Abstract
Chronic exposure to mouse allergen may contribute greatly to the inner-city asthma burden. We hypothesized that reducing mouse allergen exposure may modulate the immunopathology underlying symptomatic pediatric allergic asthma, and that this occurs through epigenetic regulation. To test this hypothesis, we studied a cohort of mouse sensitized, persistent asthmatic inner-city children undergoing mouse allergen-targeted integrated pest management (IPM) vs education in a randomized controlled intervention trial. We found that decreasing mouse allergen exposure, but not cockroach, was associated with reduced FOXP3 buccal DNA promoter methylation, but this was unrelated to mouse specific IgE production. This finding suggests that the environmental epigenetic regulation of an immunomodulatory gene may occur following changing allergen exposures in some highly exposed cohorts. Given the clinical and public health importance of inner-city pediatric asthma and the potential impact of environmental interventions, further studies will be needed to corroborate changes in epigenetic regulation following changing exposures over time, and determine their impact on asthma morbidity in susceptible children.
Keywords: Epigenetic regulation, Allergic asthma, Mouse allergen
Introduction
As many as 25–50% of inner-city children with asthma have evidence of allergic sensitization to mouse(Matsui, Eggleston et al. 2006, Ahluwalia, Peng et al. 2013). This trend suggests that chronic exposure to mouse allergen may contribute greatly to the inner-city asthma burden, particularly in the major metropolitan areas of the Northeastern United States. The multi-faceted environmental intervention by the Inner-City Asthma Study (ICAS) group, as well as others, has shown that successful reduction of indoor allergens can lead to long-lasting decreases in asthma symptoms, asthma exacerbations, missed school and disrupted sleep in children(Morgan, Crain et al. 2004, Pongracic, Visness et al. 2008, Johnson, Ciaccio et al. 2009). Nonetheless, its ability to induce immune modulation is unknown and has implications for understanding the natural course of allergic asthma, identifying those at greater risk, and determining optimal treatment.
Environmental epigenetic regulation may induce immune modulation. This is supported by observations that measures of multiple environmental toxicants, including air pollution and allergens, are associated with altered inflammatory, allergic and regulatory gene methylation(Nadeau, McDonald-Hyman et al. 2010, Niedzwiecki, Zhu et al. 2012, Salam, Byun et al. 2012). But far less studied are the impacts of potential changes in exposures over time on changes in epigenetic marks, and whether these changes in epigenetic marks are clinically relevant. As an example, human rhinovirus infection changed global methylation in nasal epithelial cells to levels that varied by asthma diagnosis(McErlean, Favoreto et al. 2014). In the Normative Aging Study of elderly men, changes in particle numbers, levels of black carbon, and ozone over the preceding 4 weeks were associated with inflammatory gene specific changes in DNA methylation(Bind, Lepeule et al. 2014). However, comparable studies of changes in pediatric cohorts are scant. One exception is the Protection Against Allergy: Study in Rural Environments (PASTURE) study that observed significant differences in the DNA methylation of several genes assessed in cord blood and then repeated at age 4.5 years. The differences with aging during childhood also varied by whether there was prenatal exposure to a farm environment, and by whether the child was subsequently diagnosed with asthma (Michel, Busato et al. 2013).
To address this research gap, we measured prospectively and repeatedly promoter DNA methylation and expression of targeted asthma candidate genes associated with regulating allergy (T regulatory gene forkhead box P3 gene (FOXP3), and allergy suppressive gene interferon (IFN)γ). Methylation of both genes previously was found to occur in association with ambient environmental exposures(Liu, Ballaney et al. 2008, Brand, Kesper et al. 2012, Kohli, Garcia et al. 2012, Niedzwiecki, Zhu et al. 2012, Runyon, Cachola et al. 2012, Tang, Levin et al. 2012) and with allergy and asthma(Liu, Ballaney et al. 2008, Brand, Kesper et al. 2012, Runyon, Cachola et al. 2012). We utilized pyrosequencing to capture and quantify small differences predicted to underlie potential changes in the exposure-outcome relationships(Murphy, Adigun et al. 2012, Michel, Busato et al. 2013, Richmond, Simpkin et al. 2015, Clifford, Jones et al. 2017). This observational substudy combined measures from mouse sensitized, moderate to severe asthmatic inner-city children (n=200; 6–17yr) undergoing mouse allergen-targeted integrated pest management (IPM) vs education in a randomized control intervention. We sampled buccal cells that comprise the aerodigestive track epithelium because they are accessed easily in children(Breton, Byun et al. 2011, Kuriakose, Torrone et al. 2011, Lovinsky-Desir, Ridder et al. 2014), and undergo molecular alterations following environmental exposures(Bhutani, Pathak et al. 2008, Salam, Byun et al. 2012, Wan, Qiu et al. 2014), and in association with airway inflammation(Breton, Byun et al. 2011, Salam, Byun et al. 2012). We postulated that reducing allergen exposure would modulate the immunopathology underlying persistent pediatric allergic asthma through epigenetic regulation. Specifically, we hypothesized that changes in mouse allergen exposure would be associated with changes in buccal cell methylation and expression, and that these would alter mouse-specific immunoglobulin (Ig)E.
Materials and methods
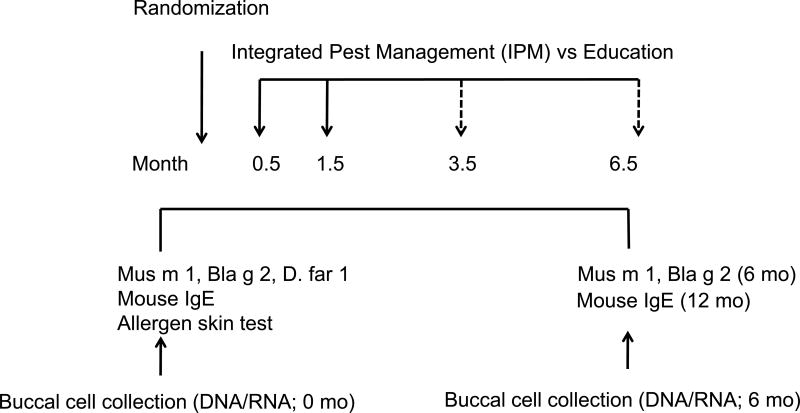
Children with persistent asthma and an exacerbation in the previous year underwent measurement of home settled bed dust and bed floor dust for mouse allergen (Mus m 1) and bed dust for cockroach (Bla g2) and dust mite allergen (Der f 1) by ELISA (Indoor Biotechnologies, Charlottesville, VA). Mouse-specific IgE (against mouse urine proteins, e72) was tested by Immunocap (ThermoFisher, Uppsala, Sweden) as described(Matsui, Eggleston et al. 2006, Sedaghat, Matsui et al. 2016). Mouse exposed (determined by bed dust mouse allergen concentration of ≥0.4 µg/g or a bedroom floor dust mouse allergen concentration of ≥0.5 µg/g) and sensitized children were enrolled in a home-based Mouse Allergen and Asthma Intervention Trial (MAAIT; Figure 1)(Sedaghat, Matsui et al. 2016). The pest management education that was delivered as a control included information about setting mouse traps, sealing holes and cracks, and housekeeping practices. The IPM was delivered in treatments. The first treatment included targeted cleaning to remove allergen reservoirs, placement of traps, application of rodenticide, sealing holes and cracks, installation of allergen-proof mattress and pillow encasements (CleanBrands, LLC, Warwick, Rhode Island), and two portable air purifiers (Filtrete™ Room Air Purifier, 3M, St. Paul, Minnesota). This was followed by a visit 1–2 weeks later to reset traps and complete any work remaining from the primary visit. Each treatment was delivered by a licensed pest technician and by study staff.
Figure 1.
Study algorithm
Extracted DNA underwent bisulfite conversion and pyrosequencing of the upstream enhancer (FOXP3 only) and promoter (both genes) areas(Lovinsky-Desir, Ridder et al. 2014). The FOXP3 CpG loci within the gene promoter area were selected based on our previous associations with Treg function and asthma(Runyon, Cachola et al. 2012) and their relatively large range in methylation levels across individuals (data not shown). The IFNγ CpG loci within the gene promoter were selected based on their conservation in mice (Niedzwiecki, Zhu et al. 2012, Collison, Siegle et al. 2013), specific roles in regulating gene expression(Gonsky, Deem et al. 2009, Brand, Kesper et al. 2012, Kohli, Garcia et al. 2012, Belsky, Sears et al. 2013), susceptibility to allergen and air pollution(Liu, Ballaney et al. 2008, Brand, Kesper et al. 2012, Niedzwiecki, Zhu et al. 2012), and previously implicated role in allergy and asthma(Kohli, Garcia et al. 2012, Runyon, Cachola et al. 2012).
Total RNA was extracted using Trizol (Molecular Research Center, Inc., Cincinnati, OH; Supplemental Tables 1A,B). qRT-PCR was carried out using the SuperScript First-Strand Synthesis System and the Applied Biosystems® 7500 Real-Time PCR Systems. The housekeeper gene cystatin A (CSTA) was selected based on its high and specific expression in the buccal mucosa(Magister and Kos 2013). Mouse and cockroach allergen levels and buccal cells were collected at baseline and 6 months later. Dust mite allergen levels were collected at baseline only. Mouse IgE was assessed at baseline and 12 months later. The study was approved by the Institutional Review Boards of Columbia University Medical Center, Johns Hopkins University, and Harvard University.
The distribution of each variable was examined and an appropriate transformation was applied as necessary to meet parametric model assumptions. Analyses were conducted separately for each CpG gene site. Allergen levels and RNA values were natural log transformed. Values (eg, mouse allergen) below a limit of detection (LOD) were assigned a value at half the LOD. Methylation levels within the FOXP3 enhancer highly correlated with each other (Spearman correlation coefficients 0.90–0.94) at each visit, and in subsequent analyses the average of the 4 values were used as a single variable.
The signed rank test was used to compare differences in allergen and mouse IgE levels between visits. The Kruskal-Wallis test was to used to detect bivariate associations between the methylation or gene expression levels and categorical variables including sex, site (Boston, Boston Children’s Hospital, Harvard Medical School; Baltimore, Johns Hopkins University), race/ethnicity, insurance type, any reported exposure to second hand smoke, maternal allergy status and particulate matter less than 10 microns or not, at each visit. The spearman correlation coefficient was used to indicate bivariate association between quantitative variables, specifically the correlations among individual buccal biomarkers and between the arithmetic change in allergen and change in methylation. To examine concurrent associations between mouse allergen and each of the methylation or gene expression outcome variables with repeated measures over time, we used marginal linear models with mouse allergen as the predictor. The generalized estimating equations (GEE) approach, taking into account within-person correlation in repeated measures, was used to estimate model parameters and make statistical inference. Because a sex difference in the FOXP3 enhancer and association between age and gene expression was evident, we controlled for sex (age) in the marginal models. To determine whether a decrease in mouse (or cockroach) allergen changed FOXP3 or IFNγ methylation or gene expression, linear regression models were built with the change in the methylation or gene expression as the dependent variables, and the change in mouse (or cockroach) allergen as the primary independent variable. Models were controlled for mouse allergen, CpG methylation or gene expression at baseline. Sex and age were not related to the change in FOXP3 or IFNγ methylation over time, so the models exhibited in Tables 2,3 controlled only for baseline levels. Linear models were used to examine whether a change in methylation (over 6 months) was associated with a change in mouse IgE (over 12 months), controlling for mouse–specific IgE and CpG methylation or gene expression at baseline.
Table 2.
The association between the change in mouse allergens over time and change in FOXP3 and IFNγ promoter methylation
| Outcome: change in FOXP3 | Outcome: change in IFNγ | |||||||
|---|---|---|---|---|---|---|---|---|
| Mouse allergen predictor | Enhancer | CpG−138 | CpG−126 | Expression | CpG−186 | CpG−54 | Expression | |
| n | 191 | 198 | 198 | 198 | 186 | 186 | 198 | |
| B (SE) | B (SE) | B (SE) | B (SE) | B (SE) | B (SE) | B (SE) | ||
| Bed dust | −0.15 (0.37) | 1.78* (0.70) | −0.45 (0.37) | 0.16 (0.10) | −0.33 (0.40) | 0.30 (0.37) | 0.04 (0.09) | |
| Bed floor dust | −0.25 (0.32) | 0.90 (0.60) | −0.25 (0.31) | 0.09 (0.08) | −0.51 (0.33) | 0.36 (0.31) | 0.05 (0.07) | |
B: Estimated regression coefficient
SE: Standard error.
Enhancer defined as average methylation at CpG−4506, CpG−4500, CpG−4494, CpG−4484.
Relative expression of FOXP3 was measured using log transformation (variable+0.1).
Relative expression of IFNγ was measured using log transformation (variable+0.5).
Analyses controlled for both mouse allergen at baseline (natural log transformed) and CpG methylation or gene expression at baseline. Sensitivity analyses of gene expression including only those with qRT-PCR cycle length <37 did not alter the results. Missing samples occurred when DNA or RNA or allergen specimens were inadequate or when pyrosequencing reactions failed.
p=0.01.
Table 3.
Change in methylation and expression over 6 month was not associated with change in mouse IgE over 12 month
| Change in FOXP3 |
Enhancer | CpG−138 | CpG−126 | Expression |
|---|---|---|---|---|
| B | −0.01 | −0.01 | −0.02 | −0.03 |
| SE | 0.01 | 0.004 | 0.01 | 0.03 |
| Change in IFNγ | CppG-186 | CpG-54 | Expression | |
| B | −0.002 | 0.004 | 0.02 | |
| SE | 0.01 | 0.01 | 0.04 |
B: Estimated regression coefficient. SE: Standard error
Relative expression of FOXP3 was measured using log transformation (variable+0.1)
Relative expression of IFNγ was measured using log transformation (variable+0.5)
Analyses controlled for both mouse allergen at baseline (natural log transformed) and CpG methylation or gene expression at baseline. Sensitivity analyses of gene expression including only those with qRT-PCR cycle length <37 did not alter the results.
N=179 for all analyses, with specimens excluded due to lack of 12 month blood specimen (n=12 samples from 11 individuals), or inadequate or pyrosequencing reactions failed (up to n=15 for IFNγ methylation analyses).
Results and Discussion
Participant baseline characteristics are displayed in Table 1. The age of the participants ranged 5.2–17.5 years (mean 9.7±3.4 standard deviation (SD)). The average number of positive skin tests in addition to mouse was 4.7± 2.8. In our cohort of n=200, similar to that previously published (Matsui et al. JAMA, in press), mouse allergen in the bed (median 0.9, IQR 2.7 to median 0.6, IQR 1.3, p<0.0001) and floor (median 4.2, IQR 16.0 to median 1.8, IQR 6.4, p<0.0001), and cockroach allergen in the bed (median 0, IQR 0.02 to median 0, IQR 14.7, p=0.04), decreased overall between visits. Also, in our subset of n=187 with paired data for mouse IgE analyses, similar to what we previously published (Matsui et al. JAMA, in press), mouse IgE decreased overall (median 10.7 kU/L, IQR 33.9 to median 7.0, IQR 22.6, p<0.0001).
Table 1.
Participant baseline characteristics (n=200)
| Number | % | |
|---|---|---|
| Site Baltimore (vs. Boston) | 168 | 84 |
| Income < $25,000 annual | 117 | 58.5 |
| Male gender | 117 | 58.5 |
| Race/ethnicity | ||
| Black | 158 | 79.0 |
| Non-black | 42 | 21.0 |
| Public medical insurance | 178 | 89.0 |
| Smoke exposure* | 98 | 47.0 |
| Maternal allergy** | ||
| Yes | 70 | 35.0 |
| No | 119 | 59.5 |
| Unknown | 11 | 5.5 |
| Number positive skin tests (in addition to mouse) | ||
| 0 | 9 | 4.5 |
| 1–2 | 28 | 14.0 |
| 3+ | 163 | 81.5 |
The age of the participants ranged 5.2–7.5 years (mean 9.7±3.4 standard deviation (SD)).
Defined as any smoker in the home
Based on maternal report
Boys had lower baseline buccal DNA methylation than girls at the FOXP3 enhancer at both visits (mean±SD: visit 1-boys: 4.96 ±5.38% vs. girls: 40.59±7.61%; visit 2-boys 4.39±5.01 vs. girls: 41.02±6.11; p <0.0001 for both visits). Baseline sex effects were apparent in the opposite direction at the two FOXP3 promoter sites (mean±SD FOXP3 CpG−138: visit 1-boys: 66.38±11.28% vs. girls: 46.03±11.03%; visit 2-boys: 69.12±12.57 vs girls: 47.40±10.70; p <0.0001 at both visits; mean±SD CpG−126: visit 1-boys: 86.15±7.10% vs. girls: 82.67±8.25%, p=0.003; visit 2-boys: 88.28±7.38 vs. girls: 84.23.40±6.60, p<0.0001). Significant sex effects for IFNγ were not observed. FOXP3 and IFNγ gene expression were associated inversely with age at visit 1 only (Spearman correlation coefficient r=−0.19, r=−0.22 respectively, p<0.05; Supplemental Table 2). Methylation at IFNγ CpG−186 also was lower with age (r=−0.18, p=0.02) at visit 2, Supplemental Table 2).
Using marginal models that accounted for within-subject correlation in repeated measures, higher levels of mouse allergen in bed dust were associated with lower methylation in the FOXP3 promoter at CpG−126 (Estimated regression coefficient B= −0.62, p=0.004), but not buccal DNA methylation at other sites or regulatory gene expression. None of the methylation variables were associated with mouse-specific IgE.
In prospective analyses of the changes in methylation or gene expression levels between study visits, we found the opposite association between allergen and methylation. Decreases in mouse allergen in the bed dust borderline correlated with decreases in methylation at the FOXP3 enhancer sites (r=0.14, p=0.05). Further, to assess whether change in mouse allergen over time predicted change in DNA methylation, regression models were run that controlled for baseline levels of mouse allergen in the bed. We found that decreases in mouse allergen predicted decreases in FOXP3 promoter methylation, in this case at the CpG−138 site (estimated regression coefficient B=1.78, p=0.01, Table 2). Controlling for FOXP3 CpG−138 and mouse allergen level at baseline, a 50% reduction of mouse allergen in bed dust was associated with mean reduction of 1.23 percent methylation in FOXP3 CpG−138.
In addition, we restricted the analyses to only those with undetectable dust mite allergen at baseline and we found the same association between change in mouse allergen and change in methylation, suggesting that there was not confounding by dust mite allergen. We reran the main models using cockroach allergen (Bla g 2 in bed dust) as the predictor on the same epigenetic outcomes. No significant association between the change in cockroach allergen over time and change in DNA methylation were detected, with and without further control for mouse allergen variable at baseline. Adding the cockroach variable at baseline did not significantly alter the associations between change in mouse allergen over time with the change in DNA methylation. We also did not find any significant changes in gene expression, including during sensitivity analyses that excluded any samples that required a high number of cycles to amplify (i.e, those with presumed very small amounts of RNA) (Table 2).
In further analyses, the changes in FOXP3 and IFNγ methylation and expression over the 6 month sampling period were not associated with a change in mouse IgE during these same 6 months plus 6 months afterwards (i.e. by the 12 month period; Table 3). These results suggest that these examples of DNA methylation did not induce sustained effects on mouse sensitization. In addition, the change in the mouse allergen in the bed was not associated directly with a change in mouse IgE levels over time. This latter finding contrasts with that from the parent study (Matsui et. al., JAMA, in press) that used random effects model and repeat measures of IgE, a different period for change in mouse allergen (6 vs 12 mo), and a larger sample.
Quantifying small differences in specific CpG targets allows us to compare methylation levels across sites. Of the two neighboring FOXP3 promoter sites (CpG−126, CpG−138), while positively correlated with each other, correlated negatively with CpG methylation levels in the FOXP3 enhancer (Supplemental Table 3). Buccal methylation levels at the FOXP3 CpG−126 site correlated negatively and weakly with IFNγ gene expression at visit 2 only. Buccal methylation at IFNγ CpG−54 and IFNγ CpG−186 correlated positively and weakly with IFNγ relative gene expression, also only at visit 2.
Overall, these findings suggest that a reduction in mouse allergen, but not cockroach, observed in the setting of IPM vs education intervention trial may have reduced FOXP3 promoter DNA methylation. The decrease in FOXP3 methylation in buccal cells may be consistent with upregulated T suppressive activity, presumably following activation of the FOXP3 gene; although, this was not detected in the buccal RNA. These findings with respect to methylation point to the gene regulatory effects of reducing allergen levels, albeit small, even if there was not a statistically significant effect on mouse specific IgE.
We also identified a strong effect of sex on buccal methylation level in the FOXP3 enhancer. The levels of FOXP3 methylation in the enhancer region in males were markedly lower than in females. In contrast, the sex-related differences were less marked in the FOXP3 promoter, and in the opposite direction. The significance of these differences is uncertain, but one could speculate that it relates to epigenetic regulation of X-chromosome inactivation (XCI). For example, female cells undergo XCI, and DNA methylation of the inactive X is known to contribute to the maintenance of its inactive state; levels of methylation within a gene silenced by XCI can vary by region. Specifically, levels of promoter methylation of genes silenced by XCI have been correlated with susceptibility to XCI, and CpG islands tend to be sites where there is greater methylation on the inactivated X chromosome compared to other areas (Sharp, Stathaki et al. 2011). Because the FOXP3 gene is subject to XCI (Tommasini, Ferrari et al. 2002), one could speculate that the higher methylation levels measured in the FOXP3 enhancer of females in this cohort reflects regulation by DNA methylation of XCI to some extent. Although this speculation would not explain the lower methylation levels found in females in the FOXP3 promoter region, other than demonstrating that variation in methylation levels across genes applies to genes silenced by XCI(Sharp, Stathaki et al. 2011).
In comparison, we previously found that methylation at the IFNγ promoter varied by sex in CD4+ lymphocyte but not buccal cell DNA (Lovinsky-Desir, Ridder et al. 2014). Here too the underlying mechanism still needs to be elucidated. For both genes, differences in hormone levels or unmeasured environmental exposures that vary by behaviors among boys and girls also may explain differential induction of DNA methylation. The sex differences in FOXP3 we observed suggest that sex effects on methylation, including its variation by CpG region, should be considered when designing studies of epigenetic biomarkers in asthma.
These findings support the premise that during an intervention against mouse allergen, the changing environment over time may drive some plasticity in the level of the DNA methylation, as reported in other studies(Bind, Lepeule et al. 2014, Borsch-Haubold, Montero et al. 2014, McErlean, Favoreto et al. 2014). The direction of these results appear different than observed in one report in pollen-allergic patients where local challenge of the nasal mucosal with pollen extract led to an increase in the FOXP3 expressing T cells(Skrindo, Scheel et al. 2011). But other studies have not examined epigenetic regulation in symptomatic children following chronic, daily allergen exposure. Further, it could be consistent with other environmental exposures like respiratory syncytial virus infection that reduced T reg function, thereby promoting susceptibility to allergic asthma (Krishnamoorthy, Khare et al. 2012). We believe these associations are gene specific as there is substantial evidence that epigenetic regulation of both FOXP3 and IFNγ is associated with clinically relevant outcomes (Liu, Ballaney et al. 2008, Brand, Kesper et al. 2012, Kohli, Garcia et al. 2012, Niedzwiecki, Zhu et al. 2012, Runyon, Cachola et al. 2012, Tang, Levin et al. 2012). There is more limited evidence to suggest that differences in global methylation instead may explain outcomes related to exposure to cigarettes and air pollution (Breton, Byun et al. 2009, Lee, Jaffar et al. 2015, Breton, Yao et al. 2016) and allergic sensitization(Sordillo, Lange et al. 2013) in some, but not other (Chi, Liu et al. 2016), studies.
Hence, with this prospective and repeat sampling design, we may have identified novel conditions for which a change in the indoor environment altered the promoter methylation profile of an immunoregulatory gene, in this case suggesting that the maintenance of T regulatory function may have become further upregulated. The results may reveal that highly exposed, allergic and asthmatic children could be resistant to usual allergen-induced regulatory mechanisms due to undefined molecular impairments(Yamamoto, Negoro et al. 2011), or the contribution of other genes or CpG sites within the same genes, or even other epigenetic pathways like histone acetylation or hydroxymethylation(Planell-Saguer, Lovinsky-Desir et al. 2014). However, the results also do not support the paradigm that decreased FOXP3 methylation is part of the causal pathway leading to changes in mouse IgE, at least according to this timeline for a clinical allergic response. Rather, FOXP3 methylation may act more as a marker of effective changes in allergen exposure, or potentially of functional changes in other immune cells.
The relevance of the expression of FOXP3 and IFNγ in the buccal mucosa described here and elsewhere(Scadding, Shamji et al. 2010), and their exact roles following allergen remediation, are still not elucidated from these results. FOXP3 methylation, perhaps because it may capture changes over a longer time period than buccal gene expression, may contribute more to the immune response to changed allergen exposure. Indeed, altered buccal DNA methylation has been associated with airway inflammation in previous pediatric cohorts(Breton, Byun et al. 2011). Striking variation by CpG site within the promoter was detected and may suggest that individual CpG site may function independently, as reported(Clifford, Jones et al. 2017). The marginal models with repeated measures that demonstrated patterns opposite to our prospective findings may have been susceptible to bias due to unmeasured confounding factors. Nonetheless, we also acknowledge that the effect sizes were generally small and we were working with a mixed population of cells. Buccal epithelial cells were used in large part because the process of obtaining them is noninvasive, which is critical in pediatric cohorts studies. We do not yet know whether the changes are universal or similar across tissues, as several asthma studies have shown differences in some gene methylation patterns by cell type(Stefanowicz, Hackett et al. 2012, Lovinsky-Desir, Ridder et al. 2014). But buccal cells have revealed epigenetic patterns relevant to allergy, when collected using comparable techniques (Breton, Byun et al. 2011, Kuriakose, Torrone et al. 2011, Salam, Byun et al. 2012, Lovinsky-Desir, Ridder et al. 2014). Our focus on the cytokine profile in buccal epithelial cells is novel, so additional studies would be beneficial to understand more fully their translation. Moreover, cockroach allergen was detected only in 25% of the homes at baseline, with borderline significant decreases over time. This observation, plus our analyses suggesting that this relatively infrequent exposure did not predict epigenetic outcomes, makes confounding by cockroach allergen less likely.
In conclusion, the longitudinal assessment of DNA methylation in this unique pediatric cohort suggests that changing environmental exposure can induce epigenetic regulation, and T suppressive activity may even increase when high levels of mouse allergen are diminished. They also support the epidemiological paradigm that findings in cross-sectional analyses may not replicate under prospective evaluations. Our results place us one step further towards understanding immunoregulation and its susceptibility to change following changes in environmental exposures. Given the high public health burden associated with frequent sensitization to mouse in inner city asthma(Matsui, Eggleston et al. 2006, Ahluwalia, Peng et al. 2013), these results may provide a foundation for pursuing the hypothesis further that allergen interventions have the potential to modify the natural course of disease among susceptible children through epigenetic mechanisms.
The IPM group received two IPM visits at 0.5 and 1.5 months (solid arrows). If there was evidence of mouse infestation at 3, 6, and 9 months, a subsequent IPM visit occurred at 3.5, 6.5 (see dashed arrows) and 9.5 months (not shown). The second mouse IgE level was measured at 12 month timepoint, in contrast to the allergen (Mus m 1, Bla g 2) and epigenetic biomarkers, each measured at baseline and 6.0 months.
Supplementary Material
Supplemental Table 1A. Pyrosequencing primers; 1B. qRT-PCR primers.
Supplemental Table 2. Correlations between buccal biomarkers and predictors, by visit.
Supplemental Table 3. Correlations among individual buccal biomarkers, by visit.
Highlights.
Decreasing mouse allergen exposure may reduce FOXP3 buccal DNA promoter methylation.
Decreasing mouse allergen exposure did not alter mouse specific IgE.
Epigenetic regulation may occur following changing allergen exposures.
Acknowledgments
Funding Sources
This work was supported by the National Institutes of Health (1R21 AI101296, P30 ES009089, U01 AI083238, R01 ES023447, K24 AI 114769 and R01ES026170).
This study has been approved by Columbia University Human Subjects protocol AAAJ6908, Johns Hopkins University SOM protocol number NA_00021358, and Harvard University protocol number 08-09-0404.
Abbreviations
- FOXP3
forkhead box P3 gene
- Ig
immunoglobulin
- ICAS
Inner-City Asthma Study
- IPM
integrated pest management
- IFN
interferon
- IQR
interquartile range
- MAAIT
Mouse Allergen and Asthma Intervention Trial
- PASTURE
Protection Against Allergy: Study in Rural Environments
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahluwalia S, Peng R, Breysse P, Diette G, Curtin-Brosnan J, Aloe C, Matsui E. Mouse allergen is the major allergen of public health relevance in Baltimore City. J Allergy Clinical Immunology. 2013;132(4):830–835. doi: 10.1016/j.jaci.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky D, Sears M, Hancox R, Harrington H, Houts R, Moffitt T, Sugden K, Williams B, Poulton R, Caspi A. Polygenic risk and the development and course of asthma: Evidence from a 4-decade longitudinal study. Lancet Respir Med. 2013;1(6):453–461. doi: 10.1016/S2213-2600(13)70101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani M, Pathak A, Fan Y, Liu D, Lee J, Tang H, Kurie J, Morice R, Kim E, Hong W, Mao L. Oral epithelium as a surrogate tissue for assessing smoking-induced molecular alterations in the lungs. Cancer Prev Res. 2008;1(1):39–44. doi: 10.1158/1940-6207.CAPR-08-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bind M-A, Lepeule J, Zanobetti A, Gasparrini A, Baccarelli A, Coull BA, Tarantini L, Vokonas PS, Koutrakis P, Schwartz J. Air pollution and gene-specific methylation in the Normative Aging Study: Association, effect modification, and mediation analysis. Epigenetics. 2014;9(3):448–458. doi: 10.4161/epi.27584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsch-Haubold AG, Montero I, Konrad K, Haubold B. Genome-wide quantitative analysis of histone H3 lysine 4 trimethylation in wild house mouse liver: Environmental change causes epigenetic plasticity. PlosOne. 2014;9(5):e97568. doi: 10.1371/journal.pone.0097568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand S, Kesper A, Teich Re, Kilic-Niebergall E, Pinkenburg O, Bothur E, Lohoff M, Garn H, Pfefferle PI, Renz H. DNA methylation of TH1/TH2 cytokine genes affects sensitization and progress of experimental asthma. J of Allergy Clin. Immunol. 2012;129:1602–1610. doi: 10.1016/j.jaci.2011.12.963. [DOI] [PubMed] [Google Scholar]
- Breton C, Yao J, Millstein J, Gao L, Siegmund K, Mack W, Whitfield-Maxwell L, Lurmann F, Hodis H, Avol E, Gilliland F. Prenatal air pollution exposures, DNA methyl transferase genotypes, and associations with newborn LINE1 and Alu methylation and childhood blood pressure and carotid intima-media thickness in the Children's Health Study. Environ Health Perspect. 2016;124(12):1905–1912. doi: 10.1289/EHP181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton CV, Byun H-M, Wang X, Salam MT, Siegmund K, Gilliland FD. DNA methylation in the ARG-NOS pathway is associated with exhaled nitric oxide in asthmatic children. American J of Respiratory and Critical Care Medicine. 2011;184(2):191–197. doi: 10.1164/rccm.201012-2029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton CV, Byun H-M, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. American J. Respiratory and Critical Care Medicine. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi G, Liu Y, MacDonald J, Barr R, Donohue K, Hensley M, Hou L, McCall C, Reynolds L, Siscovick D, Kaufman J. Long-term outdoor air pollution and DNA methylation in circulating monocytes: results from the Multi-Ethnic Study of Atherosclerosis (MESA) Environ Health. 2016;15(1):119. doi: 10.1186/s12940-016-0202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford RL, Jones MJ, MacIsaac JL, McEwen LM, Goodman SJ, Mostafavi S, Kobor MS, Carlsten C. Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. J Allergy Clin Immunol. 2017;139(1):112–121. doi: 10.1016/j.jaci.2016.03.046. [DOI] [PubMed] [Google Scholar]
- Collison A, Siegle J, Hansbro N, Kwok C-T, Herbert C, Mattes J, Hitchins M, Foster P, Kumar R. Epigenetic changes associated with disease progression in a mouse model of childhood allergic asthma. Disease Models and Mechanisms. 2013;6(4):993–1000. doi: 10.1242/dmm.011247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsky R, Deem RL, Targan SR. Distinct methylation of IFNG in the gut. Journal of Interferon and Cytokine Research. 2009;29(7):407–414. doi: 10.1089/jir.2008.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Ciaccio C, Barnes C, Kennedy K, Forrest E, Gard L, Pacheco F, Dowling P, Portnoy J. Low-cost interventions improve indoor air quality and children's health. Allergy Asthma Proc. 2009;30(4):377–385. doi: 10.2500/aap.2009.30.3257. [DOI] [PubMed] [Google Scholar]
- Kohli A, Garcia M, Maher C, Balmes J, Tager I, Hammond K, Miller R, Nadeau K. Secondhand smoke in combination with ambient air pollution exposure is associated with increased CpG methylation and decreased expression of IFN-g in T effector cells and FoxP3 in T regulatory cells. Journal of Clinical Epigenetics. 2012;4(1):17. doi: 10.1186/1868-7083-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy N, Khare A, Oriss T, Raundhal M, Morse C, Yarlagadda M, Wenzel S, Moore M, Peebles R, Ray A, Ray P. Early infection with respiratory syncytial virus impairs regulatory T cell function and increases susceptibility to allergic asthma. Nat Med. 2012;18(10):1525–1530. doi: 10.1038/nm.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose J, Torrone D, Gauvey-Kern K, Hsu S, Niedzwiecki M, Jiang H, Perera F, Miller R. Intra-individual variation in DNA methylation of IFN-γ and IL-4 over days. J. Allergy and Clin. Immunol. 2011;127(2):AB271. [Google Scholar]
- Lee J, Jaffar Z, Pinkerton K, Porter V, Postma B, Ferrini M, Holian A, Roberts K, Cho Y. Alterations in DNA methylation and airway hyperreactivity in response to in utero exposure to environmental tobacco smoke. Inhal Toxicol. 2015;27(13):724–730. doi: 10.3109/08958378.2015.1104402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ballaney M, Al-alem U, Quan C, Jin X, Perera F, Chen L-C, Miller RL. Combined inhaled diesel exhaust particles and allergen exposure alter methylation of T helper genes and IgE production in vivo. Toxicological Sciences. 2008;102(1):76–81. doi: 10.1093/toxsci/kfm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovinsky-Desir S, Ridder R, Torrone D, Maher C, Narula S, Scheuerman M, Merle D, Kattan M, DiMango E, Miller RL. DNA methylation of the allergy regulatory gene, interferon gamma, varies by age, sex, and tissue type in asthmatics. Journal of Clinical Epigenetics. 2014;6:9. doi: 10.1186/1868-7083-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magister S, Kos J. Cystatins in immune system. J. Cancer. 2013;4(1):45–56. doi: 10.7150/jca.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui E, Eggleston P, Buckley T, Krishnan J, Breysse P, Rand C, Diette G. Household mouse allergen exposure and asthma morbidity in inner-city preschool children. Ann Allergy Asthma Immunol. 2006;97(4):514–520. doi: 10.1016/S1081-1206(10)60943-X. [DOI] [PubMed] [Google Scholar]
- McErlean P, Favoreto S, Costa FF, Shen J, Quraishi J, Biyasheva A, Cooper JJ, Scholtens DM, Vanin EF, Bonaldo MFd, Xie H, Soares MB, Avila PC. Human rhinovirus infection causes different DNA methylation changes in nasal epithelial cells from healthy and asthmatic subjects. BMC Medical Genomics. 2014;7(37):1–10. doi: 10.1186/1755-8794-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel S, Busato F, Genuneit J, Pekkanen J, Dalphin J, Riedler J, Mazaleyrat N, Weber J, Karvonen A, Hirvonen M, Braun-Fahrländer C, Lauener R, Mutius Ev, Kabesch M, Tost J. Farm exposure and time trends in early childhood may influence DNA methylation in genes related to asthma and allergy. Allergy. 2013;68(3):355–364. doi: 10.1111/all.12097. [DOI] [PubMed] [Google Scholar]
- Morgan WJ, Crain EF, Gruchalla RS, O’Connor GT, Kattan M, Evans R, Stout J, Malindzak G, Smartt E, Plaut M, Walter M, Vaughn B, Mitchell H. Results of a home-based environmental intervention among urban children with asthma. The New England Journal of Medicine. 2004;351(1068–80) doi: 10.1056/NEJMoa032097. [DOI] [PubMed] [Google Scholar]
- Murphy S, Adigun A, Huang Z, Overcash F, Wang F, Jirtle R, Schildkraut J, Murtha A, Iversen E, Hoyo C. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene. 2012;494(1):36–43. doi: 10.1016/j.gene.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau K, McDonald-Hyman C, Noth E, Pratt B, Hammond S, Balmes J, Tager I. Ambient air pollution impairs regulatory T-cell function in asthma. J. Allergy and Clin. Immunol. 2010;126(4):845–852. doi: 10.1016/j.jaci.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Niedzwiecki M, Zhu H, Corson L, Grunig G, Factor PH, Chu S, Jiang H, Miller RL. Prenatal exposure to allergen and DNA methylation, allergy in grandoffspring mice. Allergy. 2012;67:904–910. doi: 10.1111/j.1398-9995.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planell-Saguer Md, Lovinsky-Desir S, Miller RL. Epigenetic regulation: The interface between prenatal and early-life exposure and asthma susceptibility. Environ Mol Mutagen. 2014;55(3):231–243. doi: 10.1002/em.21836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongracic J, Visness C, Gruchalla R, Evans R, Mitchell H. Effect of mouse allergen and rodent environmental intervention on asthma in inner-city children. Ann Allergy Asthma Immunol. 2008;101(1):35–41. doi: 10.1016/S1081-1206(10)60832-0. [DOI] [PubMed] [Google Scholar]
- Richmond R, Simpkin A, Woodward G, Gaunt T, Lyttleton O, McArdle W, Ring S, Smith A, Timpson N, Tilling K, Smith D, Relton C. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC) Hum. Mol. Genet. 2015;24:8. doi: 10.1093/hmg/ddu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runyon RS, Cachola LM, Rajeshuni N, Hunter T, Garcia M, Ahn R, Lurman F, Krasnow R, Jack L, Miller RL, Swan G, Jacobson AC, Nadeau KC. Asthma discordance in twins is mediated by epigenetic modifications of T cells. PLoS One. 2012;7(11):e48796. doi: 10.1371/journal.pone.0048796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salam MT, Byun H-M, Lurmann F, Breton CV, Wang X, Eckel SP, Gilliland FD. Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J of Allergy Clin. Immunol. 2012;129:232–239. doi: 10.1016/j.jaci.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadding GW, Shamji MH, Jacobson MR, Lee DI, Wilson D, Lima MT, Pitkin L, Pilette C, Nouri-Aria K, Durham SR. Sublingual grass pollen immunotherapy is associated with increases in sublingual Foxp3 expressing cells and elevated allergen-specific immunoglobulin G4, immunoglobulin A and serum inhibitory activity for immunoglobulin E-facilitated allergen binding to B cells. Clinical and Experimental Allergy. 2010;40:598–606. doi: 10.1111/j.1365-2222.2010.03462.x. [DOI] [PubMed] [Google Scholar]
- Sedaghat AR, Matsui E, Baxi S, Bollinger M, Miller R, Perzanowski M, Phipatanaku W. Mouse sensitivity is an independent risk factor for rhinitis in children with asthma. Journal of Allergy and Clinical Immunology-In Practice. 2016;4(1):82–88. doi: 10.1016/j.jaip.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, Antonarakis SE. DNA methylation profiles of human active and inactive X chromosomes. Genome Research. 2011;21:1592–1600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrindo I, Scheel C, Johansen F, Jahnsen F. Experimentally induced accumulation of Foxp3+ T cells in upper airway allergy. Clin Exp Allergy. 2011;41(7):954–962. doi: 10.1111/j.1365-2222.2011.03710.x. [DOI] [PubMed] [Google Scholar]
- Sordillo J, Lange N, Tarantini L, Bollati V, Zanobetti A, Sparrow D, Vokonas P, Schwartz J, Baccarelli A, Demeo D, Litonjua A. Allergen sensitization is associated with increased DNA methylation in older men. Int Arch Allergy Immunol. 2013;161(1):37–43. doi: 10.1159/000343004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanowicz D, Hackett T, Garmaroudi F, Günther O, Neumann S, Sutanto E, Ling K, Kobor M, Kicic A, Stick S, Paré P, Knight D. DNA methylation profiles of airway epithelial cells and PBMCs from healthy, atopic and asthmatic children. PloSOne. 2012;7(9):e44213. doi: 10.1371/journal.pone.0044213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W-y, Levin L, Talaska G, Cheung YY, Herbstman J, Miller RL, Perera F, Ho S-M. Maternal exposure to polycyclic aromatic hydrocarbons is associated with methylation at a 5’ -CpG island of interferon-γ in cord white blood cells. Environ Health Perspect. 2012;120:1195–1200. doi: 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini A, Ferrari S, Moratto D, Badolato R, Boniotto M, Pirulli D, Notarangelo D, Andolina M. X-chromosome inactivation analysis in a female carrier of FOXP3 mutation. Clin. Expo. Immunol. 2002;130:127–113-. doi: 10.1046/j.1365-2249.2002.01940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan E, Qiu W, Carey V, Morrow J, Bacherman H, Foreman M, Hokanson J, Bowler R, Crapo J, DeMeo D. Smoking associated site specific differential methylation in buccal mucosa in the COPDGene study. Am J Respir Cell Mol Biol. 2014;53(2):246–254. doi: 10.1165/rcmb.2014-0103OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Negoro T, Hoshi A, Wakagi A, Shimizu S, Banham A, Ishii M, Akiyama H, Kiuchi Y, Sunaga S, Tobe T, Roncador G, Itabashi K, Nakano Y. Impaired Ca2+ regulation of CD4+ CD25+ regulatory T cells from pediatric asthma. Int Arch Allergy Immunol. 2011;156(2):148–158. doi: 10.1159/000322845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1A. Pyrosequencing primers; 1B. qRT-PCR primers.
Supplemental Table 2. Correlations between buccal biomarkers and predictors, by visit.
Supplemental Table 3. Correlations among individual buccal biomarkers, by visit.