Abstract
The BRCA2 gene plays an important role in cancer carcinogenesis, and polymorphisms in this gene have been associated with cancer risk. The BRCA2 rs144848 polymorphism has been associated with several cancers, but results have been inconsistent. In the present study, a meta-analysis was performed to assess the association between the rs144848 polymorphism and cancer risk. Literature was searched from the databases of PubMed, Embase and Google Scholar before April 2016. The fixed or random effects model was used to calculate pooled odd ratios on the basis of heterogeneity. Meta-regression, sensitivity analysis, subgroup analysis and publication bias assessment were also performed using STATA 11.0 software according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses 2009. A total of 40 relevant studies from 30 publications including 34,911 cases and 48,329 controls were included in the final meta-analysis. Among them, 22 studies focused on breast cancer, seven on ovarian cancer, five on non-Hodgkin lymphoma, and the remaining six studies examined various other cancers. The meta-analysis results showed that there were significant associations between the rs144848 polymorphism and cancer risk in all genetic models. Stratified by cancer type, the rs144848 polymorphism was associated with non-Hodgkin lymphoma. Stratified by study design, the allele model was associated with breast cancer risk in population-based studies. The meta-analysis suggests that the BRCA2 rs144848 polymorphism may play a role in cancer risk. Further well-designed studies are warranted to confirm these results.
Keywords: meta-analysis, BRCA2, cancer, polymorphism, susceptibility
INTRODUCTION
Cancer is one of the most common diseases causing considerable morbidity and mortality worldwide. Environmental and genetic factors together contribute to the development of cancers [1–4]. It has been reported that DNA damage and repair is an important factor in carcinogenesis [5–7]. BRCA2 is a well-known cancer susceptibility gene involved in the repair of double-stranded DNA breaks which functions by regulating the intracellular shuttling and activity of RAD51, another critical protein in homologous recombination [8–10]. Studies have shown that cancer carcinogenesis is related to abnormalities in DNA repair mechanisms partially caused by a change in gene function which can result from genetic polymorphisms [11, 12].
Within the last few years, many studies have focused on the association between BRCA2 gene polymorphisms and cancer risk, including breast cancer, ovarian cancer, non-Hodgkin lymphoma, prostate cancer and others [13–18]. The rs144848 is the only common non-synonymous polymorphism in exon 10 of the BRCA2 gene [19]. The change from A to C in the rs144848 polymorphism results in an asparagine-to-histidine transition (N372H) which may affect BRCA2 structure at residues 290-453, a region which has been determined to interact with the histone acetyltransferase P/CAF prior to transcriptional activation of target genes [20]. Over the past decade, many association studies have been conducted to explore the role of the rs144848 N372H polymorphism in cancer risk [13, 15, 17, 18, 21–40], but it is still inconclusive whether this polymorphism in the BRCA2 gene is associated with susceptibility to cancer. Therefore, we performed a systematic review and meta-analysis of published studies focused on the association between the rs144848 polymorphism and cancer risk. Our in-depth analysis may drive a more precise estimation of risk which could in turn help identify additional genetic targets for future therapeutic interventions.
RESULTS
Study characteristics
A flow diagram for the search strategy is shown in Figure 1. Based on the search strategy, 2,174 articles were identified in the initial search. After reading titles and abstracts, 1,788 articles were excluded and 386 articles were reviewed for full text. According to the study inclusion/exclusion criteria, 40 relevant studies from 30 publications including 34,911 cases and 48,329 controls were used for the final meta-analysis [13–15, 17, 18, 21, 23–40, 46–52]. Nine studies were medium quality and 31 studies were high quality. The main characteristics of these included studies are shown in Table 1.
Figure 1. Study flow diagram.

Table 1. Characteristics of included studies that contributed to associations between rs144848 and cancer risk.
| Study [ref] per SNP | Year | Race/ethnicity | Sourcea | Cases | Controls | Allele frequencies | NOS assessment | Cancer type | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | NN | NH | HH | Total | NN | NH | HH | Casesb | Controlsb | ||||||
| Healey et al. [12] | 2000 | Caucasian | PB | 234 | 116 | 99 | 19 | 266 | 138 | 115 | 13 | 0.71 | 0.73 | 7 | Breast |
| Healey et al. [12] | 2000 | Caucasian | PB | 1667 | 858 | 664 | 145 | 1201 | 631 | 493 | 77 | 0.71 | 0.73 | 7 | Breast |
| Healey et al. [12] | 2000 | Caucasian | PB | 450 | 236 | 180 | 34 | 228 | 124 | 94 | 10 | 0.72 | 0.75 | 7 | Breast |
| Healey et al. [12] | 2000 | Caucasian | PB | 659 | 325 | 285 | 49 | 866 | 433 | 373 | 60 | 0.71 | 0.72 | 7 | Breast |
| Healey et al. [12] | 2000 | Caucasian | PB | 449 | 270 | 154 | 25 | 453 | 277 | 152 | 24 | 0.77 | 0.78 | 7 | Breast |
| Spurdle et al. [45] | 2002 | Caucasian | PB | 1397 | 720 | 548 | 129 | 775 | 417 | 308 | 50 | 0.71 | 0.74 | 7 | Breast |
| Ishitobi et al. [22] | 2003 | Asian | HB | 149 | 97 | 47 | 5 | 144 | 85 | 56 | 3 | 0.81 | 0.78 | 7 | Breast |
| Menzel et al. [24] | 2004 | Caucasian | PB | 211 | 104 | 91 | 16 | 912 | 482 | 361 | 69 | 0.71 | 0.73 | 7 | Breast |
| Menzel et al. [24] | 2004 | Caucasian | PB | 94 | 53 | 35 | 6 | 152 | 84 | 57 | 11 | 0.75 | 0.74 | 7 | Breast |
| Cox et al. [44] | 2005 | Caucasian | Nested | 1285 | 695 | 501 | 89 | 1660 | 884 | 647 | 129 | 0.74 | 0.73 | 7 | Breast |
| Millikan et al. [25] | 2005 | African | PB | 762 | 564 | 183 | 15 | 675 | 510 | 153 | 12 | 0.86 | 0.87 | 7 | Breast |
| Millikan et al. [25] | 2005 | Caucasian | PB | 1265 | 662 | 521 | 82 | 1135 | 579 | 467 | 89 | 0.73 | 0.72 | 7 | Breast |
| Garcia-Closas et al. [21] | 2006 | Caucasian | PB | 3161 | 1617 | 1278 | 266 | 2701 | 1412 | 1057 | 232 | 0.71 | 0.72 | 7 | Breast |
| Garcia-Closas et al. [21] | 2006 | Caucasian | PB | 1968 | 1007 | 826 | 135 | 2276 | 1239 | 897 | 140 | 0.72 | 0.74 | 7 | Breast |
| Johnson et al. [47] | 2007 | Caucasian | NA | 473 | 233 | 201 | 39 | 2461 | 1278 | 993 | 190 | 0.71 | 0.72 | 6 | Breast |
| Palli et al. [48] | 2007 | Caucasian | PB | 91 | 48 | 31 | 12 | 261 | 127 | 107 | 27 | 0.70 | 0.69 | 6 | Breast |
| Baynes et al. [46] | 2007 | Caucasian | PB | 4537 | 2306 | 1892 | 339 | 4339 | 2182 | 1824 | 333 | 0.72 | 0.71 | 7 | Breast |
| Seymour et al. [49] | 2008 | Caucasian | HB | 252 | 127 | 111 | 14 | 100 | 50 | 44 | 6 | 0.72 | 0.72 | 6 | Breast |
| Dombernowsky et al. [19] | 2009 | Caucasian | PB | 1200 | 604 | 503 | 93 | 4119 | 2129 | 1677 | 313 | 0.71 | 0.72 | 6 | Breast |
| Juwle et al. [23] | 2012 | Asian | NA | 100 | 68 | 28 | 4 | 50 | 39 | 8 | 3 | 0.82 | 0.86 | 6 | Breast |
| Hasan et al. [11] | 2013 | African | HB | 100 | 38 | 33 | 29 | 100 | 33 | 32 | 35 | 0.55 | 0.49 | 6 | Breast |
| Jumaah et al. [50] | 2014 | African | NA | 36 | 26 | 10 | 0 | 10 | 10 | 0 | 0 | 0.86 | 1.00 | 6 | Breast |
| Auranen et al. [26] | 2003 | Caucasian | PB | 680 | 355 | 272 | 53 | 1546 | 819 | 629 | 98 | 0.72 | 0.73 | 7 | Ovarian |
| Auranen et al. [26] | 2003 | Caucasian | PB | 441 | 222 | 176 | 43 | 1097 | 578 | 445 | 74 | 0.70 | 0.73 | 7 | Ovarian |
| Wenham et al. [28] | 2003 | Caucasian | PB | 312 | 169 | 128 | 15 | 398 | 227 | 146 | 25 | 0.75 | 0.75 | 7 | Ovarian |
| Beesley et al. [32] | 2007 | Caucasian | PB | 492 | 249 | 203 | 40 | 948 | 502 | 383 | 63 | 0.71 | 0.73 | 8 | Ovarian |
| Beesley et al. [32] | 2007 | Caucasian | PB | 930 | 460 | 401 | 69 | 825 | 461 | 296 | 68 | 0.71 | 0.74 | 8 | Ovarian |
| Ramus et al. [36] | 2008 | Mixed | Nested | 4174 | 2196 | 1655 | 323 | 7402 | 3859 | 2979 | 564 | 0.72 | 0.72 | 7 | Ovarian |
| Quaye et al. [37] | 2009 | Caucasian | PB | 1459 | 779 | 569 | 111 | 2294 | 1200 | 925 | 169 | 0.73 | 0.72 | 7 | Ovarian |
| Shen et al. [30] | 2006 | Mixed | PB | 476 | 250 | 191 | 35 | 555 | 301 | 220 | 34 | 0.73 | 0.74 | 7 | NHLc |
| Scott et al. [33] | 2007 | Caucasian | PB | 757 | 387 | 307 | 63 | 676 | 375 | 253 | 48 | 0.71 | 0.74 | 7 | NHL |
| Shen et al. [34] | 2007 | Caucasian | PB | 556 | 271 | 236 | 49 | 498 | 246 | 203 | 49 | 0.70 | 0.70 | 7 | NHL |
| Hill et al. [16] | 2006 | Mixed | PB | 1116 | 577 | 441 | 98 | 926 | 505 | 361 | 60 | 0.71 | 0.74 | 7 | NHL |
| Salagovic et al. [39] | 2012 | Caucasian | HB | 107 | 62 | 34 | 11 | 127 | 82 | 40 | 5 | 0.74 | 0.80 | 7 | NHL |
| Hu et al. [27] | 2003 | Asian | PB | 120 | 69 | 39 | 12 | 231 | 126 | 95 | 10 | 0.74 | 0.75 | 6 | Esophageal |
| Wu et al. [31] | 2006 | Caucasian | PB | 604 | 306 | 246 | 52 | 595 | 332 | 223 | 40 | 0.71 | 0.75 | 8 | Bladder |
| Debniak et al. [35] | 2008 | Caucasian | Nested | 627 | 288 | 280 | 59 | 3819 | 1994 | 1580 | 245 | 0.68 | 0.73 | 6 | Melanoma |
| Agalliu et al. [15] | 2010 | Caucasian | PB | 1269 | 655 | 498 | 116 | 1243 | 654 | 500 | 89 | 0.71 | 0.73 | 8 | Prostate |
| Agalliu et al. [15] | 2010 | African | PB | 142 | 104 | 36 | 2 | 79 | 59 | 18 | 2 | 0.86 | 0.86 | 8 | Prostate |
| Kotnis et al. [38] | 2012 | Asian | HB | 109 | 35 | 56 | 18 | 186 | 81 | 70 | 35 | 0.58 | 0.62 | 7 | Multiple |
a Source in control, PB population-based study, HB hospital-based study
b Major allele frequency
c non-Hodgkin lymphoma
Association between BRCA2 rs144848 polymorphism and cancer risk
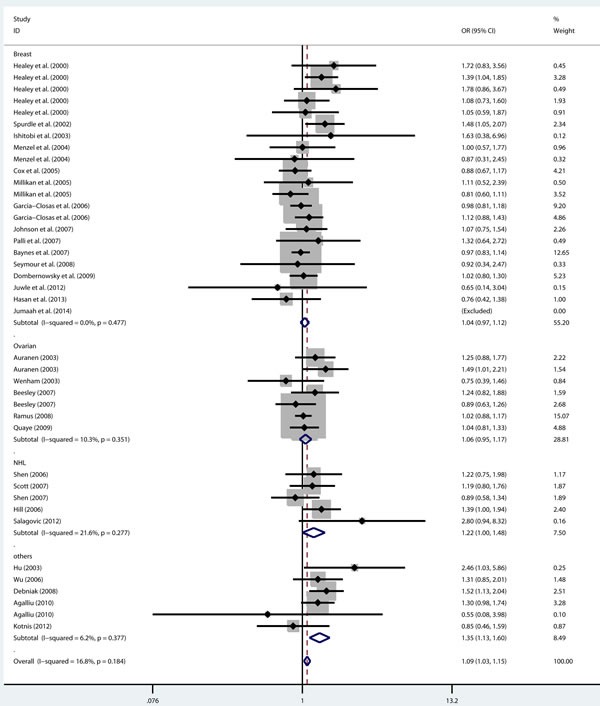
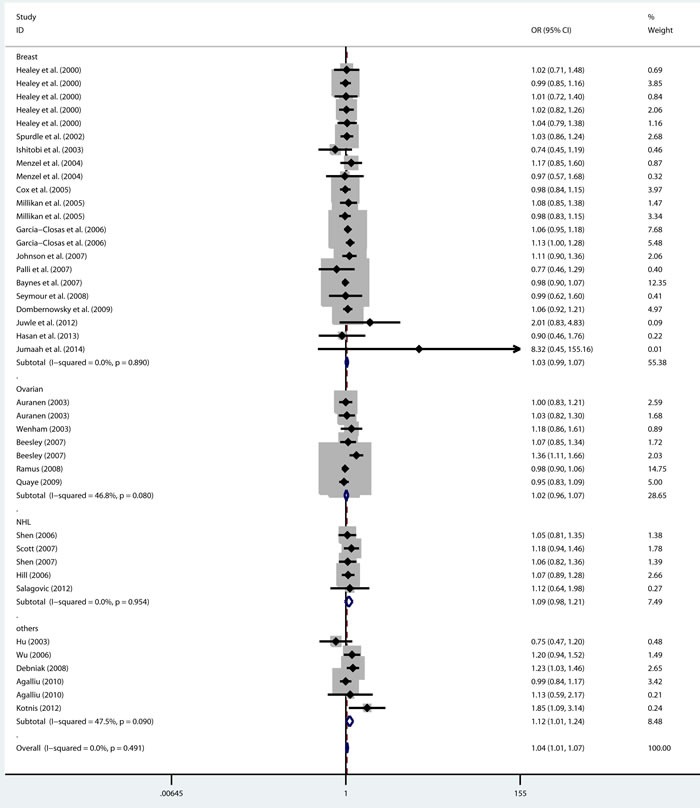
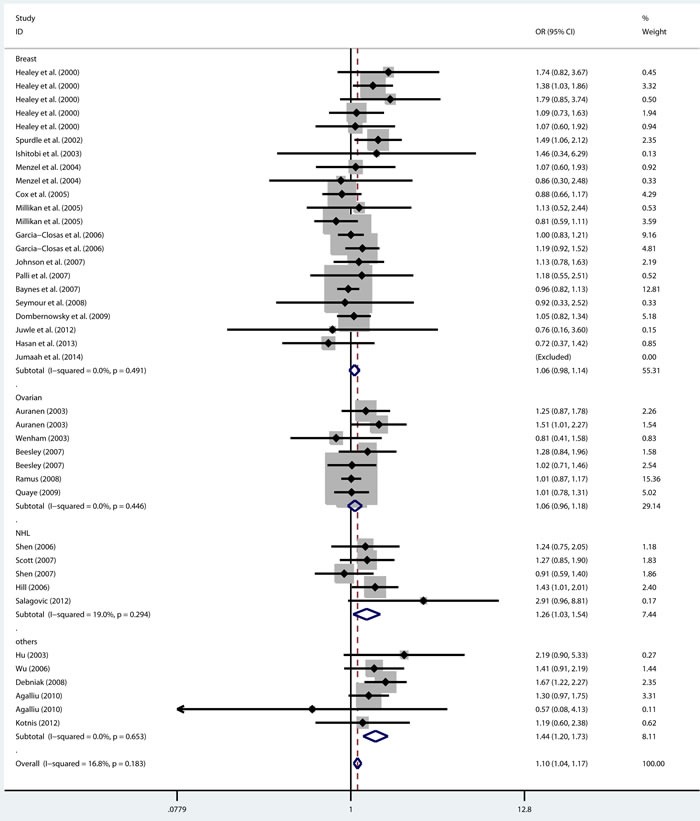
As shown in Table 2, there was no heterogeneity in any genetic model. The meta-analysis results showed that there were significant associations between the rs144848 polymorphism and cancer risk in all genetic models (H allele vs. N allele, OR = 1.044, 95% CI = 1.021-1.068, p < 0.001; NH vs. NN, OR = 1.037, 95% CI = 1.006-1.069, p = 0.018; HH vs. NN, OR = 1.104, 95% CI = 1.044-1.168, p = 0.001; dominant model, OR = 1.047, 95% CI = 1.018-1.078, p = 0.002; recessive model, OR = 1.086, 95% CI = 1.028-1.146, p = 0.003; Figure 2–6).
Table 2. Summary of OR and 95%CI for association between rs144848 polymorphism and susceptibility to cancer.
| Variable per SNP | I2 (%) | p for heterogeneity | OR (95% CI) | p value | p for publication bias | Effects model | Sensitive analysis | |||
|---|---|---|---|---|---|---|---|---|---|---|
| exclude | OR (95% CI) | p value | p for publication bias | |||||||
| H allele vs N allele | 7.0 | 0.345 | 1.044 (1.021-1.068) | <0.001a | 0.045 | fixed | [36] | 1.053 (1.028-1.080) | <0.001a | 0.143 |
| NH vs NN | 0.0 | 0.491 | 1.037 (1.006-1.069) | 0.018a | 0.147 | fixed | [36] | 1.048 (1.014-1.082) | 0.005a | 0.352 |
| HH vs NN | 16.8 | 0.183 | 1.104 (1.044-1.168) | 0.001a | 0.066 | fixed | [46] | 1.125 (1.060-1.194) | <0.001a | 0.148 |
| Dominant model | 0.0 | 0.470 | 1.047 (1.018-1.078) | 0.002a | 0.069 | fixed | [36] | 1.059 (1.026-1.092) | <0.001a | 0.069 |
| Recessive model | 16.8 | 0.184 | 1.086 (1.028-1.146) | 0.003a | 0.114 | fixed | [46] | 1.102 (1.040-1.168) | 0.001a | 0.214 |
a Statistically significant
Figure 2. Forest plot for pooled ORs for the associations between allele model (H allele vs N allele) of rs144844 and cancer risk in the overall population.
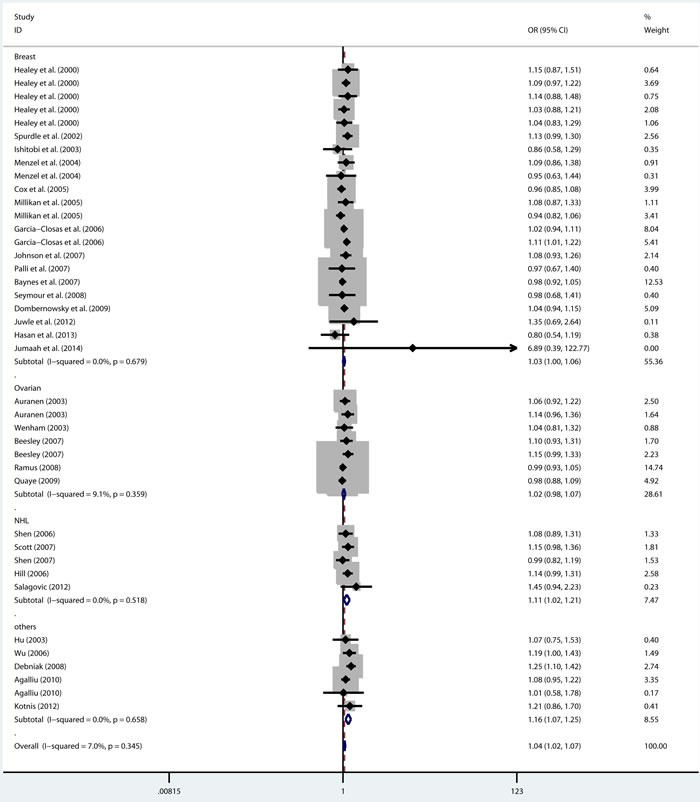
Each square is proportional to the study-specific weight.
Figure 6. Forest plot for pooled ORs for the associations between recessive model (HH vs NH+NN) of rs144844 and cancer risk in the overall population.
Each square is proportional to the study-specific weight.
Figure 3. Forest plot for pooled ORs for the associations between additive model (NH vs NN) of rs144844 and cancer risk in the overall population.
Each square is proportional to the study-specific weight.
Figure 4. Forest plot for pooled ORs for the associations between additive model (HH vs NN) of rs144844 and cancer risk in the overall population.
Each square is proportional to the study-specific weight.
Figure 5. Forest plot for pooled ORs for the associations between dominant model (NH+HH vs NN) of rs144844 and cancer risk in the overall population.
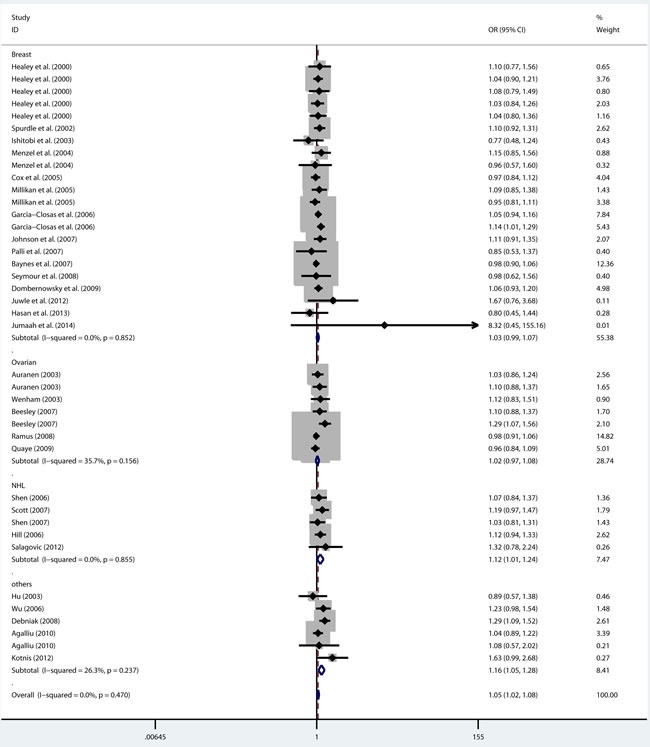
Each square is proportional to the study-specific weight.
Meta-regression analysis
The following covariates were considered for meta-regression: ethnicity, study design and cancer type. The results showed that cancer type contributed to effect in the meta-analysis (H allele vs. N allele, p = 0.011; HH vs. NN, p = 0.006; dominant model, p = 0.039; recessive model, p = 0.011).
Subgroup analysis by cancer type stratification
Based on cancer type, four groups were included in the meta-analysis: breast cancer group, ovarian cancer group, non-Hodgkin lymphoma group and other cancers group. The results showed that the rs144848 polymorphism was not associated with breast cancer or ovarian cancer in any model. However, the rs144848 polymorphism was associated with non-Hodgkin lymphoma in four models (H allele vs. N allele, OR = 1.110, 95% CI = 1.023-1.205, p = 0.012; HH vs. NN, OR = 1.263, 95% CI = 1.035-1.542, p = 0.022; dominant model, OR = 1.118, 95% CI = 1.008-1.240, p = 0.035; recessive model, OR = 1.216, 95% CI = 1.002-1.476, p = 0.048) and with other cancers in all genetic models (Table 3).
Table 3. Summary of OR and 95% CI for association of rs144848 polymorphism with cancer risk by cancer type stratification.
| Subgroup | p for heterogeneity | I2 (%) | OR (95% CI) | p value | Effects model |
|---|---|---|---|---|---|
| N allele vs H allele | |||||
| Breast cancer | 0.679 | 0.0 | 1.028 (0.997-1.060) | 0.075 | fixed |
| Ovarian cancer | 0.359 | 9.1 | 1.024 (0.981-1.068) | 0.280 | fixed |
| NHL | 0.518 | 0.0 | 1.110 (1.023-1.205) | 0.012a | fixed |
| Others | 0.658 | 0.0 | 1.158 (1.074-1.249) | <0.001a | fixed |
| NH vs NN | |||||
| Breast cancer | 0.890 | 0.0 | 1.029 (0.988-1.072) | 0.166 | fixed |
| Ovarian cancer | 0.080 | 46.8 | 1.015 (0.959-1.074) | 0.604 | fixed |
| NHL | 0.954 | 0.0 | 1.090 (0.977-1.215) | 0.122 | fixed |
| Others | 0.090 | 47.5 | 1.117 (1.009-1.236) | 0.033a | fixed |
| HH vs NN | |||||
| Breast cancer | 0.491 | 0.0 | 1.056 (0.978-1.139) | 0.162 | fixed |
| Ovarian cancer | 0.446 | 0.0 | 1.063 (0.957-1.180) | 0.253 | fixed |
| NHL | 0.294 | 19.0 | 1.263 (1.035-1.542) | 0.022a | fixed |
| Others | 0.653 | 0.0 | 1.439 (1.199-1.726) | <0.001a | fixed |
| Dominant model | |||||
| Breast cancer | 0.852 | 0.0 | 1.033 (0.994-1.074) | 0.097 | fixed |
| Ovarian cancer | 0.156 | 35.7 | 1.022 (0.969-1.079) | 0.420 | fixed |
| NHL | 0.855 | 0.0 | 1.118 (1.008-1.240) | 0.035a | fixed |
| Others | 0.237 | 26.3 | 1.162 (1.055-1.280) | 0.002a | fixed |
| Recessive model | |||||
| Breast cancer | 0.477 | 0.0 | 1.044 (0.969-1.124) | 0.259 | fixed |
| Ovarian cancer | 0.351 | 10.3 | 1.057 (0.954-1.170) | 0.290 | fixed |
| NHL | 0.277 | 21.6 | 1.216 (1.002-1.476) | 0.048a | fixed |
| Others | 0.377 | 6.2 | 1.346 (1.130-1.603) | 0.001a | fixed |
a Statistically significant
Association between BRCA2 rs144848 polymorphism and breast cancer risk
There were 22 breast cancer studies with different ethnicities and study designs. To assess the role of genetic background and the source of the control population in breast cancer risk, we carried out a subgroup analysis. In the analysis of genetic background, the overall population was divided into three subgroups, Caucasian, Asian, and African. The results showed that no statistically significant association was observed in any population (Table 4). In the analysis of study design, the overall population was divided into two subgroups, population-based studies and hospital-based studies. The results showed that the allele model was associated with the risk of breast cancer based on population-based studies (H allele vs. N allele, OR = 1.034, 95% CI = 1.000-1.068, p = 0.047; Table 5).
Table 4. Summary of OR and 95% CI for association of rs144848 polymorphism with breast cancer risk by ethnicity stratification.
| Subgroup | p for heterogeneity | I2 (%) | OR (95% CI) | p value | Effects model |
|---|---|---|---|---|---|
| N allele vs H allele | |||||
| Caucasian | 0.690 | 0.0 | 1.029 (0.997-1.061) | 0.075 | fixed |
| Asian | 0.262 | 20.5 | 0.974 (0.692-1.372) | 0.882 | fixed |
| African | 0.185 | 40.8 | 1.024 (0.850-1.235) | 0.801 | fixed |
| NH vs NN | |||||
| Caucasian | 0.970 | 0.0 | 1.028 (0.986-1.072) | 0.189 | random |
| Asian | 0.050 | 74.0 | 1.133 (0.427-3.006) | 0.801 | random |
| African | 0.337 | 8.1 | 1.069 (0.798-1.430) | 0.656 | random |
| HH vs NN | |||||
| Caucasian | 0.332 | 10.4 | 1.060 (0.981-1.146) | 0.138 | fixed |
| Asian | 0.551 | 0.0 | 1.086 (0.377-3.124) | 0.879 | fixed |
| African | 0.388 | 0.0 | 0.877 (0.529-1.455) | 0.612 | fixed |
| Dominant model | |||||
| Caucasian | 0.925 | 0.0 | 1.033 (0.993-1.075) | 0.106 | fixed |
| Asian | 0.101 | 62.8 | 0.955 (0.640-1.424) | 0.820 | fixed |
| African | 0.244 | 29.2 | 1.065 (0.855-1.325) | 0.575 | fixed |
| Recessive model | |||||
| Caucasian | 0.333 | 10.3 | 1.048 (0.972-1.130) | 0.220 | fixed |
| Asian | 0.395 | 0.0 | 1.078 (0.378-3.072) | 0.888 | fixed |
| African | 0.443 | 0.0 | 0.876 (0.548-1.399) | 0.579 | fixed |
Table 5. Summary of OR and 95% CI for association of rs144848 polymorphism with breast cancer risk by the study design stratification.
| Subgroup | p for heterogeneity | I2 (%) | OR (95% CI) | p value | Effects model |
|---|---|---|---|---|---|
| H allele vs N allele | |||||
| PB | 0.691 | 0.0 | 1.034 (1.000-1.068) | 0.047a | fixed |
| HB | 0.759 | 0.0 | 0.883 (0.707-1.103) | 0.273 | fixed |
| Others | 0.264 | 24.5 | 1.011 (0.923-1.108) | 0.810 | fixed |
| NH vs NN | |||||
| PB | 0.953 | 0.0 | 1.030 (0.986-1.076) | 0.182 | fixed |
| HB | 0.684 | 0.0 | 0.864 (0.638-1.171) | 0.346 | fixed |
| Others | 0.174 | 39.6 | 1.050 (0.930-1.186) | 0.428 | fixed |
| HH vs NN | |||||
| PB | 0.315 | 12.4 | 1.076 (0.991-1.168) | 0.082 | fixed |
| HB | 0.677 | 0.0 | 0.844 (0.501-1.422) | 0.525 | fixed |
| Others | 0.559 | 0.0 | 0.957 (0.763-1.200) | 0.702 | fixed |
| Dominant model | |||||
| PB | 0.916 | 0.0 | 1.037 (0.995-1.081) | 0.085 | fixed |
| HB | 0.750 | 0.0 | 0.856 (0.642-1.141) | 0.290 | fixed |
| Others | 0.195 | 36.2 | 1.035 (0.922-1.162) | 0.558 | fixed |
| Recessive model | |||||
| PB | 0.297 | 14.0 | 1.063 (0.982-1.151) | 0.132 | fixed |
| HB | 0.625 | 0.0 | 0.867 (0.538-1.398) | 0.558 | fixed |
| Others | 0.627 | 0.0 | 0.943 (0.757-1.175) | 0.600 | fixed |
a Statistically significant
Sensitivity analysis
To determine the degree to which an individual study affected the overall OR estimates, one-way sensitivity analysis was performed by excluding one study at a time and sequentially recalculating the overall effect. The results showed no influence on pooled ORs and 95% CIs as individual studies were excluded.
Publication bias
Publication bias was observed in only one model (H allele vs. N allele, p = 0.045; Table 2). However, there was no significant publication bias in any genetic model (p > 0.05) after sensitivity analysis. Trim and fill results showed that the adjusted risk estimate remained significant (H allele vs. N allele, OR = 1.028, 95% CI = 1.006-1.050, p = 0.014), which confirmed that the results of this meta-analysis were statistically robust.
DISCUSSION
The mechanisms underlying carcinogenesis are still not fully clear, but it has been suggested that genetic and environmental factors play the most important role in the development of cancer. The BRCA2 protein can regulate homologous recombination by interacting with the RAD51 recombinase, and many studies have suggested that the rs144848 polymorphism in the BRCA2 gene is a susceptibility locus for cancers [8]. However, until now, there has been no consistent result regarding the association between the rs144848 N372H polymorphism and cancer risk. To explain these contradictory results, a meta-analysis including 34,911 cases and 48,329 controls was conducted and five genetic models were utilized to assess the association between the BRCA2 rs144848 polymorphism and the risk of cancer.
In our meta-analysis, the results showed that there was no heterogeneity in any genetic model in overall population, while associations were observed between the rs144848 polymorphism and cancer risk in all genetic models. Meta-regression analysis suggested that ethnicity and study design had no influence on overall effect, but cancer type did contribute to effect (H allele vs. N allele, p = 0.011; HH vs. NN, p = 0.006; dominant model, p = 0.039; recessive model, p = 0.011). Based on cancer type, four groups were included in the meta-analysis: breast cancer group, ovarian cancer group, non-Hodgkin lymphoma group and other cancers group. The results showed that the rs144848 polymorphism was not associated with breast cancer or ovarian cancer in any model. However, the rs144848 polymorphism was associated with non-Hodgkin lymphoma in four models, and associated with other cancers in all genetic models.
The results showed a statistically significant association in all genetic models for overall population. Due to the relatively large number of research studies on breast cancer, we also did a subgroup analysis in the breast cancer group. To assess the role of genetic background in breast cancer, we stratified the population by ethnicity and found no association in Caucasian, Asian, and African subgroups. Considering that the number of publications in Asian and African populations was small, we believe our results may not be reliable due to insufficient statistical power, so additional studies should be conducted to confirm our results. However, after subgroup analysis by study design stratification, we found that the BRCA2 rs144848 N372H polymorphism was associated with increasing the risk of breast cancer in population-based studies (H allele vs. N allele, OR = 1.034, 95% CI = 1.000-1.068, p = 0.047). One-way sensitivity analysis suggested no influence of individual studies on pooled ORs and 95% CIs.
In 2006, a study from the breast cancer association consortium summarized the common breast cancer-associated polymorphisms but failed to show a significant association between the BRCA2 rs144848 polymorphism and breast cancer [53]. In 2010, Qiu et al. found in a meta-analysis that the BRCA2 rs144848 H allele may be a low-penetrant risk factor for developing breast cancer [54]. In 2014, Xue et al. conducted a meta-analysis to assess the association between the BRCA2 rs144848 polymorphism and cancer susceptibility [55]. In contrast to Qiu et al., they did not find an association between the BRCA2 rs144848 polymorphism and breast cancer, but did observe an association with ovarian cancer. Different results from Xue et al. were then obtained in 2015 by Wang et al., who found that the rs144848 polymorphism was not associated with ovarian cancer. Compared with this latter study, we updated and added several new studies which were strictly filtered by a quality assessment. In addition, we used five genetic models to assess the role of the BRCA2 rs144848 polymorphism in our meta-analysis. Another important difference from Wang et al. was that their results were based on the risk estimates obtained without the original genotype data, whereas all studies included in our meta-analysis provided genotype data, so that our results were more precise by calculating effect directly without potential deviations and biases.
The strength of this meta-analysis is that the most current literature was included. To guarantee the quality of the meta-analysis, the Newcastle-Ottawa scale was conducted to assess the quality of included studies, and a strict procedure for data extraction was performed by two investigators according to inclusion and exclusion criteria. Furthermore, no low-quality literature was included in this meta-analysis which might possibly have influenced our results. One-way sensitivity analysis and meta-regression were also performed to increase the robustness of our conclusions. Subgroup analysis by ethnicity and the source of the control population were used to explain the effect of genetic background and study design.
There are some limitations in this meta-analysis. First, the literature search strategy was limited by language, and only published papers in English were included. Second, because we excluded literature without original data, some studies were excluded. Third, other potential interactions including environment × gene, gene × gene and some potential covariates were not considered due to insufficient information.
In conclusion, our meta-analysis determined that the BRCA2 rs144848 polymorphism was associated with non-Hodgkin lymphoma, and indicated that the rs144848 H allele of the BRCA2 gene may be a low-penetrate risk factor enhancing carcinogenesis in breast cancer. Further well-designed studies are warranted to clarify the mechanism and increase comprehensive understanding of the role of the BRCA2 rs144848 polymorphism in cancer.
MATERIALS AND METHODS
Publication research
Studies were retrieved by searching PubMed, Embase and Google Scholar following the guidelines in Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2009 [41]. The last search was updated on April 2016 with the terms “cancer”, “tumor”, “BRCA”, “polymorphism”, “genetic”, “variant”, “rs144848” and “N372H”. References in potential articles were also included in order to find more relevant studies.
Inclusion criteria
All articles were reviewed by two investigators independently. Studies were included in the meta-analysis if they met the following criteria: (1) Studies were case-control or cohort studies; (2) articles were original studies of human participants; (3) genotype distributions were available; (4) studies were published in English; and (5) articles were association studies between rs144848 polymorphism and cancer risk. If studies were drawn from the same population, only the study with the largest sample size or with a sufficient quantity of useful data was included. If an article reported the results from different studies, each study was treated as a separate comparison in our meta-analysis.
Quality score assessment
The Newcastle-Ottawa scale was used to assess the quality of studies [42]. Three items including selection, comparability and exposure were used to calculate the score of studies with a maximum score of nine. Any disagreements were adjusted by a third reviewer. A total score of three or lower, four to six and seven or greater was considered to indicate low, medium and high quality studies, respectively.
Data extraction
Data were extracted from included studies using a standardized form. For each study, the following information was extracted: (1) name of first author, (2) year of publication, (3) ethnicity of population, (4) source of control population and (5) sample size and genotype distribution. Ethnicity was categorized as Caucasian, Asian or African, and the study design was categorized as population-based study, hospital-based study or nested study.
Statistical analysis
The odds ratios (ORs) with corresponding 95% confidence intervals (95% CIs) were calculated to assess the association between the rs144848 polymorphism and cancer risk. Five models were used in this meta-analysis: (1) H allele vs. N allele, (2) NH vs. NN, (3) HH vs. NN, (4) dominant model, (NH+HH vs. NN), and (5) recessive model, (HH vs. NH+NN). Statistical analysis was performed using STATA 11.0 (Stata Corporation, College Station, TX, USA). The chi-square test was conducted to evaluate if the studies deviated from Hardy-Weinberg equilibrium, and the threshold for disequilibrium was p < 0.05. Cochran's Q test and I2 statistic test were performed to assess heterogeneity across individual studies (p < 0.10 and I2 > 50% suggested heterogeneity). The fixed-effects model (the Mantel-Haenszel method) was used to estimate the pooled OR if I2 < 50%; otherwise, the random-effects model (the DerSimonian and Laird method) was used [43]. A value of p < 0.05 was accepted as the significance threshold for each genetic model.
Subgroup analysis was conducted based on ethnicity (Caucasian, Asian and African) and study design (population-based and hospital-based). If heterogeneity was present, meta-regression was conducted to explore the source of heterogeneity. One-way sensitivity analysis was used to assess the influence of the individual study set to the pooled ORs by sequential exclusion.
A funnel plot was performed to estimate the potential publication bias using Begg's test, in which the standard error of log (OR) was plotted against its log (OR) [44]. Egger's liner regression test was also used to evaluate publication bias with quantitative analysis as a supplement to the funnel plot [45]. The trim and fill method was used to adjust pooled ORs and 95% CIs if bias was detected.
PRISMA CHECKLIST
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China [81102278], the China Postdoctoral Science Foundation [20100481019], the Postdoctoral Science Special Foundation of Heilongjiang Province, China [LBH-TZ1208], the Postdoctoral Science Research Foundation of Heilongjiang Province, China [LBH-Q13128], and Wu lien-teh Youth Science Foundation of Harbin Medical University [WLD-QN1405].
Footnotes
CONFLICTS OF INTEREST
We declared that there is no duality of interest associated with this manuscript.
Submission declaration
Submission of the article implies that the work described has not been published previously.
REFERENCES
- 1.Collins A, Politopoulos I. The genetics of breast cancer: Risk factors for disease. Application of Clinical Genetics. 2011;4:11–19. doi: 10.2147/TACG.S13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Easton D. New insights into genetic susceptibility to breast cancer. Cancer Prevention Research. 2010:3. [Google Scholar]
- 3.Teraoka SN, Bernstein JL, Reiner AS, Haile RW, Bernstein L, Lynch CF, Malone KE, Stovall M, Capanu M, Liang X, Smith SA, Mychaleckyj J, Hou X, et al. Single nucleotide polymorphisms associated with risk for contralateral breast cancer in the Women's Environment, Cancer, and Radiation Epidemiology (WECARE) Study. Breast Cancer Research. 2011:13. doi: 10.1186/bcr3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narod SA. Genes, the environment, and breast cancer. The Lancet. 2010;375:2123–2124. doi: 10.1016/s0140-6736(10)60876-8. [DOI] [PubMed] [Google Scholar]
- 5.Vici P, Di Benedetto A, Ercolani C, Pizzuti L, Di Lauro L, Sergi D, Sperati F, Terrenato I, Dattilo R, Botti C, Fabi A, Ramieri MT, Mentuccia L, et al. Predictive significance of DNA damage and repair biomarkers in triple-negative breast cancer patients treated with neoadjuvant chemotherapy: An exploratory analysis. Oncotarget. 2015;6:42773–42780. doi: 10.18632/oncotarget.6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao D, Herman JG, Guo M. The clinical value of aberrant epigenetic changes of DNA damage repair genes in human cancer. Oncotarget. 2016;7:37331–37346. doi: 10.18632/oncotarget.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGuire SE. DNA Damage and Repair Pathway Profiles as Biomarkers in High-Risk Prostate Cancer. JAMA oncology. 2016;2:1–2. doi: 10.1001/jamaoncol.2015.4952. [DOI] [PubMed] [Google Scholar]
- 8.Lord CJ, Ashworth A. RAD51, BRCA2 and DNA repair: a partial resolution. Nature structural & molecular biology. 2007;14:461–462. doi: 10.1038/nsmb0607-461. [DOI] [PubMed] [Google Scholar]
- 9.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nature structural & molecular biology. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 11.Khan SG, Muniz-Medina V, Shahlavi T, Baker CC, Inui H, Ueda T, Emmert S, Schneider TD, Kraemer KH. The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic acids research. 2002;30:3624–3631. doi: 10.1093/nar/gkf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mumbrekar KD, Goutham HV, Vadhiraja BM, Bola Sadashiva SR. Polymorphisms in double strand break repair related genes influence radiosensitivity phenotype in lymphocytes from healthy individuals. DNA repair. 2016;40:27–34. doi: 10.1016/j.dnarep.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Healey CS, Dunning AM, Teare MD, Chase D, Parker L, Burn J, Chang-Claude J, Mannermaa A, Kataja V, Huntsman DG, Pharoah PD, Luben RN, Easton DF, et al. A common variant in BRCA2 is associated with both breast cancer risk and prenatal viability. Nature genetics. 2000;26:362–364. doi: 10.1038/81691. [DOI] [PubMed] [Google Scholar]
- 14.Hasan TN, Shafi G, Syed NA, Alsaif MA, Alsaif AA, Alshatwi AA. Lack of association of BRCA1 and BRCA2 variants with breast cancer in an ethnic population of Saudi Arabia, an emerging high-risk area. Asian Pacific journal of cancer prevention. 2013;14:5671–5674. doi: 10.7314/apjcp.2013.14.10.5671. [DOI] [PubMed] [Google Scholar]
- 15.Auranen A, Spurdle AB, Chen X, Lipscombe J, Purdie DM, Hopper JL, Green A, Healey CS, Redman K, Dunning AM, Pharoah PD, Easton DF, Ponder BAJ, et al. BRCA2 Arg372His polymorphism and epithelial ovarian cancer risk. International journal of cancer. 2003;103:427–430. doi: 10.1002/ijc.10814. [DOI] [PubMed] [Google Scholar]
- 16.Meeks HD, Song H, Michailidou K, Bolla MK, Dennis J, Wang Q, Barrowdale D, Frost D, Embrace McGuffog L, Ellis S, Feng B, Buys SS, et al. BRCA2 Polymorphic Stop Codon K3326X and the Risk of Breast, Prostate, and Ovarian Cancers. Journal of the National Cancer Institute. 2016:108. doi: 10.1093/jnci/djv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agalliu I, Kwon EM, Salinas CA, Koopmeiners JS, Ostrander EA, Stanford JL. Genetic variation in DNA repair genes and prostate cancer risk: results from a population-based study. Cancer causes & control. 2010;21:289–300. doi: 10.1007/s10552-009-9461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill DA, Wang SS, Cerhan JR, Davis S, Cozen W, Severson RK, Hartge P, Wacholder S, Yeager M, Chanock SJ, Rothman N. Risk of non-Hodgkin lymphoma (NHL) in relation to germline variation in DNA repair and related genes. Blood. 2006;108:3161–3167. doi: 10.1182/blood-2005-01-026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. Polygenic susceptibility to breast cancer and implications for prevention. Nature genetics. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- 20.Fuks F, Milner J, Kouzarides T. BRCA2 associates with acetyltransferase activity when bound to P/CAF. Oncogene. 1998;17:2531–2534. doi: 10.1038/sj.onc.1202475. [DOI] [PubMed] [Google Scholar]
- 21.Dombernowsky SL, Weischer M, Freiberg JJ, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG. Missense polymorphisms in BRCA1 and BRCA2 and risk of breast and ovarian cancer. Cancer epidemiology, biomarkers & prevention. 2009;18:2339–2342. doi: 10.1158/1055-9965.EPI-09-0447. [DOI] [PubMed] [Google Scholar]
- 22.Freedman ML, Penney KL, Stram DO, Le Marchand L, Hirschhorn JN, Kolonel LN, Altshuler D, Henderson BE, Haiman CA. Common variation in BRCA2 and breast cancer risk: a haplotype-based analysis in the Multiethnic Cohort. Human molecular genetics. 2004;13:2431–2441. doi: 10.1093/hmg/ddh270. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Closas M, Egan KM, Newcomb PA, Brinton LA, Titus-Ernstoff L, Chanock S, Welch R, Lissowska J, Peplonska B, Szeszenia-Dabrowska N, Zatonski W, Bardin-Mikolajczak A, Struewing JP. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Human genetics. 2006;119:376–388. doi: 10.1007/s00439-006-0135-z. [DOI] [PubMed] [Google Scholar]
- 24.Ishitobi M, Miyoshi Y, Ando A, Hasegawa S, Egawa C, Tamaki Y, Monden M, Noguchi S. Association of BRCA2 polymorphism at codon 784 (Met/Val) with breast cancer risk and prognosis. Clinical cancer research. 2003;9:1376–1380. [PubMed] [Google Scholar]
- 25.Juwle A, Saranath D. BRCA1/BRCA2 gene mutations/SNPs and BRCA1 haplotypes in early-onset breast cancer patients of Indian ethnicity. Medical oncology. 2012;29:3272–3281. doi: 10.1007/s12032-012-0294-9. [DOI] [PubMed] [Google Scholar]
- 26.Menzel HJ, Sarmanova J, Soucek P, Berberich R, Grunewald K, Haun M, Kraft HG. Association of NQO1 polymorphism with spontaneous breast cancer in two independent populations. British journal of cancer. 2004;90:1989–1994. doi: 10.1038/sj.bjc.6601779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millikan RC, Player JS, Decotret AR, Tse CK, Keku T. Polymorphisms in DNA repair genes, medical exposure to ionizing radiation, and breast cancer risk. Cancer epidemiology, biomarkers & prevention. 2005;14:2326–2334. doi: 10.1158/1055-9965.EPI-05-0186. [DOI] [PubMed] [Google Scholar]
- 28.Hu N, Li WJ, Su H, Wang C, Goldstein AM, Albert PS, Emmert-Buck MR, Kong LH, Roth MJ, Dawsey SM, He LJ, Cao SF, Ding T, et al. Common genetic variants of TP53 and BRCA2 in esophageal cancer patients and healthy individuals from low and high risk areas of northern China. Cancer detection and prevention. 2003;27:132–138. doi: 10.1016/s0361-090x(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 29.Wenham RM, Schildkraut JM, McLean K, Calingaert B, Bentley RC, Marks J, Berchuck A. Polymorphisms in BRCA1 and BRCA2 and risk of epithelial ovarian cancer. Clinical cancer research. 2003;9:4396–4403. [PubMed] [Google Scholar]
- 30.Rudd MF, Sellick GS, Webb EL, Catovsky D, Houlston RS. Variants in the ATM-BRCA2-CHEK2 axis predispose to chronic lymphocytic leukemia. Blood. 2006;108:638–644. doi: 10.1182/blood-2005-12-5022. [DOI] [PubMed] [Google Scholar]
- 31.Shen M, Zheng T, Lan Q, Zhang Y, Zahm SH, Wang SS, Holford TR, Leaderer B, Yeager M, Welch R, Kang D, Boyle P, Zhang B, et al. Polymorphisms in DNA repair genes and risk of non-Hodgkin lymphoma among women in Connecticut. Human genetics. 2006;119:659–668. doi: 10.1007/s00439-006-0177-2. [DOI] [PubMed] [Google Scholar]
- 32.Beesley J, Jordan SJ, Spurdle AB, Song H, Ramus SJ, Kjaer SK, Hogdall E, DiCioccio RA, McGuire V, Whittemore AS, Gayther SA, Pharoah PD, Webb PM, et al. Association between single-nucleotide polymorphisms in hormone metabolism and DNA repair genes and epithelial ovarian cancer: results from two Australian studies and an additional validation set. Cancer epidemiology, biomarkers & prevention. 2007;16:2557–2565. doi: 10.1158/1055-9965.EPI-07-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Gu J, Grossman HB, Amos CI, Etzel C, Huang M, Zhang Q, Millikan RE, Lerner S, Dinney CP, Spitz MR. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. American journal of human genetics. 2006;78:464–479. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott K, Adamson PJ, Barrans SL, Worrillow LJ, Willett EV, Allan JM. RAG1 and BRCA2 polymorphisms in non-Hodgkin lymphoma. Blood. 2007;109:5522–5523. doi: 10.1182/blood-2006-12-061572. [DOI] [PubMed] [Google Scholar]
- 35.Shen M, Purdue MP, Kricker A, Lan Q, Grulich AE, Vajdic CM, Turner J, Whitby D, Chanock S, Rothman N, Armstrong BK. Polymorphisms in DNA repair genes and risk of non-Hodgkin's lymphoma in New South Wales, Australia. Haematologica. 2007;92:1180–1185. doi: 10.3324/haematol.11324. [DOI] [PubMed] [Google Scholar]
- 36.Ramus SJ, Vierkant RA, Johnatty SE, Pike MC, Van Den Berg DJ, Wu AH, Pearce CL, Menon U, Gentry-Maharaj A, Gayther SA, Dicioccio RA, McGuire V, Whittemore AS, et al. Consortium analysis of 7 candidate SNPs for ovarian cancer. International journal of cancer. 2008;123:380–388. doi: 10.1002/ijc.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debniak T, Scott RJ, Gorski B, Cybulski C, van de Wetering T, Serrano-Fernandez P, Huzarski T, Byrski T, Nagay L, Debniak B, Kowalska E, Jakubowska A, Gronwald J, et al. Common variants of DNA repair genes and malignant melanoma. European journal of cancer. 2008;44:110–114. doi: 10.1016/j.ejca.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Kotnis A, Namkung J, Kannan S, Jayakrupakar N, Park T, Sarin R, Mulherkar R. Multiple pathway-based genetic variations associated with tobacco related multiple primary neoplasms. PloS one. 2012;7:e30013. doi: 10.1371/journal.pone.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quaye L, Tyrer J, Ramus SJ, Song H, Wozniak E, DiCioccio RA, McGuire V, Hogdall E, Hogdall C, Blaakaer J, Goode EL, Schildkraut JM, Easton DF, et al. Association between common germline genetic variation in 94 candidate genes or regions and risks of invasive epithelial ovarian cancer. PloS one. 2009;4:e5983. doi: 10.1371/journal.pone.0005983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salagovic J, Klimcakova L, Ilencikova D, Kafkova A. Association of follicular lymphoma risk with BRCA2 N372H polymorphism in Slovak population. Medical oncology. 2012;29:1173–1178. doi: 10.1007/s12032-011-9925-9. [DOI] [PubMed] [Google Scholar]
- 41.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Bmj. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 43.Fleiss JL. The statistical basis of meta-analysis. Statistical methods in medical research. 1993;2:121–145. doi: 10.1177/096228029300200202. [DOI] [PubMed] [Google Scholar]
- 44.Begg CB, Berlin JA. Publication bias and dissemination of clinical research. Journal of the National Cancer Institute. 1989;81:107–115. doi: 10.1093/jnci/81.2.107. [DOI] [PubMed] [Google Scholar]
- 45.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox DG, Hankinson SE, Hunter DJ. No association between BRCA2 N372H and breast cancer risk. Cancer epidemiology, biomarkers & prevention. 2005;14:1353–1354. doi: 10.1158/1055-9965.EPI-04-0848. [DOI] [PubMed] [Google Scholar]
- 47.Spurdle AB, Hopper JL, Chen X, Dite GS, Cui J, McCredie MRE, Giles GG, Ellis-Steinborner S, Venter DJ, Newman B, Southey MC, Chenevix-Trench G. The BRCA2 372 HH genotype is associated with risk of breast cancer in Australian women under age 60 years. Cancer Epidemiology Biomarkers and Prevention. 2002;11:413–416. [PubMed] [Google Scholar]
- 48.Baynes C, Healey CS, Pooley KA, Scollen S, Luben RN, Thompson DJ, Pharoah PD, Easton DF, Ponder BA, Dunning AM. study Sbc. Common variants in the ATM, BRCA1, BRCA2, CHEK2 and TP53 cancer susceptibility genes are unlikely to increase breast cancer risk. Breast cancer research. 2007;9:R27. doi: 10.1186/bcr1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson N, Fletcher O, Palles C, Rudd M, Webb E, Sellick G, dos Santos Silva I, McCormack V, Gibson L, Fraser A, Leonard A, Gilham C, Tavtigian SV, et al. Counting potentially functional variants in BRCA1, BRCA2 and ATM predicts breast cancer susceptibility. Human molecular genetics. 2007;16:1051–1057. doi: 10.1093/hmg/ddm050. [DOI] [PubMed] [Google Scholar]
- 50.Jumaah S, Ali NA, Abdul-Majid BA, Tobal K. Detection of BRCA2 exon 10 genetic variations in Iraqibreast cancer patients. Iraqi Journal of Biotechnology. 2014;13:14–21. [Google Scholar]
- 51.Palli D, Falchetti M, Masala G, Lupi R, Sera F, Saieva C, D'Amico C, Ceroti M, Rizzolo P, Caligo MA, Zanna I, Ottini L. Association between the BRCA2 N372H variant and male breast cancer risk: a population-based case-control study in Tuscany, Central Italy. BMC cancer. 2007;7:170. doi: 10.1186/1471-2407-7-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seymour IJ, Casadei S, Zampiga V, Rosato S, Danesi R, Falcini F, Strada M, Morini N, Naldoni C, Paradiso A, Tommasi S, Schittulli F, Amadori D, Calistri D. Disease family history and modification of breast cancer risk in common BRCA2 variants. Oncology reports. 2008;19:783–786. [PubMed] [Google Scholar]
- 53.Breast Cancer Association C Commonly studied single-nucleotide polymorphisms and breast cancer: results from the Breast Cancer Association Consortium. Journal of the National Cancer Institute. 2006;98:1382–1396. doi: 10.1093/jnci/djj374. [DOI] [PubMed] [Google Scholar]
- 54.Qiu LX, Yao L, Xue K, Zhang J, Mao C, Chen B, Zhan P, Yuan H, Hu XC. BRCA2 N372H polymorphism and breast cancer susceptibility: a meta-analysis involving 44,903 subjects. Breast cancer research and treatment. 2010;123:487–490. doi: 10.1007/s10549-010-0767-5. [DOI] [PubMed] [Google Scholar]
- 55.Xue WQ, He YQ, Zhu JH, Ma JQ, He J, Jia WH. Association of BRCA2 N372H polymorphism with cancer susceptibility: a comprehensive review and meta-analysis. Scientific reports. 2014;4:6791. doi: 10.1038/srep06791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


