited the cancer growth by targeting TOPK both in vitro and in vivo.
Keywords: Ilaprazole, PPI, cancer, drug repurposing, TOPK inhibitor
INTRODUCTION
TOPK (T-lymphokine-activated killer cell-originated protein kinase) is a Ser/Thr protein kinase, also known as PBK or PDZ-binding kinase, which belongs to the MEK protein family [1, 2]. The PBK/TOPK protein is up-regulated in certain types of cancer cells such as breast, colon, lung and glioma cancer cells [3–5], but is difficult to be detected in normal tissues except several fetal tissues and germ cells of the testis [6]. In molecular studies, over-expression of TOPK involves in mitotic checkpoint of tumor cells, cell apoptosis and inflammation [7]. In addition, TOPK was also reported to increase cell migration by modulating a PI3K/PTEN/AKT-dependent signaling pathway [8]. Furthermore, histone H3 could be phosphorylated at Ser10 by TOPK both in vivo and in vitro, which could mediate cell growth-promoting effects [9]. Based on these findings, TOPK has become a novel target for cancer treatment attracting more and more attentions [10]. Some TOPK inhibitors, such as HI-TOPK-032 [11] and OTS964 [12] have been reported in 2012 and 2014, respectively. However, none of them had been in clinical trials because of their toxicities or poor pharmacokinetic properties.
Proton pump inhibitors (PPIs), like omeprazole and esomeprazole, are standard treatments for gastric acid-related diseases, acting as potent inhibitors of the gastric acid pump [13]. A few studies showed that PPIs could enhance antitumor effects of cytotoxic drugs in drug-resistant tumors by inhibiting V-ATPase activities or affecting homeostasis, especially acidic vesicles trafficking in cancer cells [14–16]. In our previous study, pantoprazole (PPZ), a PPI was identified as a TOPK inhibitor which could suppress growth of colorectal cancer cells [17]. This is the first report that PPIs can directly inhibit cancer growth via a proton pump independent mechanism.
Inspired by our results of pantoprazole, we speculated that other PPIs may also be developed as TOPK inhibitors. Based on the availability, another six proton pump inhibitors (PPIs) in clinical use were screened against TOPK by virtual ligand screening method, microscale thermophoresis (MST) and cytotoxic activity assay. Ilaprazole, a novel proton pump inhibitor, was found to show potent antitumor activities through the inhibition of TOPK-specific kinase function.
RESULTS
Identification of ilaprazole as TOPK inhibitors
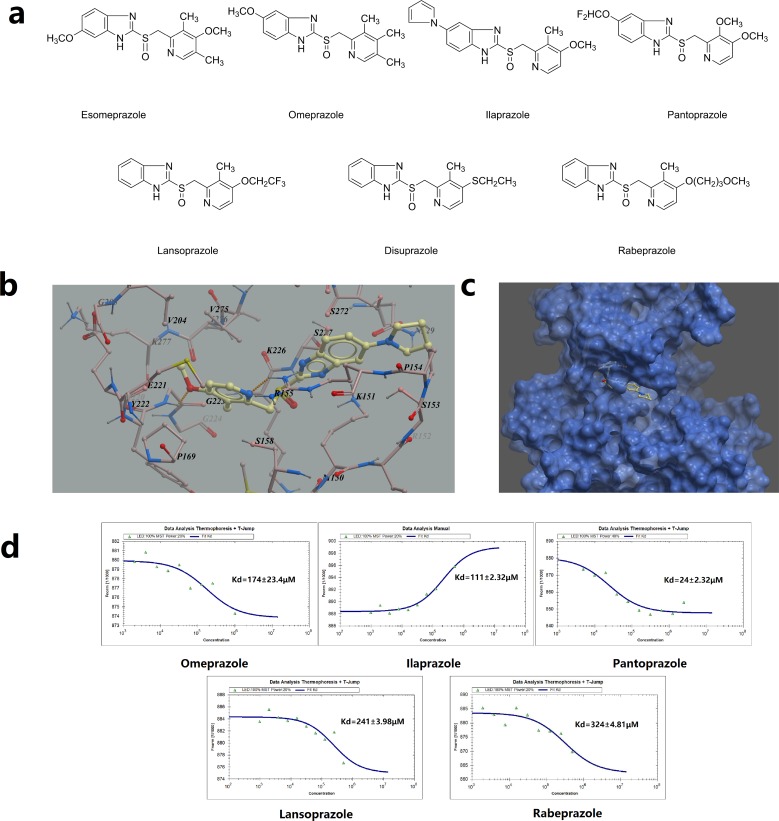
In our previous study, we identified proton pump inhibitor pantoprazole as a potent TOPK inhibitor [13]. It suppressed cancer cell growth both in vitro and in vivo. Encouraged by this finding, we then sought to determine if any drugs from the same class could also potentially inhibit TOPK activities. The homology model of human TOPK was constructed by using the X-ray structure of the mixed-lineage kinase MLK1 (PDB code: 3DTC) as the template, which has 29% of sequence identities to the human TOPK. The docking was performed by using ICM 3.8.1 modeling software (MolSoft LLC, San Diego, CA). As is shown in the Figure 1a, all of the PPIs compounds are structural analogues, weakly basic 2-pyridylmethyl-sulfinyl-benzimidazoles derivatives. The virtual screening results of seven prazoles were shown in Table 1. The top three compounds with highest ICM scores were ilaprazole, lansoprazole and omeprazole. From the generated docking model of ilaprazole, hydrogen bonds were predicted between the methoxyl oxygen of ilaprazole with Gly223, pyridine nitrogen with Arg155, respectively. Also, π-π stacking interactions may form between the pyridine ring of ilaprazole with the pyrrole ring of Pro169, the pyrrole ring of ilaprazole with the pyrrole ring of Pro154 (Figure 1b, 1c).
Figure 1. Virtual ligand screening and MST assay identifies the binding of ilaprazole to TOPK.
(a) Chemical structures of PPIs. (b) Detailed view of ilaprazole binding in the ligand binding pocket. (c) Overlay of one monomer of the TOPK–ilaprazole binary complex. (d) Measurement of affinity between screened drugs with TOPK by MST, and the resulting binding curves were shown.
Table 1. Predicted binding free energies and inhibitory activities of screening hits.
| Prazoles | ICM scores a | Kd (μM) b | IC50 for HCT116 cell activities (μM) |
|---|---|---|---|
| Esomeprazole | −26.24 | n.bc | >500 |
| Omeprazole | −26.24 | 174.3 ± 3.1 | 348.3 ± 7.3 |
| Ilaprazole | −31.88 | 111.0 ± 2.3 | 40.0 ± 0.6 |
| Pantoprazole | −25.65 | 24.2 ± 2.3 | 153.0 ± 8.5 |
| Lansoprazole | −27.35 | 241.6 ± 5.2 | 102.9 ± 4.1 |
| Disuprazole | −19.36 | n.bc | >500 |
| Rabeprazole | −8.63 | 324.2 ± 6.3 | >500 |
a. Docking score/interaction potential of compounds with TOPK (kcal/mol).
b. The Kd value is automatic calculated by the curve fitting, and presents as means ± SD for three experiments.
c. n.b. is no clear binding detected in the MST measurement.
To validate results of the virtual ligand screening, we employed microscale therm ophoresis method (MST) to assay the binding affinity between compounds and TOPK. This technology detected fluorescent changes of molecules during thermophoresis, to quantify protein-protein interactions or protein-small molecule interactions with high sensitivity and low sample cost. Among PPIs assayed, except pantoprazole, ilaprazole exhibited the lowest equilibrium dissociation constant (Kd) of 111.0 ± 2.3 μM (Table 1, Figure 1d), which meant the strongest binding.
Attempts were then made to determine whether PPIs could affect growth of colon cancer cells. As shown in Table 1, seven PPIs exhibited certain levels of inhibition for TOPK over-expressed HCT116 cells with IC50 values from 43.0 to 348.3 μM. The activity ranking was ilaprazole > lansoprazole > pantoprazole for the best three prazoles. Ilaprazole, the one with strongest inhibitory effects, was chosen for further investigations.
TOPK expression in cancer cell lines
TOPK protein levels were measured by western blot in cancer cell lines derived from various cancer types, such as lung cancer (A549), colon cancer (HCT116), human ovarian cancer (ES-2) and pancreatic cancer cell (SW1990). High expression levels of TOPK were observed in all these cell lines (Figure 2a, 2b). The growth inhibitory effects of ilaprazole on each cell line were measured, and obtained IC50 values were ranging from 30.10 to 148.21 μM (Figure 2c). Ilaprazole exhibited potent inhibitory activities against the growth of HCT116 cells (IC50 = 40.0 ± 0.6 μM) and ES-2 cells (IC50 = 33.2 ± 1.0 μM) (Figure 2d, 2e).
Figure 2. In vitro antiproliferative activity of ilaprazole.
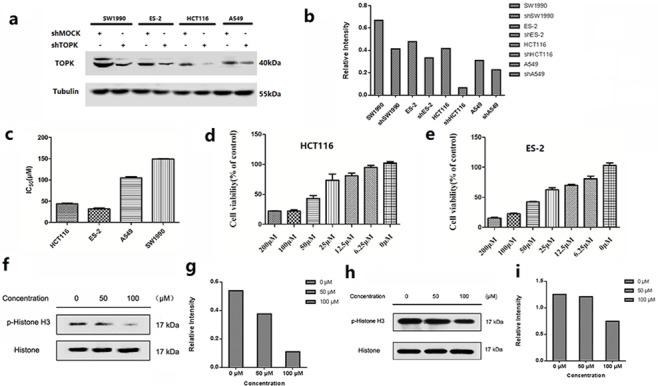
(a, b) Expression of TOPK in lung cancer (A549), colon cancer (HCT-116), human ovarian cancer (ES-2) and pancreatic cancer cell (SW1990) lines. Tubulin was examined to serve as a loading control. (c) Graph indicates IC50 values of ilaprazole in 4 TOPK-positive cancer cell lines. (d, e) The cell proliferation rate of HCT116 and ES-2 were remarkably decreased by ilaprazole. (f, g) Ilaprazole inhibits TOPK activity in HCT116 cells. The expression level of phosphorylated histone H3 (Ser10) and histone H3 was detected by western blot analysis. (h, i) Ilaprazole inhibits TOPK activity in ES-2 cells. The expression level of phosphorylated histone H3 (Ser10) and histone H3 was detected by western blot analysis. The relative intensity of p-Histone and Histone was calculated by an image J software.
Ilaprazole suppresses TOPK activities in vitro and induces apoptosis
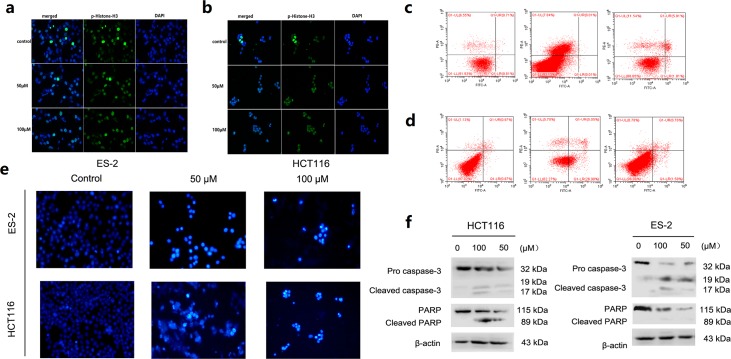
TOPK proteins were expressed mainly around chromosomal surfaces in mitotic cells, particularly at prophase and metaphase, so the histone was chosen as the substrate for TOPK activity evaluation. Some reports confirmed that TOPK could phosphorylate histone H3 in vitro and in vivo. Hence, we examined whether ilaprazole could suppress TOPK activities in HCT116 and ES-2 cells by western blot analysis. Results showed that the phosphorylation of histone H3 (Ser10) was substantially attenuated in a dose-dependent manner after treatment with ilaprazole (Figure 2f–2i). These results suggested that TOPK was a direct target for ilaprazole to suppress cancer cell growth. Immunocytochemical staining results showed that TOPK-mediated p-Histone H3 was substantially decreased dose-dependently with ilaprazole treatment. (Figure 3a, 3b, Supplementary Figure 1a, 1b)
Figure 3. Suppression of TOPK activities and induces apoptosis after treatment with Ilaprazole.
(a, b) Immunocytochemical staining analysis of p-Histone H3 in ES-2 and HCT116 cells. (c, d) ES-2 and HCT116 cells were treated for 24 h with 50 or 100μM of ilaprazole and then processed for FACS by using Annexin V/propidium iodide staining. (e) The cells were treated with ilprazole (100 and 50 μM) for 24 h, then stained with Hoechst 33342 and observed through a fluorescence microscope (×200 magnification). Bar = 20 μm. (f) The cells were incubated with increasing concentrations (0, 50, 100 μM) of ilprazole for 24 h, followed by western blot analysis for the detection of PARP and caspase-3 levels. Data are represented as mean ± standard deviation from triplicate experiments.
Effects of ilaprazole on the cellular apoptosis of HCT116 and ES-2 cells were further investigated. Cells were pre-treated with different concentrations of ilaprzole for 24 h, and then the cellular apoptosis was measured by using flow cytometry. Normalization to the DMSO-treated controls revealed that ilaprazole at 50 μM induced 32.03% (HCT116) and 7.62% (ES-2) apoptotic death compared with 2% of the control (Figure 3c, 3d).
In order to determine effects of cell apoptosis induced by ilaprazole, the cells treated with 100 and 50 μM ilaprazole were stained with Hoechst 33342 and the typical morphological features of later stage apoptosis increased condensation of chromatin material and fragmentation of the nuclei were observed (Figure 3e).
Ilaprazole was also found to induce the cleavage of poly-(ADP-ribose) polymerase (PARP), a DNA repair regulatory protein. Furthermore, the active cleavage forms of caspase-3 were observed after treating with ilaprazole for 24 hours, indicating that it might induce apoptotic cell death by activation of caspases in a dose-dependent manner (Figure 3f). These results suggested that ilaprazole might induce apoptosis in HCT116 and ES-2 cells. In addition, the cell cycle distributions of HCT116 cells were examined after treating with increasing doses of ilaprazole for 48 hours. Flow cytometric analysis demonstrated that ilaprazole induced cell cycle arrested at the G1 phase in HCT116 cells, as shown in Supplementary Figure 2.
The inhibition of TOPK by ilaprazole is dependent on the abundance of TOPK
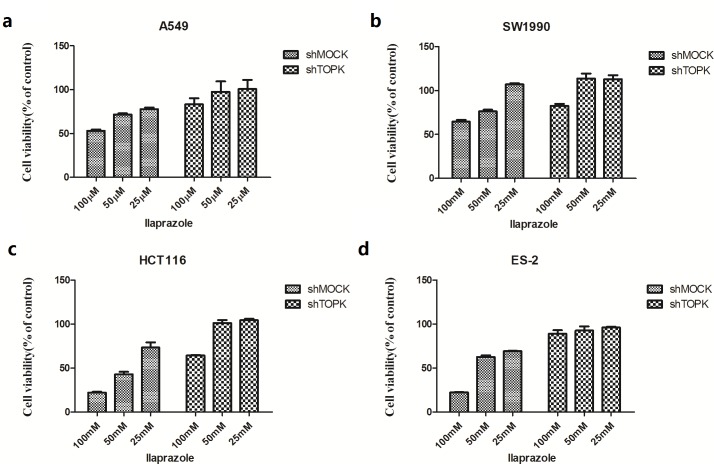
To investigate the growth-promoting role of TOPK in cancer cells, the expression of endogenous TOPK was knocked down in four cancer cells. By lentiviral infection, HCT116, ES-2, A549 and SW1990 cell lines stably expressing low levels of TOPK (shTOPK) were established. As the control, cells were infected with shMock. The abundance of TOPK in these cells was confirmed by western blot analysis (Figure 2a).
Effects of ilaprazole on the growth of shMock and shTOPK infected colon cancer cells were assessed by CCK8 assay. Results indicated that cells infected with shTOPK were resistant to the inhibitory effect of ilaprazole on the growth compared with cells infected with shMock (Figure 4). These findings suggested that anticancer activities induced by ilaprazole were dependent on the TOPK expression.
Figure 4. The inhibition of TOPK by ilaprazole is dependent on the abundance of TOPK.
Ilaprazole has less effect on anchorage-independent cell growth of shTOPK transfected cells than that of shMOCK cells. The effect of ilaprazolewas examined in shMock and knockdown cell lines with shTOPK. Cells were incubated for 48 hours and growth was determined by cck8 assay.
Ilaprazole suppresses tumor growth by inhibiting TOPK activity in vivo
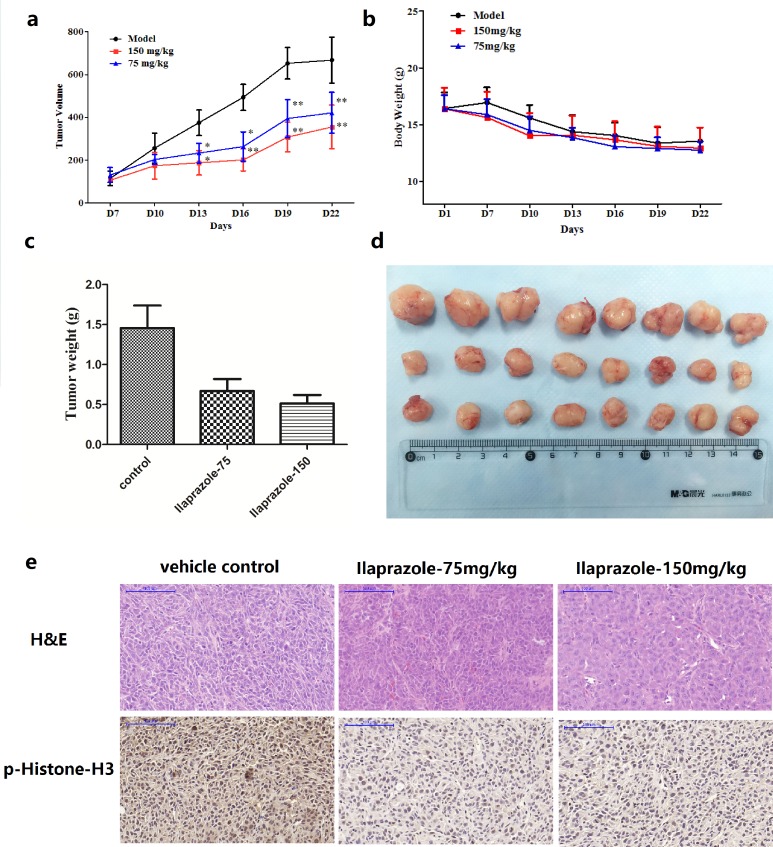
After injected with HCT 116 cells, CB-17/Icr-scid mice were divided into three matched groups including a vehicle control and two ilaprazole treating groups (75, 150 mg/kg). After establishing palpable tumors, mice were administrated with ilaprazole or vehicle via gavage daily. Estimated tumor volumes of both treating groups were less than that of the control group throughout the treatment. Significant decreases of the tumor volume were observed after 2 weeks' treatment (Figure 5a, Figure 5d). Animals were then scarified and the average tumor weights were measured and calculated. The tumor weight of the control group was 1.45 ± 0.33 g, whereas the tumor weights of low-dose and high-dose treated groups were 0.67 ± 0.21g and 0.52 ± 0.11g, respectively (Figure 5c), representing significant reduction. Moreover, no obvious hepatotoxicity and nephrotoxicity, or differences of average body weights among the three groups were observed throughout the study (Figure 5b, Supplementary Figure 3), which revealed that the dosage of ilaprazole used in this experiment had no obvious side effects on animals' growth. To further determine whether antitumor effects of ilaprazole were associated with its inhibition of TOPK activities, tumor extracted from each group were prepared and analyzed for the phosphorylation of histone H3 by immunohistochemistry analysis. Results showed that expression levels of phosphorylated histone H3 were substantially decreased in ilaprazole-treated groups compared with the vehicle group (Figure 5e). Overall, the results indicated that ilaprazole could suppress tumor growth by inhibiting TOPK activities in vivo.
Figure 5. Effect of ilaprazole on colon cancer growth in an HCT116 xenograft mouse model.
(a) The average tumor volume of vehicle-treated control mice (n = 8) and ilaprazole treated mice (n = 8, 75 or 150 mg/kg per day via gavage) plotted over 21 days after tumor cell injection. (b) Ilaprazole has no effect on mouse body weight. (c, d) After 2 weeks treatment, differences in tumor weight and size are shown. The asterisk * indicates a significant increased tumor size (P < 0.05) in the vehicle-treated group compared with the ilaprazole-treated group as determined by one-way analysis of variance. (e) Ilaprazole inhibits expression of phosphorylated histone H3 in vivo. Immunohistochemistry analysis was used to determine the level of phosphorylated histone H3 in tumor tissues.
DISCUSSION
Previous studies have shown that TOPK, a novel serine-threonine kinase, belongs to the MAPKK family. Many groups have reported that TOPK was associated with oncogenic cellular functions including tumor development, cancer growth, and anti-apoptotic effects [1–2, 7–9]. These findings suggest that TOPK is a novel target for development of anticancer agents.
Based on our previous finding that pantoprazole was the inhibitor of TOPK, we screened another 6 prazoles for potential TOPK inhibitory activities, and identified ilaprazole as another potent TOPK inhibitor. According to our results, PPIs' inhibitory effects on TOPK were affected by the presence of steric hindrance near pyridine group and substituents on the benzimidazole. Apparently, disuprazole and rabeprazole exhibited lower ICM scores, presumably because of the larger substituents ethylthio group and methoxypropoxy group respectively, which revealed that the substituent at 4-position of pyridine ring influenced the inhibitory potential as a result of steric hindrance. Results of MST assay and activities on HCT116 cells also supported this hypothesis that steric hindrance of substituents of pyridine ring was one of the crucial factors affecting inhibitory activities. Another important factor affecting inhibitory activities was the substituent at 5-position of the benzimidazole. As an electron-donating group, methoxyl group at 5-position may promote the ability to inhibit TOPK. Consequently, pantoprazole and omeprazole showed a stronger affinity with TOPK. By forming π-π stacking interaction with the pyrrole ring of Pro154, pyrrole ring at 5-position also promoted the ability to inhibit TOPK, consequently making ilaprazole a promising inhibitor against TOPK with lower IC50 value. When physicochemical and pharmacokinetic properties are taken into consideration, it is not difficult to illuminate that ilaprazole exhibited the strongest inhibitory effects in vitro and in vivo. The logP value of ilaprazole is larger than that of pantoprazole according to the data from SciFinder Academic database (logP value of ilaprazole: 2.882 ± 1.045; logP value of pantoprazole: 1.571 ± 1.065). This may offer parts of the explanation for the reason why ilaprazole showed stronger inhibitory effects in vitro and in vivo, while its binding affinity with TOPK was weaker than pantoprazole.
Use of ilaprazole has been shown to inhibit the growth of cancer cells both in vitro and in vivo. Our results indicated that TOPK highly expressed cancer cells were strongly inhibited by ilaprazole and the anticancer activity induced by ilaprazole was dependent on the TOPK expression. The phosphorylations of histone H3 (Ser10), the downstream substrate of TOPK, were strongly reduced in cancer cells by treating with ilaprazole. Notably, the phosphorylations of histone H3 (Ser10) were significantly inhibited in ilaprazole-treated tumor tissues.
As we know, long-term and high-dose PPIs treatment has been shown to be well tolerated in patients with few side effects. [18–20] Therefore, repositioning PPIs as anticancer drugs will be unlikely to have added toxicities. Although PPIs has been proposed as the adjuvant drugs for cancer treatment, and even a clinical trial was performed to evaluate the use of pantoprazole in combination with doxorubicin for advanced cancer patients with solid tumors [21, 22]. However, whether PPIs directly suppress tumor growth remained to be determined.
Ilaprazole is a new PPI that was developed by Livon Pharmaceutical Group, approved in China for the treatment of duodenal ulcer in 2007. The toxicological data showed that the LD50 of ilaprazole was more than 5000 mg/kg in rats [23]. In this study, we have demonstrated that oral administration of ilaprazole 75 mg/kg and 150 mg/kg had no obvious hepatotoxicity and nephrotoxicity, or difference of average body weights. Considering the low toxicities of ilaprazole and effective anti-tumor activities observed in this study, it would be suitable to expand its applications to targeted cancer therapy.
In conclusion, we discovered ilaprazole as a novel and specific TOPK inhibitor both in vitro and in vivo. These findings should be useful for further development of drugs targeting TOPK by employing ilaprazole as the lead compound. Future studies will focus on the efficacy of ilaprazole and characterize its therapeutic potential against other cancers with high expression levels of TOPK.
MATERIALS AND METHODS
Reagents and antibodies
Antibodies against phospho-Histone H3, TOPK, Histone H3, PAPR, caspase 3 were obtained from “Cell Signaling Technology” (USA), β-actin and horseradish peroxidase (HRP) conjugated secondary antibody from rabbit and mouse were purchased from “Santa Cruz” (USA) and “Protein Tech Group” (USA), respectively. The chemiluminescence's detection kit ECL Plus was from “GE Healthcare” (USA). The Dulbecco's Modified Eagle medium (DMEM), Roswell Park Memorial Institute medium (RPMI 1640), phosphate buffered saline (PBS), trypsin, fetal bovine serum (FBS) were purchased from Thermo Scientific. Hoechst 33342 was purchased from Sigma-Aldrich (St. Louis, MO, USA). All other common chemicals, solvents and reagents were of highest grade available from various commercial sources.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), MDC, Rhodamine 123, and Hoechst 33258 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Antibodies against PARP, Bcl-xL, Bax, cytochrome c, caspase-8, caspase-9, β-actin, p62, LC3-II, Beclin-1, p-p38, p-38, p-JNK, JNK, p-Erk, Erk, Histone H3 and NF-κB were purchased from Cell Signaling Technology (CST, Beverly, MA). NF-κB SN50 was from Merck Millipore (USA).
Cell culture
Human cancer cells lines, HCT116 (ATCC # CCL-247™), ES-2 (ATCC # CRL-1978™), A549 (ATCC # CRM-CCL-185™) and SW1990 (ATCC # CRL-2172™), were cultured under the recommendations of their respective depositors. All cell lines were authenticated by STR analysis from Shanghai Zhong Qiao Xin Zhou Biotechnology Co.,Ltd.
The cells were grown for 2-3 days, after reaching 90% of confluence, were then harvested by exposure to 0.25% Trypsin-EDTA solution and then passed into new T-25 tissue culture flasks. The cell cultures were performed following the instructions of ATCC.
Protein expression, purification
Human TOPK gene (Accssion number: NM_018492) was cloned into the pET26b vector (Novagen). The expression construct pET26b-TOPK was transformed into Escherichia coli strain BL21 (DE) (Invitrogen) and selected on kanamycin plates. The transformed cells were cultivated in Luria-Bertani (LB) media at 37°C in the presence of kanamycin until the optical density (OD) reached 0.8. Cells were then induced with 0.4 mM IPTG (isopropyl-β-D-1-thiogalactopyranoside) for 16 h at 20°C. The cells harvested by centrifugation were lysed by ultrasonication on ice in a buffer containing 20 mM Tris, pH 8.5, 200 mM NaCl, 5 mM mercaptoethanol, 0.1% TritonX-100, and 5% glycerol.
Soluble hexa-histidine tagged TOPK was bound to Ni-agarose affinity resin (Qiagen), washed with buffer A (20 mM Tris, pH 8.5, 200mM NaCl, and 10 mM imidazole), and eluted with buffer B (20 mM Tris, pH 8.8, 250 mM NaCl, and 150 mM imidazole).
Microscale thermophoresis (MST)
Recombinant TOPK was labeled with the Monolith NT™ Protein Labeling Kit RED (Cat # L001) according to the supplied labeling protocol. Labeled TOPK was used in a concentration of 50 nM. The samples were diluted in a 20 mM HEPES (pH 7.4) and 0.5 (v/v) % Tween-20. The prazoles stocks were dissolved in 10% DMSO in a concentration of 1 mM. 1mM prazoles were used as the highest concentration for serial dilutions. After 10minutes incubation at room temperature, the samples were loaded into MonolithTM standard-treated capillaries and the thermophoresis was measured at 25°C after 30 minutes incubation on a Monolith NT.115 instrument (NanoTemper Technologies, München, Germany). Laser power was set to 20% using 30 seconds on-time. The LED power was set to 100%. The dissociation constant Kd values were fitted by using the NTAnalysis software (NanoTemper Technologies, München, Germany) [24].
Homology modeling and molecular docking
With the crystal structure of the mixed-lineage kinase MLK1 (PDB code: 3DTC) as the template, a homology model of human TOPK (accession number: NP_060962) was constructed by using MODELLER (an automated homology modeling program) [25, 26]. Chemical structures of prazoles were input as mol2 files for docking [21]. The docking was performed by using ICM 3.8.1 modeling software on an Intel i7 4960 processor (MolSoft LLC, San Diego, CA) [27]. Ligand binding pocket residues were selected by using graphical tools in the ICM software, to create the boundaries of the docking search. In the docking calculation, potential energy maps of the receptor were calculated using default parameters. The compounds were imported into ICM and an index file was created. The conformational sampling was based on the Monte Carlo procedure, and finally the lowest-energy and the most favorable orientation of the ligand was selected.
Cell viability assay
To estimate cell viability, human colon cancer HCT 116, ES-2, A549 and SW1990 cells (5000 cells/well) were seeded in 96-well plates for 24 h at 37°C in a 5% CO2 incubator. The attached cells were fed with fresh medium containing various concentrations of ilaprazole (0-100 μM) for additional 24 h and 48 h. After culturing for various times, the cytotoxicity of ilaprazole was measured using a cck8 assay kit according to the manufacturer's instructions. All experiments were performed in triplicate, and the mean absorbance values were calculated. The results are expressed as the percentage of inhibition that produced a reduction in absorbance by ilaprazole treatment compared with the non-treated cells (control).
Isolation of histone H3
Histones were extracted from cells by disrupting the cells with NETN buffer [150 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl (pH 8.0), 0.5% nonionic detergent IGEPAL CA 630(NP-40), Sigma]. The insoluble fraction was pelleted for 5 minutes in a microcentrifuge (8,400 rpm). Nuclei were extracted with 0.1 M HCl to isolate the total histones. The samples were precipitated with 1 M Tris- HCl (pH 8.0) and then resuspended in ddH2O.
Western blot analysis
The harvested cells were lysed with lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10 mg/mL aprotinin, 10 mg/mL leupeptin, 5 mM phenylmethanesulfonyluoride (PMSF), 1 mM dithiolthreitol (DTT) containing 1% Triton X-100). Insoluble debris was removed by centrifugation at 12000 rpm for 15 minutes, and protein's content was determined using Bradford reagent (Bio-Rad, USA). Lysate protein (20-40 μg) was subjected to 10% SDS-PAGE and electrophoretically transferred to polyvinylidene difluoride membranes (PVDF) (Millipore, USA). The membranes were blocked with 5% non-fat milk or 5% BSA for 1 hour and then incubated with the specific primary antibodies respectively at 4°C overnight. Protein bands were visualized using an enhanced chemiluminescence reagent (ECL Plus) (GE Healthcare, USA) after hybridization with a HRP conjugated secondary antibody. Band density was quantified using the Image J software program (NIH).
Apoptosis analysis
Apoptotic cells were assessed by staining PBS washed cells with phycoerythrin (PE)-conjugated annexin-V (BD Pharmingen) for 10-15 minutes in incubation buffer (Annexin-V-FLUOS Staining Kit, Roche). Fluorescence was recorded by flow cytometry using the FACS Aria (BD Bio sciences).
Hoechst 33342 staining
After incubation with ilaprazole for 24 h, the cells were stained with Hoechst 33342 at 37°C for 30 min, and then the morphology was observed by fluorescence microscopy (Zeiss, OBSERVER D1/AX10 cam HRC).
Lentiviral infection
The lentiviral expression vectors, including Gipz-shTOPK, and packaging vectors, including pMD2.0G and psPAX, were purchased from Addgene Inc. To prepare TOPK viral particles, each viral vector and packaging vectors (pMD2.0G and psPAX) were transfected into HEK293T cells using jetPEI following the manufacturer's suggested protocols. “The target sequences of oligo siRNAs were as follows: 5′-GTGTGGCTTGCGTAAATAA -3′ for TOPK to produce lentiviral particles.”
The transfection medium was changed at 4 hours after transfection and then cells were cultured for 36 hours. The viral particles were harvested by filtration using a 0.45-mm syringe filter, then combined with 8 mg/mL of polybrene (Millipore) and infected into 60% confluent HCT-116 cells overnight. The cell culture medium was replaced with fresh complete growth medium for 24 hours and then cells were selected with puromycin (1.5 mg/mL) for 36 hours. The selected cells were used for experiments [28, 29].
In vivo xenograft mouse model
CB-17/Icr-scid mice were purchased from Beijing HFK Bioscience CO., LTD (Beijing, China). The animals were maintained under ‘specific pathogen free’ conditions. The mice were randomly divided into three groups: (i) vehicle group (n = 8); (ii) 150 mg/kg ilaprazole-treated group (n = 8); (III) 75 mg/kg ilaprazole-treated group. HCT 116 cells were inoculated subcutaneously (2 × 106 cells) into the left flank of each mouse in all groups. Treatment was started after 4 days of cell injection. The duration of the animal study was 19 days. The tumor volume was calculated from measurements of 3 diameters of the individual tumor based on the following formula:
tumor volume (mm3) = (length × width × height × 0.52). The mice were monitored until tumors reached 1 cm3 total volume, at which the mice were euthanized and the tumors were extracted. The tumors were dissected and sent for immune-histochemical analysis.
Statistical analysis
Statistical analysis of the data was performed using Graph Pad Prism 5.0 software. The data were expressed as the means ± SD. Values were analyzed using SPSS version 12.0 software by one-way analysis of variance (ANOVA), and p < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIALS FIGURES
Abbreviations
- PPIs
Proton Pump Inhibitors
- TOPK
T-cell-originated protein kinase
- PPZ
pantoprazole
- MST
microscale thermophoresis
- CCK8
Cell Counting Kit-8
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- PMSF
phenylmethanesulfonyluoride
- PVDF
Polyvinylidene
- H&E
Hematoxylin-eosin
Footnotes
Authors' contributions
HL, LXC and XMH designed experiments and provided material support. MZZ and SSL performed experiments, analyzed the data and drafted the manuscript. SYG, LC, BH, JCL and YC carried out parts of the experiments. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Abe Y, Matsumoto S, Kito K, Ueda N. Cloning and expression of a novel MAPKK-like protein kinase, lymphokine-activated killer T-cell-originated protein kinase, specifically expressed in the testis and activated lymphoid cells. J Biol Chem. 2000;275:21525–21531. doi: 10.1074/jbc.M909629199. [DOI] [PubMed] [Google Scholar]
- 2.Gaudet S, Branton D, Lue RA. Characterization of PDZ-binding kinase, a mitotic kinase. Proc Natl Acad Sci U S A. 2000;97:5167–5172. doi: 10.1073/pnas.090102397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park JH, Nishidate T, Nakamura Y, Katagiri T. Critical roles of T-LAK cell-originated protein kinase in cytokinesis. Cancer Sci. 2010;101:403–411. doi: 10.1111/j.1349-7006.2009.01400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simons-Evelyn M, Bailey-Dell K, Toretsky JA, Ross DD, Fenton R, Kalvakolanu D, Rapoport AP. PBK/TOPK is a novel mitotic kinase which is up regulated in Burkitt's lymphoma and other highly proliferative malignant cells. Blood Cells Mol Dis. 2001;27:825–829. doi: 10.1006/bcmd.2001.0452. [DOI] [PubMed] [Google Scholar]
- 5.Park JH, Jeong YJ, Won HK, Choi SY, Park JH, Oh SM. Activation of TOPK by lipopolysaccharide promotes induction of inducible nitric oxide synthase through NF-κB activity in leukemia cells. Cell Signal. 2014;26:849–856. doi: 10.1016/j.cellsig.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Park JH, Lin ML, Nishidate T, Nakamura Y, Katagiri T. PDZ-binding kinase/T-LAK cell-originated protein kinase, a putative cancer/testis antigen with an oncogenic activity in breast cancer. Cancer Res. 2006;66:9186–9195. doi: 10.1158/0008-5472.can-06-1601. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto S, Abe Y, Fujibuchi T, Takeuchi T, Kito K, Ueda N, Shigemoto K, Gyo K. Characterization of a MAPKK-like protein kinase PBK/TOPK. Biochem Biophys Res Commun. 2004;325:997–1004. doi: 10.1016/j.bbrc.2004.10.133. [DOI] [PubMed] [Google Scholar]
- 8.Shih MC, Chen JY, Wu YC, Jan YH, Yang BM, Lu PJ, Cheng HC, Huang MS, Yang CJ, Hsiao M, Lai JM. TOPK/PBK promotes cell migration via modulation of the PI3K/PTEN/AKT pathway and is associated with poor prognosis in lung cancer. Oncogene. 2012;31:2389–2400. doi: 10.1038/onc.2011.419. [DOI] [PubMed] [Google Scholar]
- 9.Kwon HR, Lee KW, Dong Z, Lee KB, Oh SM. Requirement of T-lymphokine-activated killer cell-originated protein kinase for TRAIL resistance of human HeLa cervical cancer cells. Biochem Biophys Res Commun. 2010;391:830–834. doi: 10.1016/j.bbrc.2009.11.147. [DOI] [PubMed] [Google Scholar]
- 10.Hu F, Gartenhaus RB, Eichberg D, Liu Z, Fang HB, Rapoport AP. PBK/TOPK interacts with the DBD domain of tumor suppressor p53 and modulates expression of transcriptional targets including p21. Oncogene. 2010;29:5464–5474. doi: 10.1038/onc.2010.275. [DOI] [PubMed] [Google Scholar]
- 11.Kim DJ, Li Y, Reddy K, Lee MH, Kim MO, Cho YY, Lee SY, Kim JE, Bode AM, Dong Z. Novel TOPK inhibitor HI-TOPK-032 effectively suppresses colon cancer growth. Cancer Res. 2012;72:3060–3068. doi: 10.1158/0008-5472.can-11-3851. [DOI] [PubMed] [Google Scholar]
- 12.Matsuo Y, Park JH, Miyamoto T, Yamamoto S, Hisada S, Alachkar H, Nakamura Y. TOPK inhibitor induces complete tumor regression in xenograft models of human cancer through inhibition of cytokinesis. Sci Transl Med. 2014;6:251–259. doi: 10.1126/scitranslmed.3010277. [DOI] [PubMed] [Google Scholar]
- 13.Ayllon V, O'Connor R. PBK/TOPK promotes tumour cell proliferation through p38 MAPK activity and regulation of the DNA damage response. Oncogene. 2007;26:3451–3461. doi: 10.1038/sj.onc.1210142. [DOI] [PubMed] [Google Scholar]
- 14.Mattsson JP, Vaananen K, Wallmark B, Lorentzon P. Omeprazole and bafilomycin, two proton pump inhibitors: differentiation of their effects on gastric, kidney and bone H(+)-translocating ATPases. Biochim Biophys Acta. 1991;1065:261–268. doi: 10.1016/0005-2736(91)90238-4. [DOI] [PubMed] [Google Scholar]
- 15.Na Z, Pan S, Uttamchandani M, Y SQ. Protein-Protein Interaction Inhibitors of BRCA1 Discovered Using Small Molecule Microarrays. Methods Mol Biol. 2017;1518:139–156. doi: 10.1007/978-1-4939-6584-7_10. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Li L, Fu WT, Jiang ZY, You QD, Xu XL. Optimization and bioevaluation of Cdc37-derived peptides: An insight into Hsp90-Cdc37 protein-protein interaction modulators. Bioorg Med Chem. 2017;25:233–240. doi: 10.1016/j.bmc.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 17.Zeng X, Liu L, Zheng M, Sun H, Xiao J, Lu T, Huang G, Chen P, Zhang J, Zhu F, Li H, Duan Q. Pantoprazole, an FDA-approved proton-pump inhibitor, suppresses colorectal cancer growth by targeting T-cell-originated protein kinase. Oncotarget. 2016;7:22460–73. doi: 10.18632/oncotarget.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fako VE, Wu X, Pflug B, Liu JY, Zhang JT. Repositioning proton pump inhibitors as anticancer drugs by targeting the thioesterase domain of human fatty acid synthase. J Med Chem. 2015;58:778–84. doi: 10.1021/jm501543u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson AB, Sauve MD, Kassam N, Kamitakahara H. Safety of the long-term use of proton pump inhibitors. World J Gastroenterol. 2010;16:2323–2330. doi: 10.3748/wjg.v16.i19.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang YX, Metz DC. Safety of proton pump inhibitor exposure. Gastroenterology. 2010;139:1115–1127. doi: 10.1053/j.gastro.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 21.Brana I, Ocana A, Chen EX, Razak AR, Haines C, Lee C, Douglas S, Wang L, Siu LL, Tannock IF, Bedard PL. A phase I trial of pantoprazole in combination with doxorubicin in patients with advanced solid tumors: evaluation of pharmacokinetics of both drugs and tissue penetration of doxorubicin. Invest New Drugs. 2014;32:1269–77. doi: 10.1007/s10637-014-0159-5. [DOI] [PubMed] [Google Scholar]
- 22.Shin JS, Lee JY, Cho KH, Park HL, Kukulka M, Wu JT, Kim DY, Park SH. The pharmacokinetics, pharmacodynamics and safety of oral doses of ilaprazole 10, 20 and 40 mg and esomeprazole 40 mg in healthy subjects: a randomised, open-label crossover study. Aliment Pharmacol Ther. 2014;40:548–61. doi: 10.1111/apt.12860. [DOI] [PubMed] [Google Scholar]
- 23.Liu T, Chang Y, Han R. Action mechanism and clinical evaluation of ilaprazole. Evaluation and analysis of drug-use in hospitals of China. 2009(9):724–725. [Google Scholar]
- 24.Kato T, Inoue H, Imoto S, Tamada Y, Miyamoto T, Matsuo Y, Nakamura Y, Park JH. Oncogenic roles of TOPK and MELK, and effective growth suppression by small molecular inhibitors in kidney cancer cells. Oncotarget. 2016;7:17652–64. doi: 10.18632/oncotarget.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiser A, Šali A. Modeller: Generation and refinement of homology-based protein structure models. Methods in Enzymology. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 26.Kirubakaran P, Karthikeyan M. In silico structural and functional analysis of the human TOPK protein by structure modeling and molecular dynamics studies. Journal of Molecular Modeling. 2013;19:407–419. doi: 10.1007/s00894-012-1566-1. [DOI] [PubMed] [Google Scholar]
- 27.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52:1757–68. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abagyan R, Totrov M, Kuznetsov D. ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. Journal of Computational Chemistry. 1994;15:488–506. doi: 10.1002/jcc.540150503. [DOI] [Google Scholar]
- 29.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.