Abstract
TLR4 interactor with leucine-rich repeats (TRIL) is a brain-enriched accessory protein important in Toll-like receptor 3 and 4 signalling. Here we generated TRIL-deficient mice and examined TLR responses in vitro and in vivo. We found a role for TRIL in both TLR4 and TLR3 signalling in mixed glial cells, consistent with the high level of expression of TRIL in these cells. We also found that TRIL is a modulator of the innate immune response to LPS challenge and E.coli infection in vivo. TRIL-deficient mice produce lower levels of multiple proinflammatory cytokines and chemokines specifically within the brain following E.coli and LPS challenge. Collectively these data uncover TRIL as a mediator of innate immune responses within the brain, where it enhances neuronal cytokine responses to infection.
Introduction
Acute systemic inflammatory responses to severe infections may lead to chronic inflammatory processes in the central nervous system (CNS). Septic shock is associated with a spectrum of brain dysfunction and damage, which leads to increased morbidity and mortality (1-3).
Despite its anatomical sequestration from the circulating blood by the blood-brain-barrier (BBB), lack of a lymphatic system and low MHC expression, the brain remains an active player in the inflammatory processes occurring elsewhere in the body (4, 5). In fact, the interplay between the pheripheral immune system and the CNS has a reciprocal effect on both systems. Dysregulation of the CNS impacts on the outcome of an acute systemic infection. Equally however, severe systemic infection often leads to destructive brain inflammation (6, 7).
The systemic inflammatory response is initiated by the recognition of microbial pathogen-associated molecular patterns (PAMPs) or endogenous damage-associated molecular patterns (DAMPs), by evolutionarily conserved pathogen recognition receptors (PRRs) (8). Toll-like receptors (TLRs) are a family of PRRs, which recognize a wide range of PAMPs triggering innate immunity. To date, 10 human and 13 mouse members of the TLR family have been identified, which recognize a wide variety of PAMPs (9-11). Upon activation by PAMPs, TLRs initiate downstream signalling cascades leading to the activation of transcription factors such as NF-κB and/or interferon-regulatory factors (IRFs), which in turn induce the production of proinflammatory cytokines and chemokines as well as type I interferons (IFNs) (12).
TLR4 is the most extensively studied member of the TLR family. It is responsible for the recognition of lipopolysacharide (LPS), which is a major component of the outer membrane of Gram-negative bacteria and a key player in the pathogenesis of Gram-negative sepsis (13, 14). TLR4 is constitutively expressed within the CNS and can be found in both the parenchymal glial cells, microglia and astrocytes as well as neurons (15-19). TLR4 is also expressed in the meninges, choroid plexus and circumventricular organs (CVOs) of the brain. These structures are highly vascularized and despite the presence of peculiar epithelial barriers, lack a characteristic BBB, thus are more exposed to invading pathogens allowing for the crosstalk between the periphery and the CNS (20-23).
Binding of LPS and subsequent TLR4 activation is facilitated by a number of accessory molecules including the LPS-binding protein (LBP), glycoprotein CD14 and myeloid differentiation protein-2 (MD2) (24), all of which are central for LPS sensing by TLR4. CD14 exists in a soluble form and as a GPI-linked protein in the plasma membrane (25). Similar to TLR4 it is constitutively expressed within the CNS. In fact, CD14 is found in the meninges, choroid plexus and CVOs, mirroring the expression of TLR4 within the brain (26). In addition, CD14 is also present in microglia but is absent in astrocytes (27). Interestingly, circulating LPS causes a sequential increase in the expression of CD14, first within the highly vascularized CVOs, and then in the brain parenchyma (27, 28).
TLR4 interactor with leucine-rich repeats (TRIL) was initially characterized as a novel component of the TLR4 signalling pathway, highly expressed in the brain (29). It was shown to be required for TLR4-mediated responses in vitro via direct interaction with TLR4 and its ligand, LPS (30). In subsequent in vitro studies TRIL was also shown to play a role in the regulation of TLR3-mediated signalling. TRIL is therefore similar to CD14, which can also regulate TLR3 signalling (31).
Here we have generated TRIL-deficient mice to further investigate the role of TRIL. We confirmed the role of TRIL in mixed glial cells in TLR4 and TLR3 signalling. TRIL-deficient mice also produced less cytokines in the brain, following intracranial LPS challenge and intraperitoneal infection with E.coli. These results confirm a specific role for TRIL in the regulation of TLR4 and TLR3 signalling primarily within the brain.
Materials and Methods
Animals
C57BL/6 mice from Jackson Laboratories (Bar Harbor, ME) and generated Tril-/- mice were bred at UMASS Medical School. Mouse strains were maintained under specific pathogen-free conditions in the animal facilities at the UMASS Medical School. Mice studies were carried out in strict accordance with guidelines set forth by the American Association for Laboratory Animal Science (AALAS). The animal protocols for this work were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Massachusetts Medical School (Permit Number: A-2258-11).
TRIL-deficient mice generation
The targeting vector was designed to encode 19 kb fragment of mouse genomic Tril DNA together with the FRT-neomycin resistance cassette, flanked by two LoxP sites. Generated construct was used to transfect the embryonic stem (ES) cells in the C57BL/6 mice, to enable homologous recombination. After a number of rounds of selection with neomycin, random ES cells clones were chosen and conditional Tril-/- mice were generated. Female founders were next crossed with C57BL/6 males expressing Protamine-Cre resulting in permanent deletion of LoxP-flanked Tril alleles and generation of global TRIL-deficient mice.
Genotyping of TRIL-deficient mice
The genotypes of Tril-/- mice were determined by PCR analysis of genomic DNA, from tail biopsies. The genomic DNA was isolated using the Genomic DNA isolation Kit (Lamda Biotech) according to the manufacturer's instructions. Isolated genomic DNA was next used for genotyping by PCR with specific oligonucleotide primers for the Tril wild type and targeted allele (TRIL-F, 5′-TTC ACT TAC CAC CCT GCC AGG TTC -3′, TRIL-R1, 5′-GTC TGT ATG GGA AGA GAG GCA CAC TG -3′, TRL-R2, 5′-CAC CAG AGC GTT CTG GTC ATG C -3′). Primers F and R1 amplified wild type allele and F and R2 targeted one. The three primers were used in a PCR reaction using GoTaq (Promega) with the following amplification conditions: 95°C for 5 min and 30 cycles of 95°C for 30 s, 58°C for 30 s, and a 5 min incubation at 72°C at the end of the run. Amplification products were resolved on a 2% agarose gel.
Cell culture and stimulations
Primary murine bone marrow derived macrophages (BMDMs) and dendritic cells (BMDCs) were generated from wild type and age/sex matched TRIL-deficient mice. BMDMs were cultured in in DMEM with 10% fetal bovine serum and 20% L929 supernatants and BMDCs were maintained in RPMI 1640 (with 10% FCS, L-glutamine (2mM), 50μM β-mercaptoethanol, 1% penicillin-streptomycin solution (v/v), supplemented with granulocyte macrophage colony stimulating factor (GMCSF)(20ng/ml)). Primary murine mixed glial cells were prepared from one- to three-day-old neonatal brains of wild type and age/sex matched TRIL-deficient mice. Cells were cultured in DMEM supplemented with 10% FCS and 1% penicillin-streptomycin solution (v/v). All cells were used at 10 DIV (days in vitro) plated out and stimulated the next day. The cellular composition of primary mixed glial cells was assessed by FACS analysis using markers specific for astrocytes (GLAST-APC; Miltenyi Biotech), microglia (Cd11b-PE; eBioscience) and neurons (β-3-Tubulin; Biolegend), indicating over 83% of astrocytytes, approximately 2-3% of microglia and only trace amount of neurons within the primary mixed glial cell population. Cultured primary hippocampal neurons were generated from embryonic day 15-17 embryos using previously described method (32) and maintained in serum free Neurobasal media supplemented with B27 and GlutaMAX (Invitrogen). Primary microglia and astrocytes were isolated from mixed glial cells cultures. Generated as described above primary mixed glial cells were cultured in DMEM supplemented with 10% FCS and 1% penicillin-streptomycin solution (v/v), in the presence of 5ng/ml MCSF (R&D Systems) until fully confluent. Primary microglia were then separated from astrocyte monolayers by agitation on a rotary shaker at 125rpm for 4h. Primary astrocytes were isolated from the same cultures by trypsinization after microglia were removed as previously described (33). Obtained cells were maintained in DMEM with 10% FCS and 1% penicillin-streptomycin solution (v/v). Microglial cultures were additionally supplemented with 5ng/ml MCSF. The purity of primary microglia and astrocytes populations was assessed by FACS analysis using specific markers for astrocytes and microglia, GLAST-APC (Miltenyi Biotech) and CD11b-PE (eBioscience), revealing over 98% purity of isolated microglia and and 97% purity of isolated astrocytes (data not shown). Additionally, morphological assessment of primary mixed glial cells and isolated populations of microglia and astrocytes was carried out using phase contrast microscopy (data not shown). Immortalized microglia cells were generated by infecting primary mixed glial cells with a recombinant retrovirus J2 encoding viral oncogenes v-myc and v-raf, based on the method previously described by Blasi et al. (34). Characterization of the immortalized clonal microglial cells using FACS technique revealed them to be CD11b-PE positive and GFAP-APC negative (data not shown). For Quantitative RT-PCR analysis cells were stimulated for 5 h with 100ng/ml of LPS (Sigma-Aldrich) or 25μg/ml of Poly(I:C) (Sigma-Aldrich) prior to RNA isolation. For ELISA assay cells were treated with 10 or 100ng/ml of LPS, 25 or 50μg/ml of Poly(I:C), 100nM of Pam3CSK4 (Invivogen) or 1μg/ml of R848 (Invivogen) prior to harvesting supernatants.
ELISA
Cell culture supernatants were assayed by ELISA for CCL5/RANTES (R&D Systems), TNFα (eBiosciences) and/or IL6 (eBiosciences) in accordance with the manufacturer's instructions. A sandwich ELISA for mouse IFNβ was used as previously described (35).
Nanostring and Quantitative RT-PCR
Primary murine mixed glial cells were treated as described above followed by the RNA extraction with an RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. Brain and spleen tissues were isolated from wild type and TRIL-deficient mice 6 h post E.coli infection and RNA was purified using an RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. cDNA was synthesized from total RNA using the iScript Select cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR was performed by using iQ SYBR green supermix (Bio-Rad) and specific primers for murine Tril (forward, 5′- ACG TGC TCA CCT ACA GCC TA-3′, reverse, 5′-CAG GAC GGT CTT ACC CTT TCC-3′), Il6 (forward, 5′-AAC GAT GAT GCA CTT GCA GA-3′, reverse, 5′-GAG CAT TGG AAA TTG GGG TA -3′) and Ccl5 (forward, 5′-GCC CAC GTC AAG GAG TAT TTC TA -3′, reverse, 5′-ACA CAC TTG GCG GTT CCT TC -3′). Relative quantification was performed using standard curve analysis. The mRNA in samples was normalized to that of beta-Actin or Gapdh, and represented as the mRNA levels in arbitrary units (A.U) or as a ratio of gene copy number per 100 copies of beta-actin or Gapdh. The standard errors of means (SEM) were calculated using the Student's t test. For the Nanostring analysis, total RNA was hybridized to a custom gene expression CodeSet and analysed on an nCounter Digital Analyzer. Counts were normalized to endogenous controls per Nanostring Technologies' specifications. Values were log-transformed and displayed as a heat map (Euclidean clustering) generated using the ggplot package within the open source R software environment.
In vivo intracranial LPS challenge
6-8 weeks old age- and sex-matched C57BL/6 and Tril-/- mice were anesthetized with isoflurane and then injected intracranially with 20μl of LPS (100ng/μl, 2μg of LPS per mouse) or saline (control). 24 h post LPS injection the expression of proinflammatory cytokines Il6 and Ccl5 was examined within the brain. Generated data were analyzed by unpaired two-tailed Student's t test with Prism software. p values of less than 0.05 were considered significant.
In vivo E.coli infection
Age- and sex-matched C57BL/6 and Tril-/- mice were infected with 109 colony-forming units (CFU) of E.coli BL21 strain via the intraperitoneal route. Cytokine levels both at the mRNA and protein levels were analyzed 6 h post-infection in the brain and spleen. Data from in vivo experiments were analyzed by unpaired two-tailed Student's t test with Prism software. p values of less than 0.05 were considered significant.
Statistical analysis
Differences between groups were analyzed for statistical significance with Student's t test using Prism 6 Software (GraphPad, San Diego, CA). p < 0.05 was considered as statistically significant.
Results
Generation and characterization of TRIL knockout mice
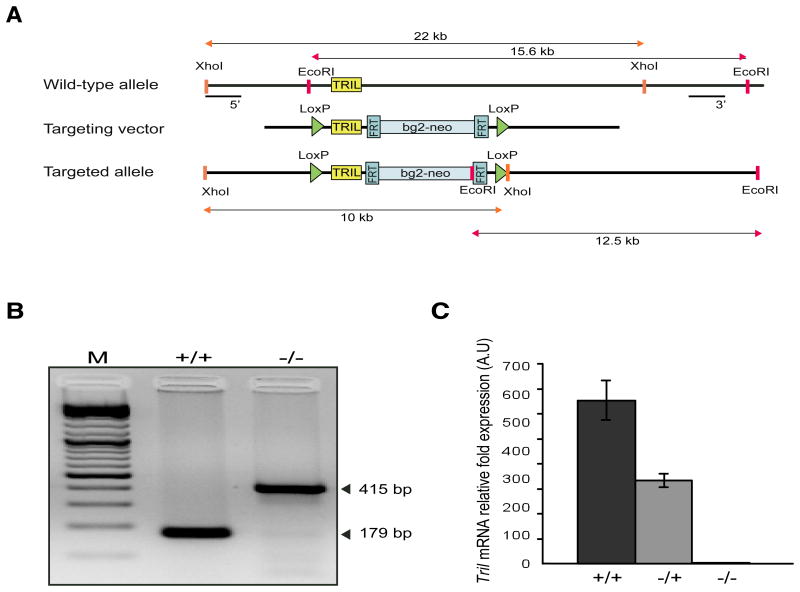
A targeting vector to generate Tril-/- mice was constructed as outlined in Fig. 1A. Conditional Tril-/- female founders were crossed with C57BL/6 males expressing Protamine-Cre resulting in permanent deletion of LoxP-flanked Tril alleles. Mice homozygous for the targeted allele were confirmed by PCR using genomic DNA isolated from tail biopsies (Fig. 1B). As TRIL is a brain enriched protein the cDNA derived from brain lysates was used to conduct RT-PCR analysis of Tril expression in wild type (+/+), heterozygous (+/-) and homozygous Tril-/- mice. As expected Tril expression was absent in TRIL-deficient mice while moderate to high expression was observed in heterozygous and WT mice, respectively (Fig. 1C). These data confirm successful deletion of Tril in generated knockout mice, which were used in subsequent experiments.
Figure 1. Generation of TRIL-deficient mice.
A, Gene targeting strategy for generation of Tril-/- mice. The structures of WT allele, targeting vector and targeted allele are represented. B, PCR analysis of genomic DNA prepared from tail biopsies using primers TRIL-F1, TRIL-R1 and TRIL-R2, detecting both wild-type (179 bp) and mutant (415 bp) alleles. C, RT-PCR analysis of Tril expression in the brain of wild-type (+/+), heterozygous (+/-) and mutant (-/-) mice. Tril mRNA levels were normalized against Gapdh and expressed relative to the lowest detectable sample. Data are presented as the mean ± SD of one experiment representative of three independent experiments.
General characteristics of TRIL-deficient mice
Tril-/- mice are viable and fertile and born at expected Mendialian ratios. To address whether TRIL deficiency affects any behavioural characteristics, we subjected Tril-/- mice and their WT littermates to a comprehensive set of behavioural tests. TRIL-deficient mice appeared healthy and displayed no phenotypic differences in general behaviour as well as in motor coordination (as determined via the rotarod), anxiety (elevated zero maze, stress induced hyperthermia, four-plate test) and pain (tail-flick, thermal sensitivity in response to inflammation) (Sup. 1A).
Test scores demonstrate no significant differences between wild type (WT), heterozygous (HET) or TRIL knockout mice (KO), in the acute pain model represented by tail-flick assay (Sup. 1B) as well as in the behavioural studies evaluating the impact of TRIL on stress-induced hyperthermic responses (Sup. 1C) and locomotor activity (Sup. 1D).
TRIL does not impact TLR4 and TLR3 mediated responses in peripheral immune cells
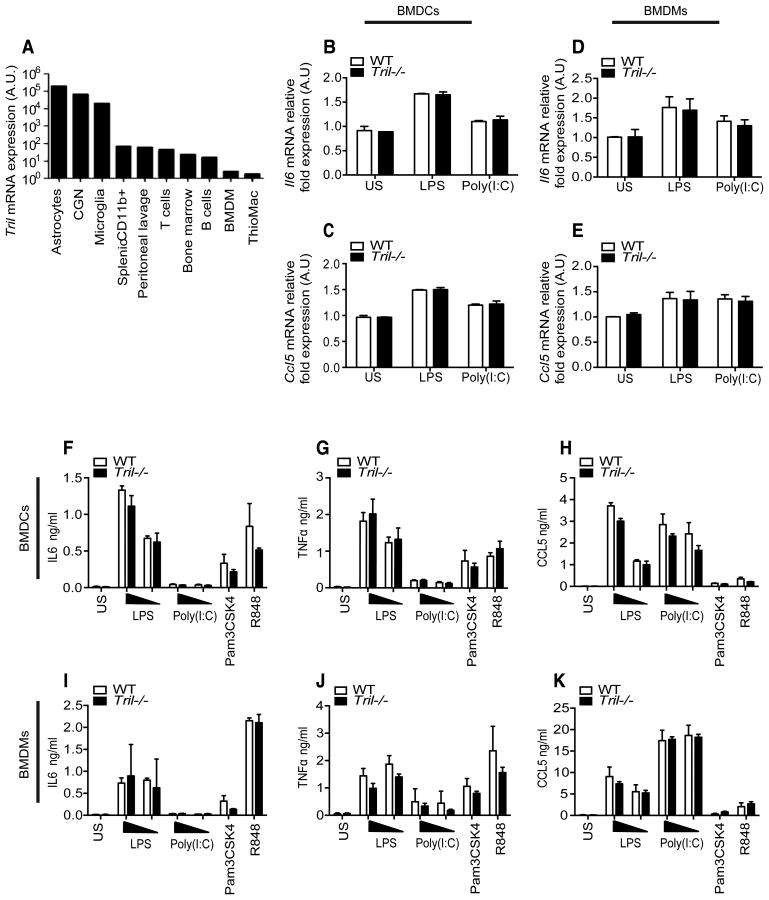
Our previous studies characterized a role for TRIL in mixed glial cells and in the astrocytoma cell line U373 in which we knocked down Tril using siRNA and shRNA (31). Using cells from Tril-/- mice, we analyzed TRIL function in multiple cell types and confirmed a high level of Tril expression in astrocytes, cerebellar granule neurons (CGN), and microglia, with lower expression evident in a range of pheriperhal immune cells (Fig. 2A). We next examined responses to various TLR agonists in primary bone marrow-derived macrophages (BMDMs) and bone marrow-derived dendritic cells (BMDCs) isolated from TRIL-deficient and WT mice. We analyzed cytokine expression following stimulation with the respective TLR4 and TLR3 ligands, LPS and Poly(I:C). Treating BMDCs with LPS led to an increase in mRNA for Il6 (Fig. 2B) and Ccl5 (Fig. 2C) and Tril deficiency had no effect on these responses, consistent with the low expression level of Tril in these cells. Poly(I:C) was a weak inductor of BMDCs. In BMDMs lack of TRIL also had no effect on the induction of Il6 (Fig. 2D) and Ccl5 (Fig. 2E) mRNA in response to stimulation with both LPS and Poly(I:C). Similar results were seen with LPS and Poly(I:C) when IL6 (F and I), TNFα (G and J) and CCL5 (H and K) production as measured by ELISA (Fig. 2F-K). Tril deficiency also had no effect on induction of IL6, TNFα and CCL5 by the TLR2 ligand Pam3CSK4 and TLR7/8 ligand R848, in either BMDCs (Fig. 2F-H) or BMDMs (Fig. 2I-K).
Figure 2. TRIL does not impact TLR mediated responses in bone marrow-derived macrophages (BMDMs) and dendritic cells (BMDCs).
A, Tril expression in various mouse cells populations measured by RT-PCR. Tril expression levels were normalized to Gapdh and expressed relative to the lowest detectable sample. B-E, Expression of Il6 (B and D) and Ccl5 (C and E) in primary BMDCs (left panel) and BMDMs (right panel) isolated from wild type (WT) and Tril-/- mice untreated or stimulated for 5 h with LPS (100ng/ml) or Poly(I:C) (25μg/ml). mRNA levels were normalized to beta-Actin and represented in arbitrary units (A.U) relative to unstimulated cells. F-K, ELISA for IL6 (F and I), TNFα (G and J) and CCL5 (H and K) measured in primary BMDCs (top panel) and BMDMs (bottom panel) derived from WT and Tril-/- mice stimulated for 24 h with LPS (10 and 100ng/ml), Poly(I:C) (25 and 50μg/ml), Pam3CSK4 (100nM) or R848 (1μg/ml). Data are presented as the mean ± SEM of two independent experiments carried out in triplicates.
TRIL modulates TLR4 and TLR3 but not TLR2 or TLR7/8 mediated responses in primary murine mixed glial cells
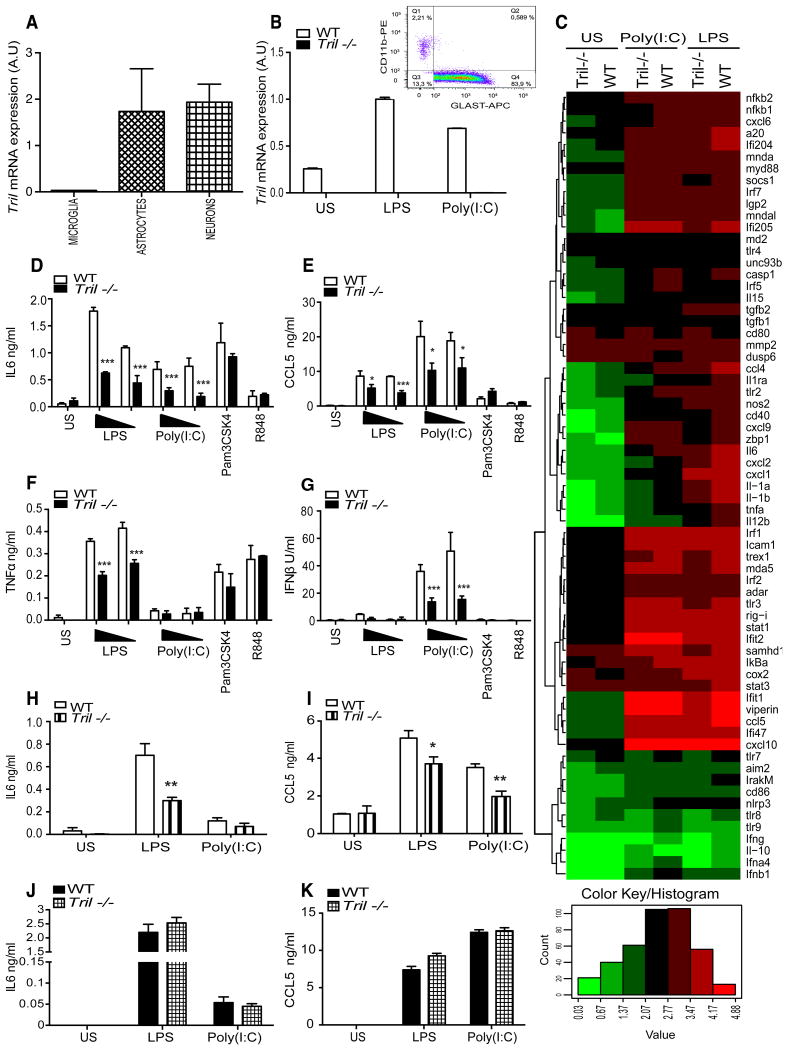
Tril is highly expressed within brain cells, notably in astrocytes and neurons compare to microglia (Fig. 3A). We therefore next investigated TLR mediated responses in mixed glial cells (which primarily consist of astrocytes, over 83% astrocytes and approximatelly 2-3% of microglia (Fig. 3B, histogram)) derived from WT and Tril-/- mice. As shown on the bar graph in Fig. 3B, Tril-/- cells are indeed devoid of Tril expression as expected, high basal level of Tril mRNA in the untreated WT mixed glial cells was further boosted following stimulation with both LPS and Poly(I:C), consistent with our previous studies (29, 31). We next analyzed the mRNA levels of 50 murine genes in WT and Tril-/- primary mixed glial cells prior to and following 5 h stimulation with LPS (100ng/ml) and Poly(I:C) (50μg/ml) (Fig. 3C) using a non-enzymatic RNA profiling technology that employs bar-coded fluorescent probes to simultaneously analyze mRNA expression levels of differentially regulated genes (nCounter, Nanostring). We found that the expression of a number of proinflammatory cytokines and chemokines were reduced in TRIL-deficient cells in response to LPS and Poly(I:C) (Fig. 3C). The mRNA levels of Il6, Ccl5, Tnfa, Il1a, Il1b and Ifnb1 were all decreased in Tril-/- cells. Additionally, the expression levels of chemokines such as the Cxcl2 and Ccl4 were also found to be significantly reduced in Tril-/- upon ligand activation. Following on from the gene expression studies we also examined cytokine production by ELISA in both WT and TRIL-deficient primary mixed glial cells following stimulation with TLR agonists (Fig. 3D-G). In agreement with the gene expression data, following 24 h treatment with two different doses of LPS (10 and 100ng/ml) and Poly(I:C) (25 and 50μg/ml) a statistically significant decrease in the IL6 and CCL5 production was observed in primary mixed glial cells derived from Tril-/- mice compared to WT controls (Fig. 3D and E). In addition, lack of TRIL affected TNFα and IFNβ protein levels in response to LPS and Poly(I:C), respectively (Fig. 3F and G). No major differences in the responses of Tril-/- and WT cells were seen following treatment with the TLR2 agonist Pam3CSK4, and TLR7/8 ligand R848 (Fig. 3D-G). Taken together, these data strongly indicate that TRIL affects both TLR3 and TLR4 signalling pathways in glial cells, but it does not impact responses of TLR2 and TLR7/8, consistent with our previous studies on the U373 cell line (31).
Figure 3. TRIL modulates TLR4 and TLR3 mediated in the primary murine mixed glial cells and astrocytes.
A, RT-PCR analysis of Tril expression in cultured microglia, astrocytes and neurons cells populations, mRNA levels of Tril were normalized to beta-Actin and expressed in arbitrary units (A.U). B, Expression of Tril at the basal level and following 5 h of stimulation with LPS (100ng/ml) or Poly(I:C) (25μg/ml) in murine mixed glial cells derived from wild type (WT) and TRIL-deficient (Tril-/-) mice, mRNA levels of Tril were normalized to beta-Actin and expressed in arbitrary units (A.U). B (histogram) FACS analysis of microglia and astrocytes composition within primary mixed glial cells. Isolated primary mixed glial cells were stained using markers specific for astrocytes (GLAST-APC; Miltenyi Biotech), microglia (Cd11b-PE; eBioscience) and neurons (β-3-Tubulin; Biolegend, not shown), graph demonstrates two main populations of cells among mixed glial cells; astrocytes and microglia, data are representative of three individual experiments. C, Gene expression analysis of primary murine mixed glial cells derived from WT and TRIL-deficient mice (Tril-/-) untreated or following 5 h of stimulation with LPS (100ng/ml) or Poly(I:C) (25μg/ml). Gene expression profiles are displayed as a heat map (log10 transformed) with hierarchical clustering indicated by dendrogram. Upregulated genes are shown in red, downregulated genes are represented in green. D-G, ELISA for IL6 (D), CCL5 (E), TNFα (F), and IFNβ (G), production in primary murine mixed glial cells derived from WT and Tril-/- mice, untreated or stimulated for 24 h with LPS (10 and 100ng/ml), Poly(I:C) (25 and 50μg/ml), Pam3CSK4 (100nM) or R848 (1μg/ml). Data are represented as the mean ± SD of one experiment representative of three independent experiments, all carried out in triplicates. ***, p<0.001, **, p<0.01, *, p<0.05, H and I, ELISA for IL6 (H) and CCL5 (I), production in the purified primary astrocyte population isolated from primary murine mixed glial cells derived from WT and TRIL-deficient mice (Tril-/-), untreated or stimulated for 24 h with LPS (100ng/ml) or Poly(I:C) (25μg/ml). Data are represented as the mean ± SEM of two independent experiments, a total of three mice were pulled in each of three experiments carried out, ***, p<0.001, **, p<0.01, *, p<0.05. J and K, ELISA for IL6 (J) and CCL5 (K), production examined in immortalized migroglia cells derived from WT and Tril-/- mice, untreated or stimulated for 24 h with LPS (100ng/ml) or Poly(I:C) (25μg/ml). Data are represented as the mean ± SEM of three independent experiments all carried out in triplicates.
As a control to confirm that LPS and Poly(I:C) were acting via TLR4 and TLR3 respectively in mixed glial cells, we also examined the TLR mediated responses in cells from Tlr4-/-, Tlr3-/- and Trif-/- mice, both at the mRNA and protein level and observed attenuated response to LPS in TLR4 and TRIF-deficient cells, and Poly(I:C) in cells lacking TRIF and TLR3 (data not shown).
In order to evaluate in more detail the cell type responsible for detected differences in the cytokine production between WT and Tril-/- mixed glilal cells, we next examined purified populations of astrocytes and microglia. In agreement with the data obtained with primary mixed glial cells comprising largely of astrocytes, deficiency of TRIL in purified astrocyte population strongly affected both IL6 and CCL5 production following LPS and Poly(I:C) stimulation (Fig. 3H and I). Due to technical difficulities in obtaining sufficient number of purified microglia and strongly restricted number of Tril-/- mice available, we were unable to examine the responses to LPS and Poly(I:C) stimulation among these cells. However, analysis of immortalised mixed glial cell population comprising primarily of microglia and completely devoid of astrcoytes revealed no significant differences in the IL6 and CCL5 production upon LPS and Poly(I:C) stimulation (Fig. 3J and K). These data clearly demonstrate that astrocyte and not microglia are primarily responsible for the reduced cytokine production in Tril-/- primary mixed glial cells following stimulation with LPS and Poly(I:C), which is in agreement with the high expression of Tril within these cells.
Investigation into the in vivo role of TRIL in E.coli induced sepsis and upon intracranial LPS challenge
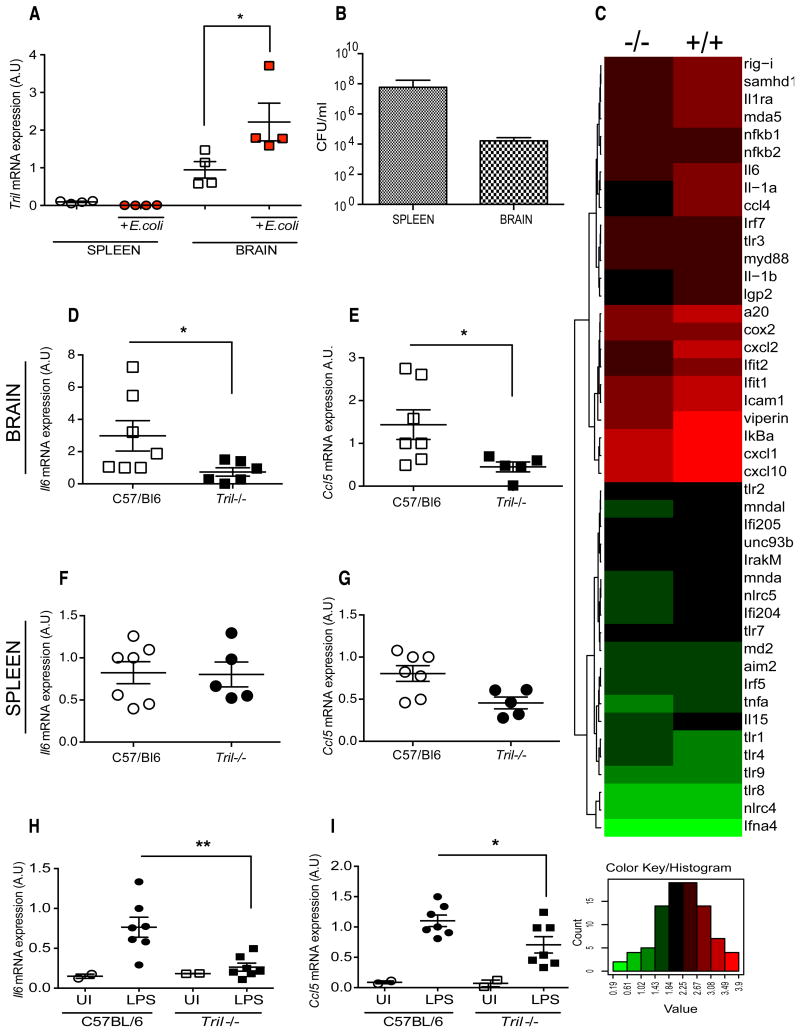
Finally we addressed the role of TRIL in vivo using an E.coli challenge model, which is dependant on TLR recognition pathways in vivo. We first examined the expression of Tril prior to and following 6 h of infection with 109 CFU of the E.coli via the intraperitoneal (IP) route. The expression of Tril was analyzed within the spleen and brain tissues by RT-PCR. As can be seen in Fig. 4A, the basal expression of Tril was significantly higher in the brain compared to spleen, and it was further enhanced following E.coli infection. Tril expression was not detected in the spleen no increase in expression was observed upon infection. Bacterial load measured within the spleen and brain revealed that both tissues contain high number of bacteria ranging from 108 to 104 CFU/ml in the spleen and brain, respectively (Fig. 4B). As shown in Fig. 4C, analysis of gene expression profile generated using total RNA isolated from the brain of WT (+/+) and TRIL-deficient (-/-) mice following intraperitoneal infection with E.coli revealed reduced expression levels of proinflammatory cytokines such as Il6, Tnfa, Il1a and Il1b as well as chemokines Ccl4, Cxcl2, Cxcl10, in the Tril-/- mice. Interestingly, a number of genes involved in the antiviral response, such as Viperin and Rig-I were also dramatically reduced in TRIL-deficient mice (Fig. 4C). An additional RT-PCR analysis using cDNA generated from spleen and brain of Tril-/- and WT mice infected with E.coli, demonstrated that the mRNA levels of Il6 and Ccl5 were significantly decreased in the brain samples derived from Tril-/- mice (Fig. 4D and E). RT-PCR of cDNA from the spleens of WT and Tril-/- mice show no significant change in Il6 and Ccl5 mRNA levels (Fig. 4F and G).
Figure 4. Investigation into the in vivo role of TRIL in the E.coli induced Gram-negative sepsis model and following intracranial LPS challenge.
A, Expression of Tril measured by RT-PCR in both spleen and brain samples derived from WT and Tril-/- mice infected with E.coli strain BL21 (109 CFU) for 6 h (n=4). mRNA levels of Tril were normalized to beta-Actin and expressed in arbitrary units (A.U). B, Bacterial dissemination in the spleen and brain of WT mice following intraperitoneal E.coli infection for 6 h. C, Gene expression profile generated using RNA isolated from the brain of WT and Tril-/- mice. Gene expression profile is displayed as a heat map (log10 transformed) with hierarchical clustering indicated by dendrogram. Upregulated genes are shown in red, downregulated genes are represented in green. D-G, Cytokine expression measured by RT-PCR in brain (top panel) and spleen (bottom panel) of WT and Tril-/- mice infected with E.coli (109 CFU, colony forming units) for 6 h (n=5-7). Data are presented as a mean ± SEM of two (A and B) or three (D-G) independent experiments, each carried out in triplicates. *, p<0.05. H and I, Cytokine expression measured by RT-PCR in brain of WT and Tril-/- mice following 24h of intracranial LPS challenge (2μg of LPS or saline control per mice) (n=2-7). Data are presented as a mean ± SEM of two independent experiments, each carried out in triplicate. **, p<0.01, *, p<0.05.
Despite the intraperitoneal administration of bacteria in the in vivo E.coli infection model tested, the significant differences in the proinflammatory cytokines production between WT and Tril-/- were detected in the brain but not the spleen (Fig. 4F and G). We decided therefore to further exmine if the observed differences were caused by the direct effect of TRIL on TLR4-mediated response within the brain. As can be seen in Fig. 4H and I, direct intracranial injection of 2μg of LPS into the brain of WT and TRIL-deficient mice leads to significant decrease in the Il6 and Ccl5 expression in the brain of Tril-/- compared to WT controls (Fig. 4H and I). TRIL is therefore involved in the modulation of TLR4 mediated response directly within the brain.
These results indicate that TRIL functions primarily within the brain, where in response to intracranial LPS challenge and intraperitoneal infection with E.coli, it mediates cytokine production from glial cells.
Discussion
Interaction between the peripheral immune system and the CNS plays a fundamental role in mounting an appropriate response to acute systemic infections (36). Brain dysfunction often actively contributes to deterioration of systemic infections however prolonged brain inflammation is frequently a consequence of the systemic inflammatory response.
TRIL was characterized as a novel accessory protein for TLR4 and TLR3 highly expressed in the brain (29, 31). In a series of in vitro studies, TRIL was shown to play a positive role in the regulation of TLR4 and TLR3 signalling pathways. However its function in vivo has never been tested. The aim of this study was to further investigate TRIL in an in vitro and more importantly in vivo, using TRIL-deficient mice.
Primary BMDMs and BMDCs derived from TRIL-deficient and WT mice produced the same level of cytokines in response to LPS or Poly(I:C). TRIL possesses a similar structure and function to CD14, therefore we previously speculated that TRIL might act as a substitute for CD14 in cells where CD14 is expressed at low levels. CD14 is abundantly expressed on BMDMs and BMDCs (37-39). However among glial cells, CD14 is highly expressed within microglia, and is not present in astrocytes or neurons (15, 16). We demonstrated that mRNA levels of various proinflammatory genes induced in response to LPS or Poly(I:C) were strongly reduced in primary mixed glial cells derived from Tril-/- mice when compared to WT controls. Since primary mixed glial cells are approximately 83-85% astrocytes and 2-3% microglia, it is possible that TRIL may indeed fulfil the role of CD14 in astrocytes, which is further supported by differences in the IL6 and CCL5 production between WT and Tril-/- purified astrocytes but not immortalized microglia cells following LPS and Poly(I:C) stimulation. In addition no difference was observed in response to Pam3CSK4 or R848, consistent with the lack of a role for TRIL in signalling mediated by TLR2 and TLR7/8 (29, 31).
Nearly one-third of all cases of sepsis are caused by Gram-negative bacterial infection, among which E.coli is considered the most causative pathogen (40). Thus, aiming to address the in vivo role of TRIL we used intraperitoneal infection with E.coli. Expression of Tril was enhanced following E.coli infection, however this occurred exclusively in the brain and not spleen, consistent with the results indicating the significance of Tril expression in mixed glial cells compared to macrophages and dendritic cells. Further analysis of the cytokine expression profile in the brain following E.coli challenge revealed reduced levels of proinflammatory cytokines and chemokines in samples derived from TRIL-deficient mice when compared to littermate controls. Similar to our earlier observation in the in vitro studies using primary mixed glial cells, levels of many inflammatory cytokines such as Il6, Tnfa, Il1a and Il1b and chemokines Ccl4, Cxcl2, Cxcl10, were all reduced in the TRIL-deficient mice upon bacterial infection. Analysis of the gene expression panel also revealed a dramatic difference in some of the interferon stimulated genes (ISGs) involved in viral recognition and the antiviral response, such as Rig-I and Viperin, respectively, in the LPS treated TRIL-deficient mice. TRIL is implicated in the regulation of TLR4 mediated signalling, therefore detected differences in ISGs expression could be explained by lower levels of IFN production in response to LPS. This data together with the previously reported involvement of TRIL in the TLR3 signalling pathway, suggest a possible role for TRIL in the antiviral immune response. However, further studies are needed in order to verify these findings.
In our sepsis model, Gram-negative E.coli was administrated via the IP route, but interestingly the main effect of the lack of TRIL was observed in the brain. As mentioned earlier, TLR4 is highly expressed within the CNS. High levels of TLR4 can be found in the meninges, choroid plexus and circumventricular organs (CVO) of rat brain (41). Constitutive expression of TLR4 and CD14 in the CVO and meninges, sites of the brain with direct access to the circulation provide for the possibility of direct TLR4-mediated LPS action in the CNS, which would also require TRIL (22, 41). Alternatively, brain inflammation can also be triggered by direct sensing of bacteria by resident glial cells, microglia and astrocytes within the brain parenchyma following disruption of the BBB. In fact, we detected the presence of bacteria in the brain of WT mice following infection with E.coli, which suggested that the integrity of BBB was breached allowing for bacteria to disseminate throughout the brain.
Astrocytes are one of the most abundant cell type in the CNS. They participate in the innate immune responses, provide support to neurons, regulate synaptic activity and contribute to the formation of the BBB (42). Both human and murine astrocytes express a wide range of TLRs. Cultured human astrocytes were reported to constitutively express TLR2, TLR3 and TLR4 (43, 44), while mouse-derived astrocytes express TLRs 1-9, with particularly high levels of TLR3 and TLR4 (45). Following activation, astrocytes produce a wide range of proinflammatory cytokines such as IL6, TNFα and IFNβ and chemokines CCL2, CCL5, CCL20, CXCL8 and CXCL10 (46-48). Our studies on primary mixed glial cells and purified primary astrocytes strongly suggest that TRIL, which is highly expressed in astrocytes, functions as a regulator of TLR mediated responses within these cells.
Of note, astrocytes are also involved in the up-regulation of inducible nitric oxide synthase (iNOS) and nitric oxide production (49). Thus, astrocytes participate in processes such as tissue damage and neurotoxicity. Given multiple functions of astrocytes, there is a possibility that TRIL might impact not only innate immune responses within these cells, but also BBB permeability and neuronal cell death.
In summary our study clearly identifies a key role for TRIL in TLR4 responses in the brain. It also provides in vivo evidence that TRIL acts as a mediator of cytokine production in response to E.coli infection within the CNS.
Supplementary Material
Supplementary Figure 1 Behavioural phenotyping of Tril-/- and HET mice relative to WT controls A, Overall evaluation accross multiple behavioural tests examining anxiety, depression, general bahaviour, motor coordination and pain sensation, carried out using a cohort of WT, HET andTRIL-deficient mice (n=7, 10 and 10, respectively). B, Tail-flick assay assessing pain sensation by measuring tail withdrawal latency. Data are represented as a mean ± SEM (n=7-10). C, Model of stress-induced hyperthermic responses examinig an anxiolytic phenotype of Tril-/- and HET mice relative to WT cohorts (n=7-10). D, Spontaneous locomotor activity assay evaluating pattern of exploratory bahaviour of Tril-/- and HET mice relative to WT cohorts (n=7-10). Data are represented as mean ± SEM (n=7-10).
Acknowledgments
The authors would like to thank A. Cerny and K. Army for animal husbandry. M. Uccellini for RNA extracts from brain cells subpopulations. J. Kaminski for his assistance with the mixed glial cells isolation. S.Sharma for helpful discussions and other members of the Fitzgerald and O'Neill labs.
This work was supported by the European Commission under the 7th Framework Programme (TranSVIR FP7-PEOPLE-ITN-2008 #238756).
Abbreviations
- TLR
Toll-like Receptor
- TRIL
TLR4 Interactor with leucine rich repeats
- CFU
colony-forming units
- DIV
days in vitro
References
- 1.Sharshar T, Annane D, de la Grandmaison GL, Brouland JP, Hopkinson NS, Francoise G. The neuropathology of septic shock. Brain pathology. 2004;14:21–33. doi: 10.1111/j.1750-3639.2004.tb00494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharshar T, Carlier R, Bernard F, Guidoux C, Brouland JP, Nardi O, de la Grandmaison GL, Aboab J, Gray F, Menon D, Annane D. Brain lesions in septic shock: a magnetic resonance imaging study. Intensive care medicine. 2007;33:798–806. doi: 10.1007/s00134-007-0598-y. [DOI] [PubMed] [Google Scholar]
- 3.Papadopoulos MC, Davies DC, Moss RF, Tighe D, Bennett ED. Pathophysiology of septic encephalopathy: a review. Critical care medicine. 2000;28:3019–3024. doi: 10.1097/00003246-200008000-00057. [DOI] [PubMed] [Google Scholar]
- 4.Galea I, Bechmann I, Perry VH. What is immune privilege (not)? Trends in immunology. 2007;28:12–18. doi: 10.1016/j.it.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Ransohoff RM, Kivisakk P, Kidd G. Three or more routes for leukocyte migration into the central nervous system. Nat Rev Immunol. 2003;3:569–581. doi: 10.1038/nri1130. [DOI] [PubMed] [Google Scholar]
- 6.Green R, Scott LK, Minagar A, Conrad S. Sepsis associated encephalopathy (SAE): a review. Front Biosci. 2004;9:1637–1641. doi: 10.2741/1250. [DOI] [PubMed] [Google Scholar]
- 7.de Bock F, Derijard B, Dornand J, Bockaert J, Rondouin G. The neuronal death induced by endotoxic shock but not that induced by excitatory amino acids requires TNF-alpha. Eur J Neurosci. 1998;10:3107–3114. doi: 10.1046/j.1460-9568.1998.00317.x. [DOI] [PubMed] [Google Scholar]
- 8.Janeway CA, Jr, Medzhitov R. Introduction: the role of innate immunity in the adaptive immune response. Semin Immunol. 1998;10:349–350. doi: 10.1006/smim.1998.0142. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter S, O'Neill LA. How important are Toll-like receptors for antimicrobial responses? Cellular microbiology. 2007;9:1891–1901. doi: 10.1111/j.1462-5822.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 10.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 13.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 14.Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nature reviews Immunology. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 15.Lehnardt S, Lachance C, Patrizi S, Lefebvre S, Follett PL, Jensen FE, Rosenberg PA, Volpe JJ, Vartanian T. The toll-like receptor TLR4 is necessary for lipopolysaccharide-induced oligodendrocyte injury in the CNS. J Neurosci. 2002;22:2478–2486. doi: 10.1523/JNEUROSCI.22-07-02478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehnardt S, Massillon L, Follett P, Jensen FE, Ratan R, Rosenberg PA, Volpe JJ, Vartanian T. Activation of innate immunity in the CNS triggers neurodegeneration through a Toll-like receptor 4-dependent pathway. Proc Natl Acad Sci U S A. 2003;100:8514–8519. doi: 10.1073/pnas.1432609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rolls A, Shechter R, London A, Ziv Y, Ronen A, Levy R, Schwartz M. Toll-like receptors modulate adult hippocampal neurogenesis. Nat Cell Biol. 2007;9:1081–1088. doi: 10.1038/ncb1629. [DOI] [PubMed] [Google Scholar]
- 18.Acosta C, Davies A. Bacterial lipopolysaccharide regulates nociceptin expression in sensory neurons. J Neurosci Res. 2008;86:1077–1086. doi: 10.1002/jnr.21565. [DOI] [PubMed] [Google Scholar]
- 19.Tu Z, Portillo JA, Howell S, Bu H, Subauste CS, Al-Ubaidi MR, Pearlman E, Lin F. Photoreceptor cells constitutively express functional TLR4. J Neuroimmunol. 2011;230:183–187. doi: 10.1016/j.jneuroim.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laflamme N, Soucy G, Rivest S. Circulating cell wall components derived from gram-negative, not gram-positive, bacteria cause a profound induction of the gene-encoding Toll-like receptor 2 in the CNS. Journal of neurochemistry. 2001;79:648–657. doi: 10.1046/j.1471-4159.2001.00603.x. [DOI] [PubMed] [Google Scholar]
- 21.Laflamme N, Echchannaoui H, Landmann R, Rivest S. Cooperation between toll-like receptor 2 and 4 in the brain of mice challenged with cell wall components derived from gram-negative and gram-positive bacteria. Eur J Immunol. 2003;33:1127–1138. doi: 10.1002/eji.200323821. [DOI] [PubMed] [Google Scholar]
- 22.Chakravarty S, Herkenham M. Toll-like receptor 4 on nonhematopoietic cells sustains CNS inflammation during endotoxemia, independent of systemic cytokines. J Neurosci. 2005;25:1788–1796. doi: 10.1523/JNEUROSCI.4268-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth J, Harre EM, Rummel C, Gerstberger R, Hubschle T. Signaling the brain in systemic inflammation: role of sensory circumventricular organs. Frontiers in bioscience : a journal and virtual library. 2004;9:290–300. doi: 10.2741/1241. [DOI] [PubMed] [Google Scholar]
- 24.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 26.Xia Y, Yamagata K, Krukoff TL. Differential expression of the CD14/TLR4 complex and inflammatory signaling molecules following i.c.v. administration of LPS. Brain Res. 2006;1095:85–95. doi: 10.1016/j.brainres.2006.03.112. [DOI] [PubMed] [Google Scholar]
- 27.Nadeau S, Rivest S. Role of microglial-derived tumor necrosis factor in mediating CD14 transcription and nuclear factor kappa B activity in the brain during endotoxemia. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:3456–3468. doi: 10.1523/JNEUROSCI.20-09-03456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacroix S, Feinstein D, Rivest S. The bacterial endotoxin lipopolysaccharide has the ability to target the brain in upregulating its membrane CD14 receptor within specific cellular populations. Brain Pathol. 1998;8:625–640. doi: 10.1111/j.1750-3639.1998.tb00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpenter S, Carlson T, Dellacasagrande J, Garcia A, Gibbons S, Hertzog P, Lyons A, Lin LL, Lynch M, Monie T, Murphy C, Seidl KJ, Wells C, Dunne A, O'Neill LA. TRIL, a functional component of the TLR4 signaling complex, highly expressed in brain. Journal of immunology. 2009;183:3989–3995. doi: 10.4049/jimmunol.0901518. [DOI] [PubMed] [Google Scholar]
- 30.Carpenter S, O'Neill LA. Recent insights into the structure of Toll-like receptors and post-translational modifications of their associated signalling proteins. The Biochemical journal. 2009;422:1–10. doi: 10.1042/BJ20090616. [DOI] [PubMed] [Google Scholar]
- 31.Carpenter S, Wochal P, Dunne A, O'Neill LA. Toll-like receptor 3 (TLR3) signaling requires TLR4 Interactor with leucine-rich REPeats (TRIL) The Journal of biological chemistry. 2011;286:38795–38804. doi: 10.1074/jbc.M111.255893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaech S, Banker G. Culturing hippocampal neurons. Nature protocols. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 33.Floden AM, Combs CK. Microglia repetitively isolated from in vitro mixed glial cultures retain their initial phenotype. Journal of neuroscience methods. 2007;164:218–224. doi: 10.1016/j.jneumeth.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F. Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. Journal of neuroimmunology. 1990;27:229–237. doi: 10.1016/0165-5728(90)90073-v. [DOI] [PubMed] [Google Scholar]
- 35.Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, Kawai T, Akira S, Savan R, van Echo D, Fitzgerald KA, Young HA, Ching LM, Vogel SN. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. The Journal of experimental medicine. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharshar T, Hopkinson NS, Orlikowski D, Annane D. Science review: The brain in sepsis--culprit and victim. Crit Care. 2005;9:37–44. doi: 10.1186/cc2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becher B, Fedorowicz V, Antel JP. Regulation of CD14 expression on human adult central nervous system-derived microglia. J Neurosci Res. 1996;45:375–381. doi: 10.1002/(SICI)1097-4547(19960815)45:4<375::AID-JNR6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 38.Haziot A, Chen S, Ferrero E, Low MG, Silber R, Goyert SM. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J Immunol. 1988;141:547–552. [PubMed] [Google Scholar]
- 39.Mahnke K, Becher E, Ricciardi-Castagnoli P, Luger TA, Schwarz T, Grabbe S. CD14 is expressed by subsets of murine dendritic cells and upregulated by lipopolysaccharide. Advances in experimental medicine and biology. 1997;417:145–159. doi: 10.1007/978-1-4757-9966-8_25. [DOI] [PubMed] [Google Scholar]
- 40.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 41.Laflamme N, Rivest S. Toll-like receptor 4: the missing link of the cerebral innate immune response triggered by circulating gram-negative bacterial cell wall components. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2001;15:155–163. doi: 10.1096/fj.00-0339com. [DOI] [PubMed] [Google Scholar]
- 42.Dong Y, Benveniste EN. Immune function of astrocytes. Glia. 2001;36:180–190. doi: 10.1002/glia.1107. [DOI] [PubMed] [Google Scholar]
- 43.Bsibsi M, Ravid R, Gveric D, van Noort JM. Broad expression of Toll-like receptors in the human central nervous system. Journal of neuropathology and experimental neurology. 2002;61:1013–1021. doi: 10.1093/jnen/61.11.1013. [DOI] [PubMed] [Google Scholar]
- 44.Bsibsi M, Persoon-Deen C, Verwer RW, Meeuwsen S, Ravid R, Van Noort JM. Toll-like receptor 3 on adult human astrocytes triggers production of neuroprotective mediators. Glia. 2006;53:688–695. doi: 10.1002/glia.20328. [DOI] [PubMed] [Google Scholar]
- 45.Gorina R, Font-Nieves M, Marquez-Kisinousky L, Santalucia T, Planas AM. Astrocyte TLR4 activation induces a proinflammatory environment through the interplay between MyD88-dependent NFkappaB signaling, MAPK, and Jak1/Stat1 pathways. Glia. 2011;59:242–255. doi: 10.1002/glia.21094. [DOI] [PubMed] [Google Scholar]
- 46.Farina C, Krumbholz M, Giese T, Hartmann G, Aloisi F, Meinl E. Preferential expression and function of Toll-like receptor 3 in human astrocytes. Journal of neuroimmunology. 2005;159:12–19. doi: 10.1016/j.jneuroim.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 47.Jack CS, Arbour N, Manusow J, Montgrain V, Blain M, McCrea E, Shapiro A, Antel JP. TLR signaling tailors innate immune responses in human microglia and astrocytes. Journal of immunology. 2005;175:4320–4330. doi: 10.4049/jimmunol.175.7.4320. [DOI] [PubMed] [Google Scholar]
- 48.Park C, Lee S, Cho IH, Lee HK, Kim D, Choi SY, Oh SB, Park K, Kim JS, Lee SJ. TLR3-mediated signal induces proinflammatory cytokine and chemokine gene expression in astrocytes: differential signaling mechanisms of TLR3-induced IP-10 and IL-8 gene expression. Glia. 2006;53:248–256. doi: 10.1002/glia.20278. [DOI] [PubMed] [Google Scholar]
- 49.Carpentier PA, Begolka WS, Olson JK, Elhofy A, Karpus WJ, Miller SD. Differential activation of astrocytes by innate and adaptive immune stimuli. Glia. 2005;49:360–374. doi: 10.1002/glia.20117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Behavioural phenotyping of Tril-/- and HET mice relative to WT controls A, Overall evaluation accross multiple behavioural tests examining anxiety, depression, general bahaviour, motor coordination and pain sensation, carried out using a cohort of WT, HET andTRIL-deficient mice (n=7, 10 and 10, respectively). B, Tail-flick assay assessing pain sensation by measuring tail withdrawal latency. Data are represented as a mean ± SEM (n=7-10). C, Model of stress-induced hyperthermic responses examinig an anxiolytic phenotype of Tril-/- and HET mice relative to WT cohorts (n=7-10). D, Spontaneous locomotor activity assay evaluating pattern of exploratory bahaviour of Tril-/- and HET mice relative to WT cohorts (n=7-10). Data are represented as mean ± SEM (n=7-10).