SUMMARY
Never-in-mitosis A-related kinase 1 (Nek1) has established roles in apoptosis and cell cycle regulation. We show that human Nek1 regulates homologous recombination (HR) by phosphorylating Rad54 at Ser572 in late G2 phase. Nek1 deficiency as well as expression of unphosphorylatable Rad54 (Rad54-S572A) cause unresolved Rad51 foci and confer a defect in HR. Phosphomimic Rad54 (Rad54-S572E), in contrast, promotes HR and rescues the HR defect associated with Nek1 loss. Although expression of phosphomimic Rad54 is beneficial for HR, it causes Rad51 removal from chromatin and degradation of stalled replication forks in S phase. Thus, G2-specific phosphorylation of Rad54 by Nek1 promotes Rad51 chromatin removal during HR in G2 phase, and its absence in S phase is required for replication fork stability. In summary, Nek1 regulates Rad51 removal to orchestrate HR and replication fork stability.
In Brief
Spies et al. uncover the participation of the kinase Nek1 during homologous recombination. Nek1 phosphorylates Rad54 in G2 to promote Rad51 removal. Untimely phosphorylation of Rad54 and subsequent removal of Rad51 in S phase causes replication fork instability. The authors hereby demonstrate the physiological relevance of Rad54 regulation.
INTRODUCTION
Two main pathways exist for the repair of two-ended double-strand breaks (DSBs), non-homologous end-joining (NHEJ), and homologous recombination (HR), the latter operating during S and G2 phase when the sister chromatid is available as a template for repair (van Gent et al., 2001; Lukas and Lukas, 2013). HR is initiated by resection of the 5′-end and Rad51 loading to single-stranded DNA (ssDNA). Later stages of HR involve homology search, DNA strand invasion, and repair synthesis to copy the missing sequence information at the break site from the donor sister chromatid (Mazón et al., 2010; Renkawitz et al., 2014). HR is finalized by the dissolution or resolution of the formed Holliday junctions (Matos and West, 2014).
In contrast to two-ended exogenously induced DSBs, which can be efficiently repaired by HR and NHEJ, HR is the predominant pathway for dealing with one-ended DSBs that arise at the replication fork (Chapman et al., 2012; Moynahan and Jasin, 2010). Such DSBs occur at appreciable frequencies endogenously when replication forks encounter spontaneous base damages and/or single-strand breaks but also arise from agents that induce such single-stranded lesions (Ensminger et al., 2014; Jeggo and Löbrich, 2015). In addition to their role in repairing one-ended DSBs, HR factors also exert important functions in protecting stalled replication forks and their absence leads to degradation of newly synthesized DNA (Branzei and Foiani, 2010; Schlacher et al., 2012). The timely completion of replication is important as its failure can lead to the occurrence of under-replicated DNA regions that give rise to chromosome breakage during mitosis (Naim et al., 2013; Ying et al., 2013).
The motor protein Rad54 has multiple roles in HR-mediated DSB repair. A critical role is thought to occur after homology search is complete, to transform the synaptic complex containing three homologously aligned DNA strands (ssDNA:Rad51:dsDNA) into heteroduplex DNA. During this process promoted by Rad54’s ATPase activity, Rad51 is removed from DNA which allows 3′-end access and subsequent repair synthesis by DNA polymerases to enable the completion of HR (Agarwal et al., 2011; Ceballos and Heyer, 2011; Wright and Heyer, 2014). In the absence of Rad54, Rad51 is not removed and HR cannot be completed. Besides its role in HR, Rad51 also functions to protect stalled replication forks from degradation (Hashimoto et al., 2010; Schlacher et al., 2011). It is unclear whether fork protection is endowed by Rad51 bound to ssDNA, dsDNA, or the synaptic complex. Notably, Rad54 is not required for fork protection (Schlacher et al., 2011), suggesting that Rad51 is not removed from stalled replication forks. This raises the conceptual question of how Rad54 is differentially regulated to remove Rad51 from DNA during HR but not during replication fork stalling.
We have previously observed that gene expression of never-in-mitosis A related kinase 1 (Nek1), a member of the mammalian Nek family with highly conserved serine/threonine (Ser/Thr) and tyrosine kinase motives (Meirelles et al., 2014), is significantly upregulated in cells exposed to ionizing radiation (IR) (Grudzenski et al., 2010). The few reports available for Nek family members explored the roles of Nek8 and Nek11 at the replication fork and during checkpoint activation, respectively (Choi et al., 2013; Melixetian et al., 2009). Nek1 is also implicated in the DNA damage response by its roles during apoptosis and cell cycle regulation (Chen et al., 2008, 2009, 2011a, 2014). More recently, Nek1 was shown to be required for proper ATR activation (Liu et al., 2013). Although Nek1-deficient cells display elevated chromosome breaks following DNA damaging agents (Chen et al., 2008), it is unclear if this phenotype results from its established role in cell cycle checkpoint regulation or represents a genuine function in a DSB repair process.
Here, we show that Nek1 phosphorylates Rad54 specifically in the G2 phase of the cell cycle. This promotes Rad51 removal from chromatin and allows the completion of HR. The absence of Rad54 phosphorylation during S phase prevents removal of Rad51 from stalled replication forks and ensures fork protection.
RESULTS
Nek1 Functions during the DNA Damage Response and Serves to Maintain Genomic Stability
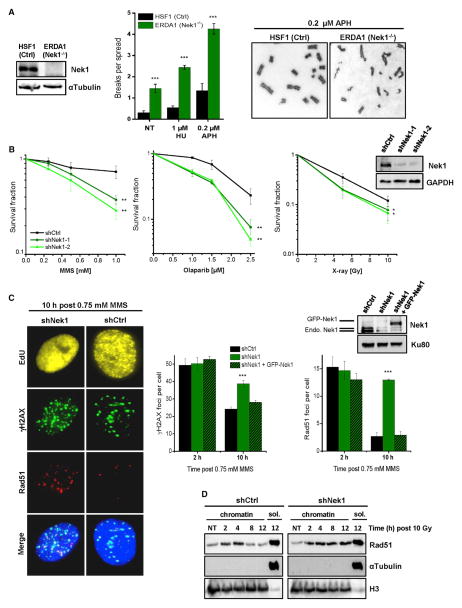
To explore the function of Nek1 during the DNA damage response, we first analyzed fibroblasts from a patient with the human disorder short-rib polydactyly syndrome (SRPS) type Majewski that harbors a nonsense mutation in Nek1 (Thiel et al., 2011). Such cells show proliferation defects following treatment with the DNA damaging agent methylmethane sulfonate (MMS) (Figure S1A) and exhibit pronounced chromosomal instability after treatment with low concentrations of hydroxyurea (HU) and aphidicolin (APH), which also induce DNA damage (Figure 1A). Since these primary cells were poorly growing, we generated Nek1-deficient HeLa cells by shRNA technology. Using these cells, we observed substantially diminished colony formation after MMS and olaparib (PARP inhibitor) treatment, and a more modest reduction after X-rays (Figure 1B), consistent with earlier findings that loss of Nek1 expression confers sensitivity to genotoxic agents (Chen et al., 2011b; Polci et al., 2004). Because these agents induce DSBs, we investigated the efficiency of DSB repair in Nek1-depleted cells by analyzing γH2AX and Rad51 foci, both markers for DSBs. We pulse-labeled growing cell populations with the thymidine analog EdU and quantified foci in EdU-positive cells, which represent cells that were in S phase at the time of MMS treatment (Figure S1B). We observed high foci levels early after MMS treatment that decreased due to repair while cells progressed into G2 (Figures 1C and S1B). Nek1-deficient cells showed foci levels similar to control cells at initial time points but substantially elevated levels at later times, suggesting that Nek1 is involved in repairing DSBs. Of note, the defect was most striking for Rad51 foci, which monitor the repair of resected DSBs by HR (Figure 1C). The elevated foci levels were rescued by expression of shRNA-resistant GFP-Nek1 (Figure 1C). Because MMS induces DSBs during replication, we wished to explore if Nek1 has a general role during HR (as opposed to a more specific role during replication) and investigated Rad51 removal from DSBs induced by IR outside of S phase. We synchronized cells in G2 and assessed chromatin-bound Rad51 levels by immunoblotting. Chromatin-bound Rad51 increased in control cells until 2–4 hr after IR and then decreased due to repair. In Nek1-deficient cells, the increase was similar but Rad51 was not released from chromatin until at least 12 hr post IR (Figures 1D and S1C). As discussed below, these data were confirmed analyzing Rad51 foci.
Figure 1. Nek1 Functions during the DNA Damage Response and Serves to Maintain Genomic Stability.
(A) Chromosome spreads of human fibroblasts. Chromatid breaks were analyzed in Nek1-deficient (ERDA1) and control (HSF1) cells both spontaneously (NT, not treated) and after a 20-hr exposure to HU or APH. Mean ± SEM (n = 3).
(B) Clonogenic survival of Nek1-deficient cells. Two independent shNek1 HeLa cell clones were generated by genomic shRNA insertion. Non-silencing shRNA was used as a control. DNA damage was induced by MMS (for 1 hr), olaparib (permanent), or X-rays. Mean ± SEM (n = 3).
(C) γH2AX and Rad51 foci in Nek1-depleted and GFP-Nek1-complemented cells. Asynchronous cells were co-treated with MMS and EdU for 1 hr. γH2AX and Rad51 foci were enumerated in EdU-positive cells (Figure S1B). Mean ± SEM (n = 3); spontaneous foci were subtracted.
(D) Chromatin fraction of Rad51 in Nek1-depleted cells. Synchronized cells were X-irradiated in G2 (Figure S1C) and chromatin fractions were analyzed for Rad51 by immunoblotting. H3 and αTubulin signals demonstrate the efficiency of chromatin fractionation (sol., soluble fraction).
Nek1 Functions during DSB Repair by HR and Interacts with Rad54
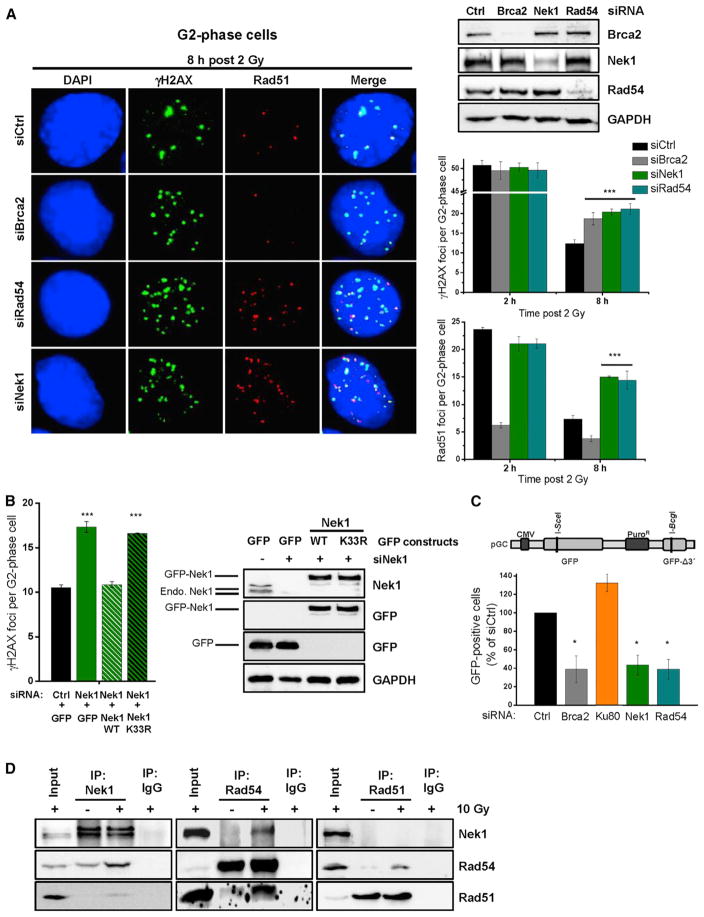
The failure of Nek1-deficient cells to remove Rad51 from DSBs suggests that Nek1 has a role during HR. We therefore investigated DSB repair kinetics after IR in G1- and G2-phase cells as previously described (Löbrich et al., 2010) (Figure S2A). IR-induced DSBs are repaired by NHEJ in G1 and by NHEJ or HR in G2 (Rothkamm et al., 2003; Riballo et al., 2004). We depleted Nek1 by siRNA and observed similar γH2AX foci levels as in control cells at all time points in G1 suggesting that Nek1 is not involved in NHEJ (Figure S2B). In G2 phase, γH2AX and Rad51 foci levels in Nek1-deficient cells were similar to control cells initially but were elevated compared to control cells at later times after IR (Figures 2A and S2B). The elevated γH2AX foci level was similar to Brca2- and Rad54-depleted cells while the elevated Rad51 foci level was similar to Rad54-deficient cells but distinct from Brca2-depleted cells (Figure 2A). This reflects the role of Brca2 in Rad51 filament formation and the function of Rad54 during Rad51 removal (Moynahan and Jasin, 2010; Shah et al., 2010). A second siRNA for Nek1 provided the same result (Figure S2C). Of note, concomitant downregulation of Nek1 and Rad54 provided no further increase than the single Nek1 or Rad54 knockdowns (Figure S2C). Furthermore, wild-type (WT) but not kinase-deficient Nek1 (Nek1-K33R) rescued the elevated foci level of Nek1-depleted cells (Figure 2B). Fibroblasts from SRPS patients also showed kinetics for γH2AX foci removal distinct to NHEJ mutants but similar to HR mutants (Figure S2D), and Nek1-depleted non-transformed G2 fibroblasts exhibited elevated Rad51 foci levels, demonstrating that the repair defect is not cell line dependent (Figure S2E). Collectively, these data support the conclusion that Nek1 operates during HR.
Figure 2. Nek1 Functions during DSB Repair by HR and Interacts with Rad54.
(A) γH2AX and Rad51 foci in Nek1-, Rad54-, and Brca2-depleted cells. HeLa cells were treated with siRNAs, EdU labeled, and X-irradiated. γH2AX and Rad51 foci were analyzed in EdU-negative G2-phase cells (Figure S2A). Mean ± SEM (n = 3); spontaneous foci were subtracted.
(B) γH2AX foci in catalytically deficient Nek1 cells. HeLa cells were treated with siNek1, transfected with siRNA resistant plasmids, X-irradiated, and γH2AX foci were enumerated 8 hr post 2 Gy in G2-phase cells identified as in (A). Mean ± SEM (n = 3); spontaneous foci were subtracted.
(C) GFP-based HR reporter assay with Nek1-, Rad54-, Brca2-, and Ku80-depleted cells. HeLa pGC cells were treated with siRNAs and transfected with an I-SceI plasmid. The number of GFP-positive cells was analyzed by IF microscopy. Mean ± SEM (n = 4).
(D) Physical interaction between Nek1, Rad54, and Rad51 in HeLa cells. Proteins were immunoprecipitated from nuclear cell extracts at 5 hr post irradiation and interactions were tested by immunoblotting.
To further substantiate this notion, we used HeLa cells containing an integrated HR reporter with two differentially mutated GFP genes (Mansour et al., 2008). Expression of the endonuclease I-SceI generates a DSB in one of the two genes that can be repaired by HR with the second gene copy serving as a template, resulting in a cell expressing functional GFP. HR frequencies assessed by the fraction of GFP-positive cells were significantly decreased after depletion of HR but not NHEJ factors. Strikingly, Nek1-depleted cells showed a reduction in GFP-positive recombinants identical to Brca2- or Rad54-depleted cells (Figures 2C and S2F). We also measured the formation of sister chromatid exchanges (SCEs), which arise due to HR. As previously described (Conrad et al., 2011), IR in G2-phase cells induces SCEs. Nek1-depleted cells showed reduced SCE levels similar to Brca2- and Rad54-depleted cells (Figure S2G). Finally, we assessed DNA synthesis occurring during later stages of HR. For this, we quantified the incorporation of the nucleotide analog EdU following irradiation of G2-phase cells. Nuclear EdU foci arise in control cells within 8 hr post IR, and depletion of HR but not NHEJ factors abolishes EdU foci formation (Beucher et al., 2009). Nek1-depleted cells exhibited the same defect as Rad54-depleted cells (Figure S2H). In summary, these data firmly establish that Nek1 is a critical HR factor.
The elevated level of unresolved Rad51 foci and the failure to perform DNA synthesis suggest that Nek1 functions after resection but before repair synthesis. This is similar to Rad54 (Essers et al., 2002) and, indeed, all assays performed in the present study provided identical results for Nek1- and Rad54-deficient cells. Therefore, we investigated if Nek1 interacts with Rad54 by co-immunoprecipitation (coIP) experiments. We confirmed the presence of Rad51 in IPs from Rad54 (Heyer et al., 2006), and detected Rad54 but not Rad51 in IPs from Nek1 and vice versa (Figure 2D). The interactions were induced by IR (Figure 2D), resisted DNase treatment suggesting that they are independent of DNA, and were confirmed in another cell system (Figure S2I).
Nek1 Phosphorylates Rad54 at Ser572 Specifically in G2 Phase
The interaction between Nek1 and Rad54 raised the possibility that Rad54 is a phosphorylation target of Nek1. To identify potential Nek1 phosphorylation sites on Rad54, we screened Rad54 for Nek1 consensus sites, Ser/Thr residues with phenylalanine at position −3 relative to Ser/Thr (Chen et al., 2009; Surpili et al., 2003). Rad54-Ser572 is such a Nek1 consensus site located in a highly conserved ATPase domain and is also surface-exposed (Thomä et al., 2005). We mutated Ser572 to the unphosphorylatable (phospho mutant) alanine (S572A) or the potentially phosphomimic glutamate (S572E) (Figure 3A). First, we performed an in vitro kinase assay with immunoprecipitated GFP-Rad54, recombinant Nek1, and radioactive ATP. GFP-Rad54-WT but not GFP-Rad54-S572A was readily phosphorylated by Nek1 (Figure 3B). To verify Nek1-dependent Rad54 phosphorylation at Ser572 (Rad54-pS572), we used a phospho-specific antibody which provided a signal for immunoprecipitated GFP-Rad54-WT but not GFP-Rad54-S572A proteins incubated with Nek1 and ATP (Figure 3C). Importantly, Rad54-pS572 was observed in vivo in whole cell extracts of HeLa and Hek293 cells in a manner dependent on Nek1 (Figure 3D). We then investigated the time-course of Rad54 phosphorylation in nuclear cell extracts of synchronized G2-phase cells and observed strongly increased Rad54-pS572 levels at 8 hr after IR, a time when Rad51 is removed from chromatin but irradiated G2-phase cells have not yet entered mitosis (Deckbar et al., 2007) (Figures 3E and S3A). We also assessed Rad54-pS572 levels in S-phase cells treated with DNA-damaging agents. Of note, Rad54-pS572 is delayed in damaged S-phase cells and does not reach its maximum level until the cells have progressed into G2 phase (Figures 3F and S3B). A slight increase in Rad54 phosphorylation from S to G2 was also observed in undamaged cells (Figure S3C). We finally aimed to assess the fraction of Rad54, which is phosphorylated by Nek1 after damage induction. We used Phos-tag gels, which allow the visualization of phosphorylation events by band shifts. We used G2-synchronized cells and detected only minor Rad54 phosphorylation events in unirradiated cells. In contrast, about half of all Rad54 proteins were phosphorylated at 8 hr after IR (Figure 3G). The fraction of phosphorylated Rad54 was reduced following phosphatase treatment or Nek1 siRNA (Figure 3G).
Figure 3. Nek1 Phosphorylates Rad54 at Ser572 Specifically in G2 Phase.
(A) Schematic diagram showing the position of Ser572 within ATPase domain V of Rad54. GFP-Rad54 plasmids with point mutations S572A and S572E were generated by site-directed mutagenesis.
(B) Detection of Rad54 phosphorylation using autoradiography. GFP-coupled Rad54-WT or Rad54-S572A was obtained from transfected Hek293 cells by IP. The in vitro kinase assay was performed with radioactive ATP and constitutively active recombinant Nek1. The presence of Rad54 in the reaction was controlled by immunoblotting. Arrows indicate phosphorylated GFP-Rad54 and autophosphorylated Nek1.
(C) Detection of Rad54 phosphorylation using a phospho-specific antibody. The in vitro kinase assay was performed as in (B) and Rad54 phosphorylation at Ser572 (Rad54-pS572) was analyzed with a phospho-specific antibody.
(D) Detection of Rad54 phosphorylation in vivo. HeLa and Hek293 cells were treated with siRNAs prior to X-irradiation. Cell extracts were obtained at 4 hr post 10 Gy and analyzed by immunoblotting using the antibody against Rad54-pS572.
(E) Time course of Rad54 phosphorylation in G2-irradiated cells. HeLa cells were synchronized in G2 (Figure S3A), X-irradiated, and Rad54-pS572 in nuclear cell extracts was analyzed by immunoblotting.
(F) Time course of Rad54 phosphorylation in S- and G2-irradiated cells. HeLa cells were synchronized in S or G2 (Figures S3A and S3B), X-irradiated, and Rad54-pS572 in nuclear cell extracts was analyzed by immunoblotting.
(G) Analysis of the phosphorylated fraction of Rad54 in G2-irradiated cells. HeLa cells were synchronized in G2, X-irradiated, and Rad54 phosphorylation was analyzed on Phos-tag gels via immunoblotting. GAPDH and Nek1 were analyzed on a regular acrylamide gel. Phosphatase treatment was applied to control phospho-specific band shifts.
Nek1 Promotes HR by Phosphorylating Rad54 at Ser572
We then investigated if Rad54-pS572 is required for efficient DSB repair. Because Rad54’s critical function during DSB repair involves its interaction with Rad51, we first investigated whether the three GFP-Rad54 variants differ in their ability to interact with Rad51. We transiently transfected Hek293 cells with GFP-Rad54-WT, -S572A or -S572E constructs, immuno-precipitated Rad51, and observed similar interaction levels in all three Rad54 variants (Figure S4A). Purified Rad54-S572A and -S572E proteins also showed similar interaction levels (Figure S7D). We then generated HeLa cell clones with stably integrated siRNA-resistant GFP-tagged Rad54-WT, Rad54-S572A, or Rad54-S572E (hereafter named 54WT for a clone with Rad54-WT, 54SA for a clone with Rad54-S572A, 54SE for a clone with Rad54-S572E, and HeLa for the uncomplemented parental cells). All three clones showed physiological Rad54 expression levels by immunoblotting and formed similar numbers of IR-induced GFP-Rad54 foci that co-localized with Rad51 foci (Figures 4A and S4B). Forty-eight hours prior to all experiments, we depleted the endogenous Rad54 by siRNA. We assessed Rad51 and γH2AX foci levels in G2-irradiated cells and revealed unrepaired foci in 54SA but not in 54SE cells (Figure 4B). The magnitude of the repair defect in 54SA cells was similar to that of HeLa cells treated with siRad54 (hereafter named 54KD). Nek1 depletion by siRNA caused similarly elevated foci levels in 54WT and 54SA cells, demonstrating epistasis between Nek1 deficiency and the inability to phosphorylate Rad54 at Ser572. Nek1 depletion in 54SE cells had little effect, demonstrating that the major function of Nek1 during DSB repair by HR is to phosphorylate Rad54 at Ser572 (Figures 4B and S4C). To use foci-independent DSB repair measurements, we assessed chromatin-bound Rad51 levels by immunoblotting in G2-sychronized cells at distinct time points after irradiation. In 54WT and 54SE cells, chromatin-bound Rad51 was increased at 4 hr after IR and then decreased due to repair. In contrast, chromatin-bound Rad51 did not decrease between 4 and 10 hr after IR in 54SA cells (Figure 4C and S4D). This is consistent with the Rad51 foci analysis and confirms the HR defect of cells with unphosphorylatable Rad54-S572A. We also analyzed chromatid breaks and SCEs as a measure for unrepaired DSBs and efficient HR events, respectively. 54SA and 54KD but not 54SE cells showed elevated chromatid breaks and a failure to form SCEs (Figures 4D and S4E).
Figure 4. Nek1 Promotes HR by Phosphorylating Rad54 at Ser572.
(A) Generation of GFP-Rad54 mutants. HeLa clones with stably integrated siRNA-resistant and GFP-tagged Rad54-WT, Rad54-S572A, or Rad54-S572E were generated (named 54WT, 54SA, and 54SE) and treated with siRad54. IF images show cells with GFP-Rad54 and Rad51 foci at 2 hr post 2 Gy.
(B) Rad51 and γH2AX foci in Rad54 mutants. HeLa clones were treated with siRNAs, X-irradiated, and Rad51 and γH2AX foci were analyzed in G2-phase cells that were identified as in Figure 2A. Mean ± SEM (n = 3); spontaneous foci numbers were subtracted. 54KD, HeLa cells treated with siRad54.
(C) Chromatin fraction of Rad51 in Rad54 mutants. HeLa clones were treated with siRad54, synchronized, irradiated with 10 Gy in G2, and chromatin fractions were analyzed by immunoblotting. The soluble fractions served as controls.
(D) SCEs and chromatid breaks in Rad54 mutants. HeLa clones were treated with siRad54, EdU labeled, and X-irradiated. SCEs and chromatid breaks were analyzed in EdU-negative mitotic spreads from G2-irradiated cells (Figure S4E). Mean ± SEM (n = 3); spontaneous SCEs and breaks were subtracted. (E) Rad51 foci in transiently transfected HeLa cells. Cells were treated with siRNAs, transfected with siRNA-resistent Rad54 plasmids and X-irradiated. Rad51 foci were counted in G2-phase cells (identified as in Figure 2A) that formed GFP-Rad54 foci (~30% of all transfected cells). Mean ± SEM (n = 4).
(F) GFP-based HR reporter assay with transiently transfected HeLa pGC cells. Cells were treated with siRNAs and transfected with RFP-Rad54 and I-SceI plasmids. The ratio between RFP-positive cells which were also positive for GFP and all RFP-positive cells was assessed by IF microscopy. Note that the impact of a deficiency in HR, exemplified by cells treated with siBrca2, is less pronounced than in Figure 2C, likely due to the modified experimental setup involving the dual transfection of I-SceI and RFP-Rad54 plasmids. Mean ± SEM (n = 4).
To independently confirm the results with the stable cell lines, we transiently transfected cells with Rad54 constructs. We depleted endogenous Rad54 and/or Nek1 in HeLa cells; complemented them with siRNA-resistant GFP-Rad54-WT, -S572A, or -S572E constructs; and confirmed that they show similar expression levels (Figure S4F). First, we measured Rad51 foci in irradiated G2 cells that formed GFP-Rad54 foci of physiological intensity. Rad54-WT and Rad54-S572E but not Rad54-S572A complemented the elevated foci level of siRad54-treated cells (Figure 4E). Moreover, the elevated foci level conferred by Nek1 depletion was rescued by the Rad54-S572E mutant, demonstrating that phosphomimic Rad54 suppresses the requirement for Nek1 function (Figure 4E). We then quantified GFP-Rad54 foci and obtained results identical as with Rad51 foci; that is, we observed elevated foci levels in Nek1-depleted cells and in cells expressing Rad54-S572A but not Rad54-WT or Rad54-S572E constructs and a rescue of the Nek1 defect by the Rad54-S572E mutant (Figure S4G). Moreover, the analysis of γH2AX foci in cells with a pan-nuclear GFP-Rad54 signal provided similar results to that of cells which formed GFP-Rad54 foci of physiological intensity (Figure S4H), demonstrating that differences in Rad54 expression levels do not substantially affect the repair capacity. Finally, we used the HR reporter assay in cells expressing RFP-tagged Rad54 constructs and observed diminished HR frequencies in the S572A mutant and a rescue of the HR defect in Nek1-depleted cells through expression of the S572E mutant (Figure 4F). In summary, these data establish that Nek1 promotes HR by phosphorylating Rad54 at Ser572.
Rad54 Phosphorylation during S Phase Causes Rad51 Removal from Stalled Replication Forks
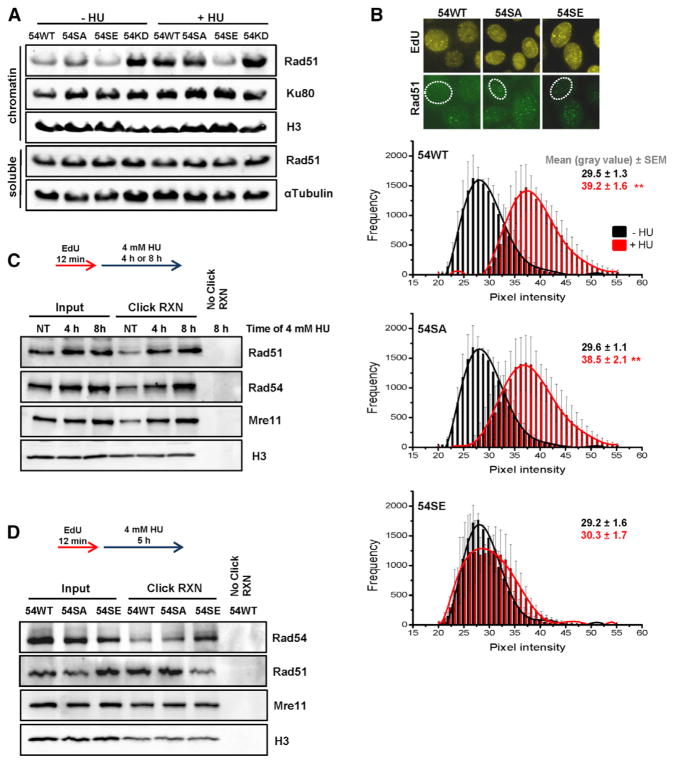
The finding that Rad54 is regulated by a specific phosphorylation event raises the possibility that permanent phosphorylation of Rad54, although being beneficial for HR, could be detrimental under specific conditions. The observation that Rad54-pS572 occurs specifically in G2 further suggests that Rad54 phosphorylation might be detrimental during S phase. To explore this possibility, we analyzed HeLa cells with the stably integrated Rad54 variants after exposure to high doses of HU that are known to cause replication fork stalling. Since Rad54-pS572 promotes Rad51 removal during late stages of HR, we speculated that it might also remove Rad51 from stalled replication forks where Rad51 is required to prevent fork degradation (Hashimoto et al., 2010; Schlacher et al., 2011). We first employed immunoblotting and observed diminished levels of chromatin-bound Rad51 after HU treatment in 54SE cells, suggesting that Rad54-pS572 removes Rad51 from stalled forks (Figure 5A). In addition, Hek293 cells overexpressing Rad54-S572E, but not cells overexpressing Rad54-WT or Rad54-S572A, showed diminished levels of chromatin-bound Rad51 after HU treatment (Figure S5A). We then assessed the level of chromatin-bound Rad51 by immunofluorescence microscopy. Rad51 bound to stalled replication forks co-localizes with newly synthesized DNA but does not form clear Rad51 foci (Petermann and Helleday, 2010; Zellweger et al., 2015). We therefore applied an extraction procedure to remove Rad51 that is not bound to chromatin and measured the total nuclear Rad51 intensity in EdU-positive S-phase cells. 54WT and 54SA cells showed an increase in Rad51 intensity after HU treatment suggesting that Rad51 binds to stalled replication forks (Figure 5B). Of note, HU-induced Rad51 binding was absent in 54SE cells (Figure 5B). We also analyzed Rad51 foci and did not detect an increase in foci number under these treatment conditions, consistent with the observation that Rad51 bound to stalled replication forks does not form foci (Petermann and Helleday, 2010; Zellweger et al., 2015) (Figure S5B). This control experiment confirms that the differences in the HU-induced total nuclear Rad51 intensity between 54WT/54SA and 54SE are not affected by differences in foci number.
Figure 5. Rad54 Phosphorylation during S Phase Causes Rad51 Removal from Stalled Replication Forks.
(A) Chromatin fraction of Rad51 in Rad54 mutants. HeLa clones were treated with siRad54 prior to HU treatment (4 mM for 5 hr), and chromatin fractions were analyzed by immunoblotting. The soluble fractions served as controls.
(B) Chromatin-bound Rad51 in Rad54 mutants. HeLa clones were treated with siRad54, co-treated with HU (0.5 mM for 2 hr) and EdU, and chromatin-bound Rad51 levels were analyzed by IF microscopy in EdU-positive nuclei. Rad51 showed a distribution of intensities with signals in the gray value range between 20 and 55 representing non-foci signals and intensities between 150 and 250 representing foci signals. The analysis was restricted to signals between 20 and 55. The mean ± SEM for each intensity is shown (n = 4).
(C) Analysis of proteins bound to stalled replication forks using iPOND. Hek293 cells were labeled with EdU, followed by different times of HU treatment. H3 signals were used to control the pull-down efficiency of EdU-labeled chromatin.
(D) Analysis of proteins bound to stalled replication forks in Rad54 mutants using iPOND. HeLa clones were treated with siRad54 prior to EdU labeling and HU treatment. H3 signals were used to control the pull-down efficiency of EdU-labeled chromatin.
To gain further insight into the processes of how untimely phosphorylation of Rad54 during S phase causes removal of Rad51 from chromatin, we applied iPOND technology (Sirbu et al., 2011). We observed that Rad54 as well as Rad51 bind to stalled replication forks, with the level of Rad54/Rad51 binding increasing with increasing periods of fork stalling. Interestingly, despite the increased abundance of Rad54 at stalled forks, Rad51 was not removed (Figure 5C), consistent with the interpretation that WT Rad54 does not remove Rad51 from chromatin during S phase. This is supported by the observation that Rad54 is not phosphorylated at Ser572 during prolonged periods of replication fork stalling (Figure S5C). We then investigated how the different Rad54 variants bind to stalled replication forks and observed that Rad54-S572E has significantly higher occupancy at stalled forks than Rad54-WT or Rad54-S572A. Importantly, the enhanced presence of Rad54-S572E lead to removal of Rad51 from stalled forks, consistent with the interpretation that phosphorylated Rad54 promotes removal of Rad51 from chromatin (Figure 5D). Thus, untimely phosphorylation of Rad54 during S phase leads to Rad51 removal from stalled replication forks.
Rad54 Phosphorylation during S Phase Causes Degradation of Stalled Replication Forks
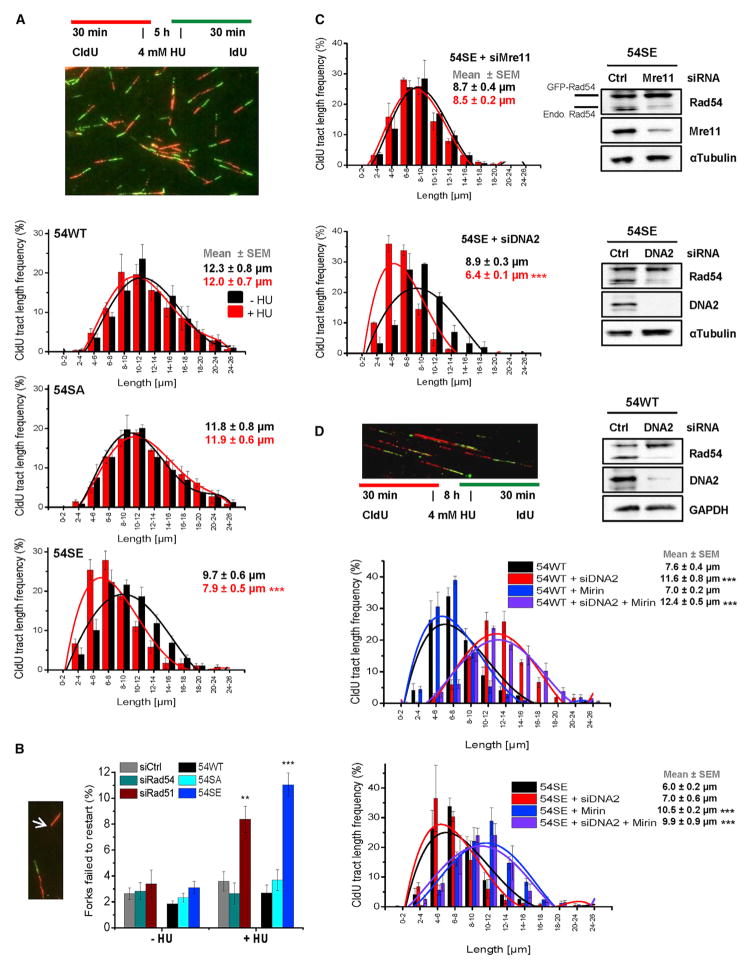
We next investigated if Rad51 removal from stalled replication forks in 54SE cells causes fork degradation. We applied the DNA fiber assay and used conditions that were described to cause fork degradation in cells with destabilized Rad51 (4 mM HU for 5 hr) (Schlacher et al., 2011). Of note, 54SE cells but not 54WT or 54SA cells exhibited degradation of newly synthesized DNA (Figure 6A). The extent of degradation in 54SE cells was similar to that of HeLa cells treated with siRad51 whereas cells treated with siRad54 (54KD cells) did not exhibit fork degradation (Figure S6A). Moreover, 54SE cells treated with siRad51 did not show more extensive degradation than HeLa cells treated with siRad51 or 54SE cells without siRad51, demonstrating an epistatic relationship between Rad51 depletion and Rad54-S572E expression for replication fork degradation (Figure S6B). We also quantified the fraction of forks that failed to restart after HU withdrawal (Petermann et al., 2010). HeLa cells treated with siRad51 and 54SE cells showed a defect in the ability to restart stalled replication forks (Figure 6B). Finally, fork degradation in 54SE cells was suppressed by Mre11 siRNA or treatment with the Mre11 nuclease inhibitor Mirin, which was previously shown to suppress fork degradation in Brca2-deficient cells (Schlacher et al., 2011), but was unaffected by DNA2 siRNA (Figures 6C and S6C). Collectively, these findings demonstrate that untimely Rad54 phosphorylation during S phase phenocopies the loss of Rad51 and severely compromises the ability to stabilize stalled replication forks.
Figure 6. Rad54 Phosphorylation during S Phase Causes Degradation of Stalled Replication Forks.
(A) DNA degradation at stalled forks in Rad54 mutants analyzed by the DNA fiber assay. CldU was added to siRad54-treated HeLa clones, followed by HU treatment and an IdU pulse. CldU-positive DNA fibers were analyzed and categorized according to size. The mean ± SEM for each category separately and for all categories together are shown (n = 5).
(B) Replication fork recovery in Rad54 mutants. HeLa clones were treated with siRad54 and HeLa cells with siCtrl, siRad54 or siRad51 prior to the experiment which was performed as in (A). CldU-positive fibers without a flanking IdU signal were scored (indicated by arrow). Mean ± SEM (n = 5).
(C) DNA degradation at stalled forks in the 54SE mutant analyzed by the DNA fiber assay. CldU was added to siRNA-treated 54SE cells, followed by HU treatment and an IdU pulse. The analysis was performed as in (A). Mean ± SEM (n = 3).
(D) DNA degradation at stalled forks in 54WT or 54SE cells analyzed by the DNA fiber assay. HeLa clones were treated with siRNAs and/or Mirin. CldU was added, followed by HU treatment and an IdU pulse. The analysis was performed as in (A). Mean ± SEM (n = 3).
To gain further insight into the process of fork degradation by Rad54 phosphorylation, we performed the fiber assay under conditions that were described to cause degradation of stalled forks even in control cells (4 mM HU for 8 hr) (Thangavel et al., 2015). Consistent with this study, we observed that this degradation in 54WT cells is diminished after DNA2 siRNA but unaffected by Mre11 siRNA (Figure 6D). Of note, fork degradation in 54SE cells was purely dependent on Mre11; i.e., Mirin but not DNA2 siRNA treatment abolished the degradation (Figure 6D). Thus, we obtained fundamentally different results with 54WT and 54SE cells. 54WT cells showed fork degradation only after 8 hr HU (and not after 5 hr), which depends on DNA2. 54SE cells showed fork degradation after 5 hr, which depends on Mre11, and the degradation after 8 hr remained dependent on Mre11. Because the DNA2-dependent fork degradation in control cells has been suggested to occur at chicken foot structures that arise after prolonged periods of replication fork stalling (Thangavel et al., 2015), our data suggest that 54SE cells fail to form such structures and remove Rad51 from stalled replication forks before these convert into chicken foot structures. This interpretation is in line with the finding that Rad51 is essential for the conversion from stalled forks into chicken foot structures (Zellweger et al., 2015).
Cells with Unregulatable Rad54 Show Genomic Instability
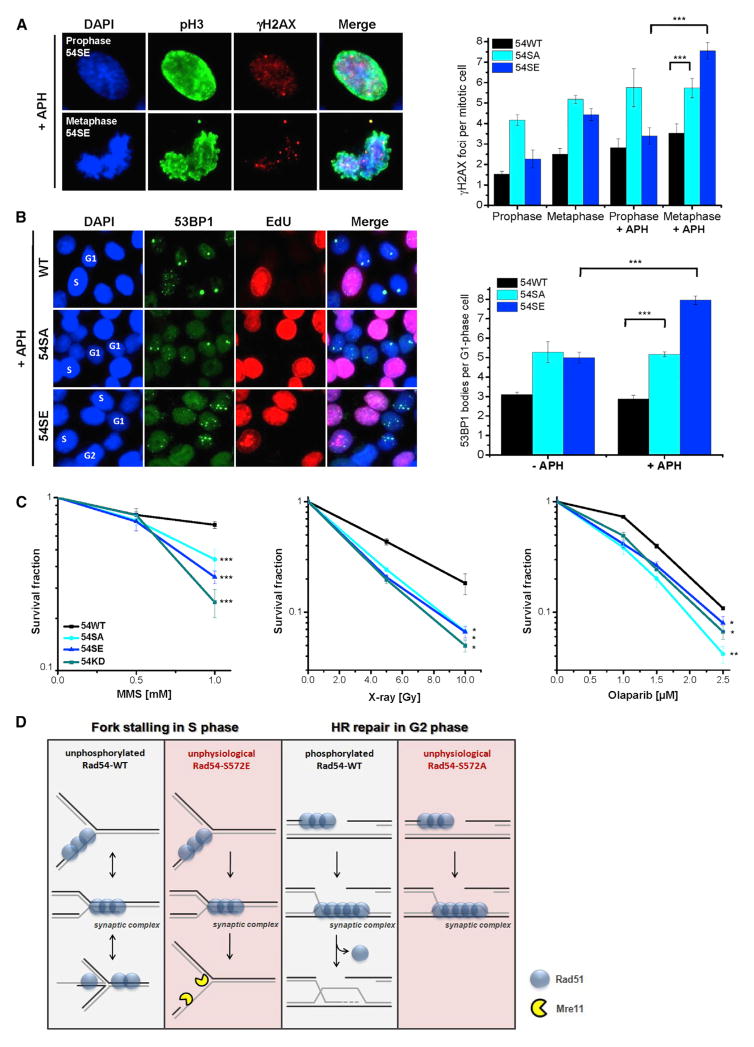
Cells with unphosphorylatable Rad54-S572A fail to efficiently repair DSBs by HR whereas cells with phosphomimic Rad54-S572E fail to protect stalled replication forks. We therefore reasoned that both cell types might be unable to cope with replication stress, which requires that cells minimize the generation of lesions at stalled replication forks and repair DSBs which inevitably arise. To explore this possibility, we initially exposed 54SA or 54SE cells to low concentrations of APH and assessed the level of γH2AX foci in mitotic cells. 54SA cells show elevated foci levels in prophase cells compared to 54SE and 54WT cells, both with and without APH treatment, likely reflecting the inability of 54SA cells to repair DSBs by HR (Figure 7A). Of note, the treatment conditions applied are known to cause under-replicated DNA regions that result during mitotic chromatin condensation in an increase in γH2AX foci numbers when cells progress from prophase to metaphase (Glover, 2006; Ying et al., 2013). We therefore also assessed foci levels in metaphase cells and observed that 54SE but not 54SA or 54WT cells show a substantial increase in γH2AX foci numbers between prophase and metaphase, both with and without APH treatment (Figure 7A). Together, this demonstrates that both cell variants with unregulatable Rad54 exhibit a diminished ability to cope with replication stress. We also quantified 53BP1 bodies in G1-phase cells, which are known to arise from under-replicated DNA regions (Lukas et al., 2011). 54SE cells show increased 53BP1 bodies after APH treatment, suggesting that Rad54 phosphorylation during S phase causes under-replicated DNA regions. 54SA cells, in contrast, show elevated 53BP1 bodies already in untreated cells (Figure 7B), consistent with the interpretation that unrepaired γH2AX foci in prophase lead to 53BP1 bodies in G1 phase. Finally, we studied the survival of cells with unregulatable Rad54 after agents inducing DSBs as well as other lesions which interfere with replication. Compared to 54WT cells, both 54SA and 54SE cells show diminished colony formation after MMS, olaparib or X-rays, confirming that regulation of Rad54 phosphorylation is important for maintaining genomic stability (Figure 7C).
Figure 7. Cells with Unregulatable Rad54 Show Genomic Instability.
(A) γH2AX foci in mitotic Rad54 mutants. HeLa clones were treated with siRad54 and exposed to low concentrations of APH (0.3 μM) for 20 hr. γH2AX foci were quantified in pH3-positive pro- and metaphases. Mean ± SEM (n = 3).
(B) 53BP1 bodies in Rad54 mutants in G1 phase. HeLa clones were treated with siRad54, exposed to low concentrations of APH (0.3 μM) for 24 hr, and EdU labeled. 53BP1 bodies were counted in EdU-negative G1-phase cells. Mean ± SEM (n = 3).
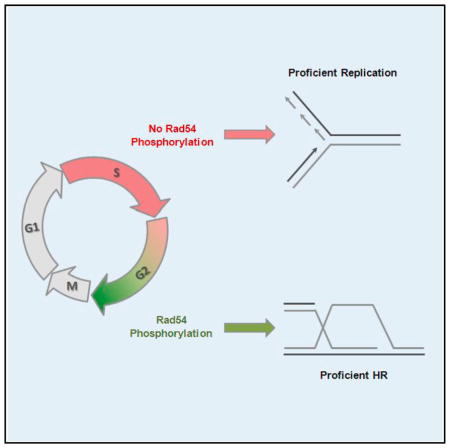
(C) Clonogenic survival of Rad54 mutants. SiRad54-treated HeLa clones were treated with MMS (for 1 hr), olaparib (permanent), or X-rays. Mean ± SEM (n = 3). (D) Model: effects of timely phosphorylation of Rad54 (boxes 1 and 3): the absence of Rad54 phosphorylation during S phase stabilizes Rad51 at stalled replication forks to prevent degradation of newly synthesized DNA. The presence of Rad54 phosphorylation in G2 phase promotes Rad51 removal and the completion of HR. Consequences of untimely phosphorylation of Rad54 (boxes 2 and 4): Rad54 phosphorylation during S phase destabilizes Rad51 at stalled replication forks causing degradation of newly synthesized DNA. The absence of Rad54 phosphorylation in G2 phase prevents Rad51 removal and the completion of HR.
DISCUSSION
We discovered that Rad54 is phosphorylated at Ser572 and generated stable cell lines expressing either unphosphorylatable Rad54-S572A or phosphomimic Rad54-S572E protein (54SA or 54SE cells, respectively). 54SA cells fail to resolve Rad51 foci during DSB repair by HR whereas 54SE cells repair DSBs by HR as efficiently as control cells with Rad54-WT (54WT cells). Strikingly, although Rad54-S572E is beneficial for HR, it is detrimental for the protection of stalled replication forks. This latter effect is associated with removal of Rad51 from stalled forks, which leads to fork degradation similar to what is observed in cells lacking Rad51. In contrast, 54SA cells are able to protect stalled replication forks (Figure 6A). Thus, Rad54 phosphorylation exerts cell cycle phase-specific positive or negative effects and hence needs to be finely tuned dependent on the cell cycle requirements. The necessity to regulate Rad54 phosphorylation is further demonstrated by the observation that both 54SA and 54SE cells show elevated DNA damage and decreased survival if exposed to agents that induce replication fork stalling as well as DSBs (Figures 7A–7C).
Rad54 removes Rad51 from DNA when the synaptic complex of ssDNA:Rad51:dsDNA is transformed into heteroduplex DNA (Solinger et al., 2002; Wright and Heyer, 2014). We observed that 54WT, 54KD, 54SA, and 54SE cells form Rad51 foci at early time points post IR with equal efficiency (Figure 4B), implying that the presence of Rad54 or its phosphorylation does not affect Rad51 binding to ssDNA at resected DSBs. Moreover, purified Rad54 does not exhibit ATPase activity on ssDNA in vitro (Swagemakers et al., 1998). The observation that Rad54 phosphorylation removes Rad51 from stalled replication forks might therefore suggest that the protective role of Rad51 at stalled forks involves the presence of a synaptic complex (Figure 7D). How might a synaptic complex arise during replication fork stalling? The prevailing evidence suggests that chicken foot structures arise during prolonged periods of replication fork stalling (Thangavel et al., 2015). Thus, it might be possible that Rad51 is initially loaded onto ssDNA by Brca2 but then promotes, via homology search, the formation of a synaptic complex which serves to stabilize the stalled replication fork until it can be converted into a chicken foot structure, or directly aids in this process (Figure 7D). In either case, Rad51 is not removed during this process (Sirbu et al., 2011), consistent with our iPOND data (Figure 5C) and the observation that Rad54 is not phosphorylated at Ser572 during replication (Figure S5C).
How might Rad54 phosphorylation promote Rad51 removal from chromatin? The Ser572 phosphorylation site of Rad54 is positioned within one of seven highly conserved ATPase domains (Ceballos and Heyer, 2011; Thomä et al., 2005) and phosphorylation events have been reported to enhance the activity of other ATPases (Alzamora et al., 2010). It is therefore tempting to speculate that Ser572 phosphorylation stimulates Rad54’s ATPase function. To test the possibility that Rad54-Ser572 phosphorylation directly affects the ATPase activity of Rad54 or its ability to promote critical HR reactions, we purified WT and mutant Rad54 proteins (Figure S7A). Surprisingly, the Rad51-stimulated ATPase activity of the Rad54-S572E mutant and its D-loop formation ability were substantially lower than those of Rad54-WT or the Rad54-S572A mutant (Figures S7B and S7C), although it retained the ability to interact with Rad51 and bind dsDNA (Figures S7D and S7E). Also contrary to expectation, the Rad54-S572A mutant protein was proficient in stimulating Rad51-mediated D-loop formation, even better than Rad54-WT under the tested conditions (Figure S7C), and displayed near WT ATPase activity on dsDNA (Figure S7B). Finally, none of the three Rad54 variants showed ATPase activity on ssDNA (Figure S7F). Lack of a defect in Rad54-S572A might suggest an alternative view that there is a factor that restrains unphosphorylated Rad54 activity in vivo. The activity of Rad54-S572E, though reduced, appears sufficient in vivo when coupled to the change effected by the phosphomimic. The inability to pinpoint biochemical differences to explain the cellular phenotypes suggests that yet unknown factors are missing in the in vitro reactions. One possibility is the Rad54 paralog Rad54B, which shows highly similar biochemical activity and partially overlapping in vivo functions. Thus, the precise mechanism of how Rad54 phosphorylation promotes Rad51 removal from chromatin awaits clarification.
We have shown that Rad54 phosphorylation following DNA damage induction is restricted to late G2 phase irrespective of the position in the cell cycle when the damage is induced. This cell-cycle-specific modulation of Rad54 allows for the timely removal of Rad51 prior to the onset of mitosis and complements previous studies by others showing that nucleases such as Mus81-Eme1 and Gen1 are under cell cycle-specific regulation to resolve late HR intermediates during mitosis (Matos and West, 2014; Ying et al., 2013). Moreover, it was described that DNA lesions that arise from replication stress can be repaired by HR uncoupled from replication in the following G2 phase and it was further suggested that such repair is promoted by cell-cycle-specific kinases (Gonzá lez-Prieto et al., 2013; Karras and Jentsch, 2010). Our discovery of the G2-specific activation of Rad54 closes the gap between damage processing that starts during S phase and is completed in mitosis and thus represents both the missing link and a mechanistic explanation for these previous findings. Collectively, our work, together with published findings, establishes that the process of HR is finely regulated during the cell cycle so that the required factors are activated when they are most needed and the least harmful. The concept that a synchronization process underlies HR has precedent from meiosis, where defined steps of HR occur at defined stages during meiotic progression (Baudat et al., 2013). However, it has to be considered that the process of HR synchronization with cell cycle progression may be lesion dependent (DSBs, gaps, stalled forks) as double Holiday junctions can be resolved by the BLM–TopoIIIα–RMI1–RMI2 (BTR) complex during S phase (Matos and West, 2014; Sarbajna et al., 2014).
In summary, our work shows that the process of HR is regulated during the cell cycle by restricting Rad54 phosphorylation to late G2 phase. On one hand, this limits Rad54 function during replication and allows Rad51 to protect stalled replication forks; on the other hand, it promotes Rad51 removal prior to the onset of mitosis and the completion of HR (Figure 7D). We identified Nek1 as the kinase regulating Rad51 removal and orchestrating HR with replication fork stability.
EXPERIMENTAL PROCEDURES
ShNek1 or shCtrl cells were generated by viral transduction. Stable cell lines expressing GFP-Rad54 variants were generated by transfection with plasmids carrying a G418 resistance cassette. SiRNA and plasmid transfections were carried out using HiPerFect and MATra-A reagents, respectively. For foci analysis, cells were categorized at the microscope in G1-, G2-, or S-phase cells by their DAPI content and EdU intensity. Foci were enumerated manually. Intensity measurements of chromatin-bound Rad51 using IF microscopy involved pre-extraction using ice cold methanol.
The EdU incorporation assay, preparation of chromosome spreads, SCE analysis, and clonogenic survival assays were carried out as described (Beucher et al., 2009; Conrad et al., 2011; Nikolova et al., 2010). For the HR reporter assay, HeLa pGC cells were siRNA treated, transfected with RFP-Rad54 and I-SceI plasmids (Mansour et al., 2008), and GFP-positive cells were counted at the microscope. For DNA fiber analysis, cells were labeled with CldU for 30 min, treated with 4 mM HU for 5 or 8 hr, incubated with IdU for 30 min, harvested, and prepared for DNA fiber spreading as described (Schlacher et al., 2011).
Isolation of nuclear cell extracts and chromatin or soluble protein fractions, protein analysis by SDS-PAGE and immunoblotting were performed as described (Barton et al., 2014). For in vitro kinase assays, constitutively active Nek1 protein was pre-incubated with 32P ATP or with unlabeled ATP before immunoprecipitated Rad54 was added. Phosphorylation signals were detected by auto-radiography or with a custom made antibody for Rad54-pSer572. Detailed descriptions of all assays including coIP and iPOND experiments are provided in the Supplemental Experimental Procedures.
P values were obtained by student’s t test and represent a comparison of all cells analyzed in the indicated cell populations (for all foci and chromosomal experiments) or a comparison of the data mean (for the HR reporter, colony formation and the DNA fiber assays and for Rad51 intensity measurements); *, p < 0.05; **, p < 0.01; ***, p < 0.001. For each experiment, protein expression levels were controlled by immunoblotting and are displayed in the corresponding figures.
Supplementary Material
Highlights.
Nek1 functions during homologous recombination
Nek1 phosphorylates Rad54 at Ser572 in late G2 phase
Unphosphorylatable Rad54 mutants are defective in homologous recombination
Phosphomimic Rad54 causes degradation of stalled replication forks
Acknowledgments
We thank Jochen Dahm-Daphi, Jörg Kobarg, Roland Kanaar, and Christian Thiel for sharing cell lines and DNA plasmids and Helle Ulrich, Cristina Cardoso, David Chen, Junjie Chen, Dale Wigley, and the M.L. lab for critical discussions. We further thank Bettina Basso, Christel Braun, and Cornelia Schmitt for technical assistance and Jessica Sneeden for pilot experiments. This work was supported by the Deutsche Forschungsgemeinschaft (GRK 1657 to M.L.), the Bundesministerium für Bildung und Forschung (02NUK037C to M.L.), and the NIH (GM58015 to W.-D.H.).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.04.032.
AUTHOR CONTRIBUTIONS
J.S. and M.L. designed the study and wrote the paper with input from W.D.W. and W.-D.H.; J.S., A.W., O.B., W.D.W., W.-D.H., and M.L. analyzed and interpreted the data. J.S., A.W., O.B., M.S., and W.D.W. conducted the experiments.
References
- Agarwal S, van Cappellen WA, Guénolé A, Eppink B, Linsen SE, Meijering E, Houtsmuller A, Kanaar R, Essers J. ATP-dependent and independent functions of Rad54 in genome maintenance. J Cell Biol. 2011;192:735–750. doi: 10.1083/jcb.201011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzamora R, Thali RF, Gong F, Smolak C, Li H, Baty CJ, Bertrand CA, Auchli Y, Brunisholz RA, Neumann D, et al. PKA regulates vacuolar H+-ATPase localization and activity via direct phosphorylation of the a subunit in kidney cells. J Biol Chem. 2010;285:24676–24685. doi: 10.1074/jbc.M110.106278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton O, Naumann SC, Diemer-Biehs R, Künzel J, Steinlage M, Conrad S, Makharashvili N, Wang J, Feng L, Lopez BS, et al. Polo-like kinase 3 regulates CtIP during DNA double-strand break repair in G1. J Cell Biol. 2014;206:877–894. doi: 10.1083/jcb.201401146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Löbrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Ceballos SJ, Heyer WD. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim Biophys Acta. 2011;1809:509–523. doi: 10.1016/j.bbagrm.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MRG, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen PL, Chen CF, Jiang X, Riley DJ. Never-in-mitosis related kinase 1 functions in DNA damage response and checkpoint control. Cell Cycle. 2008;7:3194–3201. doi: 10.4161/cc.7.20.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Craigen WJ, Riley DJ. Nek1 regulates cell death and mitochondrial membrane permeability through phosphorylation of VDAC1. Cell Cycle. 2009;8:257–267. doi: 10.4161/cc.8.2.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chen CF, Chiang HC, Pena M, Polci R, Wei RL, Edwards RA, Hansel DE, Chen PL, Riley DJ. Mutation of NIMA-related kinase 1 (NEK1) leads to chromosome instability. Mol Cancer. 2011a;10:5. doi: 10.1186/1476-4598-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chen CF, Riley DJ, Chen PL. Nek1 kinase functions in DNA damage response and checkpoint control through a pathway independent of ATM and ATR. Cell Cycle. 2011b;10:655–663. doi: 10.4161/cc.10.4.14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chen CF, Polci R, Wei R, Riley DJ, Chen PL. Increased Nek1 expression in renal cell carcinoma cells is associated with decreased sensitivity to DNA-damaging treatment. Oncotarget. 2014;5:4283–4294. doi: 10.18632/oncotarget.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJC, Lin JR, Vannier JB, Slaats GG, Kile AC, Paulsen RD, Manning DK, Beier DR, Giles RH, Boulton SJ, Cimprich KA. NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol Cell. 2013;51:423–439. doi: 10.1016/j.molcel.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad S, Künzel J, Löbrich M. Sister chromatid exchanges occur in G2-irradiated cells. Cell Cycle. 2011;10:222–228. doi: 10.4161/cc.10.2.14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckbar D, Birraux J, Krempler A, Tchouandong L, Beucher A, Walker S, Stiff T, Jeggo P, Löbrich M. Chromosome breakage after G2 checkpoint release. J Cell Biol. 2007;176:749–755. doi: 10.1083/jcb.200612047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensminger M, Iloff L, Ebel C, Nikolova T, Kaina B, Löbrich M. DNA breaks and chromosomal aberrations arise when replication meets base excision repair. J Cell Biol. 2014;206:29–43. doi: 10.1083/jcb.201312078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Houtsmuller AB, van Veelen L, Paulusma C, Nigg AL, Pastink A, Vermeulen W, Hoeijmakers JHJ, Kanaar R. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover TW. Common fragile sites. Cancer Lett. 2006;232:4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- González-Prieto R, Muñoz-Cabello AM, Cabello-Lobato MJ, Prado F. Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J. 2013;32:1307–1321. doi: 10.1038/emboj.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudzenski S, Raths A, Conrad S, Rübe CE, Löbrich M. Inducible response required for repair of low-dose radiation damage in human fibroblasts. Proc Natl Acad Sci USA. 2010;107:14205–14210. doi: 10.1073/pnas.1002213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W-D, Li X, Rolfsmeier M, Zhang X-P. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA, Löbrich M. How cancer cells hijack DNA double-strand break repair pathways to gain genomic instability. Biochem J. 2015;471:1–11. doi: 10.1042/BJ20150582. [DOI] [PubMed] [Google Scholar]
- Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Liu S, Ho CK, Ouyang J, Zou L. Nek1 kinase associates with ATR-ATRIP and primes ATR for efficient DNA damage signaling. Proc Natl Acad Sci USA. 2013;110:2175–2180. doi: 10.1073/pnas.1217781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löbrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, Jeggo PA. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9:662–669. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C. Molecular biology. Shielding broken DNA for a quick fix. Science. 2013;339:652–653. doi: 10.1126/science.1234602. [DOI] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grøfte M, Chan KL, Hickson ID, Bartek J, Lukas J. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- Mansour WY, Schumacher S, Rosskopf R, Rhein T, Schmidt-Petersen F, Gatzemeier F, Haag F, Borgmann K, Willers H, Dahm-Daphi J. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008;36:4088–4098. doi: 10.1093/nar/gkn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, West SC. Holliday junction resolution: regulation in space and time. DNA Repair (Amst ) 2014;19:176–181. doi: 10.1016/j.dnarep.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazón G, Mimitou EP, Symington LS. SnapShot: homologous recombination in DNA double-strand break repair. Cell. 2010;142:648.e1–e648.e2. doi: 10.1016/j.cell.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Meirelles GV, Perez AM, de Souza EE, Basei FL, Papa PF, Melo Hanchuk TD, Cardoso VB, Kobarg J. ‘‘Stop Ne(c)king around’’: How interactomics contributes to functionally characterize Nek family kinases. World J Biol Chem. 2014;5:141–160. doi: 10.4331/wjbc.v5.i2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melixetian M, Klein DK, Sørensen CS, Helin K. NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat Cell Biol. 2009;11:1247–1253. doi: 10.1038/ncb1969. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Wilhelm T, Debatisse M, Rosselli F. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat Cell Biol. 2013;15:1008–1015. doi: 10.1038/ncb2793. [DOI] [PubMed] [Google Scholar]
- Nikolova T, Ensminger M, Löbrich M, Kaina B. Homologous recombination protects mammalian cells from replication-associated DNA double-strand breaks arising in response to methyl methanesulfonate. DNA Repair (Amst ) 2010;9:1050–1063. doi: 10.1016/j.dnarep.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polci R, Peng A, Chen PL, Riley DJ, Chen Y. NIMA-related protein kinase 1 is involved early in the ionizing radiation-induced DNA damage response. Cancer Res. 2004;64:8800–8803. doi: 10.1158/0008-5472.CAN-04-2243. [DOI] [PubMed] [Google Scholar]
- Renkawitz J, Lademann CA, Jentsch S. Mechanisms and principles of homology search during recombination. Nat Rev Mol Cell Biol. 2014;15:369–383. doi: 10.1038/nrm3805. [DOI] [PubMed] [Google Scholar]
- Riballo E, Kühne M, Rief N, Doherty A, Smith GCM, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Krüger I, Thompson LH, Löbrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbajna S, Davies D, West SC. Roles of SLX1-SLX4, MUS81-EME1, and GEN1 in avoiding genome instability and mitotic catastrophe. Genes Dev. 2014;28:1124–1136. doi: 10.1101/gad.238303.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Zheng X, Epshtein A, Carey JN, Bishop DK, Klein HL. Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol Cell. 2010;39:862–872. doi: 10.1016/j.molcel.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Surpili MJ, Delben TM, Kobarg J. Identification of proteins that interact with the central coiled-coil region of the human protein kinase NEK1. Biochemistry. 2003;42:15369–15376. doi: 10.1021/bi034575v. [DOI] [PubMed] [Google Scholar]
- Swagemakers SMA, Essers J, de Wit J, Hoeijmakers JHJ, Kanaar R. The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J Biol Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, Zellweger R, Moore H, Lee EH, Hendrickson EA, et al. DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol. 2015;208:545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel C, Kessler K, Giessl A, Dimmler A, Shalev SA, von der Haar S, Zenker M, Zahnleiter D, Stöss H, Beinder E, et al. NEK1 mutations cause short-rib polydactyly syndrome type majewski. Am J Hum Genet. 2011;88:106–114. doi: 10.1016/j.ajhg.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomä NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JHJ, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Wright WD, Heyer WD. Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D loop formation. Mol Cell. 2014;53:420–432. doi: 10.1016/j.molcel.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Minocherhomji S, Chan KL, Palmai-Pallag T, Chu WK, Wass T, Mankouri HW, Liu Y, Hickson ID. MUS81 promotes common fragile site expression. Nat Cell Biol. 2013;15:1001–1007. doi: 10.1038/ncb2773. [DOI] [PubMed] [Google Scholar]
- Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.