Abstract
Soluble Klotho (sKlotho) is the shed ectodomain of antiaging membrane Klotho that contains 2 extracellular domains KL1 and KL2, each of which shares sequence homology to glycosyl hydrolases. sKlotho elicits pleiotropic cellular responses with a poorly understood mechanism of action. Notably, in injury settings, sKlotho confers cardiac and renal protection by down-regulating calcium-permeable transient receptor potential canonical type isoform 6 (TRPC6) channels in cardiomyocytes and glomerular podocytes. Inhibition of PI3K-dependent exocytosis of TRPC6 is thought to be the underlying mechanism, and recent studies showed that sKlotho interacts with α2-3-sialyllactose-containing gangliosides enriched in lipid rafts to inhibit raft-dependent PI3K signaling. However, the structural basis for binding and recognition of α2-3-sialyllactose by sKlotho is unknown. Using homology modeling followed by docking, we identified key protein residues in the KL1 domain that are likely involved in binding sialyllactose. Functional experiments based on the ability of Klotho to down-regulate TRPC6 channel activity confirm the importance of these residues. Furthermore, KL1 domain binds α2-3-sialyllactose, down-regulates TRPC6 channels, and exerts protection against stress-induced cardiac hypertrophy in mice. Our results support the notion that sialogangliosides and lipid rafts are membrane receptors for sKlotho and that the KL1 domain is sufficient for the tested biologic activities. These findings can help guide the design of a simpler Klotho mimetic.—Wright, J. D., An, S.-W., Xie, J., Yoon, J., Nischan, N., Kohler, J. J., Oliver, N., Lim, C., Huang, C.-L. Modeled structural basis for the recognition of α2-3-sialyllactose by soluble Klotho.
Keywords: KL1 domain, TRPC6, PI3K
Mice deficient in α-Klotho manifest multiple phenotypes resembling premature aging in humans and die at ∼3 mo after birth (1). α-Klotho is predominantly expressed in kidney, parathyroid glands, and epithelial cells of the choroids plexus. Overexpression of α-Klotho extends life span in mice, supporting its role in suppressing aging (2). α-Klotho’s role in human aging is suggested by reports that increased circulating levels of soluble Klotho (sKlotho) or certain polymorphisms of KLOTHO are correlated with life span and several aging-related phenotypes, including osteoporosis, coronary artery disease, and decline in cognitive function (3–7).
α-Klotho protein is a type 1 membrane protein with a large extracellular domain, a membrane-spanning segment, and a short intracellular carboxyl terminus (1). The full-length membranous α-Klotho plays an important role in phosphate homeostasis by physical association with fibroblast growth factor (FGF) receptors to form coreceptors for the phosphaturic hormone FGF23 (8–10). The regulation of phosphate homeostasis by membranous α-Klotho-FGF coreceptors is critical for aging suppression and normal growth of wild-type mice. Mice deficient in α-Klotho have systemic phosphate retention; dietary phosphate restriction prevents phosphate retention and rescues the growth retardation and premature death in α-Klotho-deficient mice (11).
The large extracellular domain of α-Klotho is ∼950 aa in length and consists of 2 related domains, KL1 and KL2, each sharing amino acid sequence homology to family 1 glycosyl hydrolases (12). The ectodomain of α-Klotho (sKlotho) is shed; it is believed to function as an endocrine or local paracrine hormone in systemic circulation, urine, and cerebrospinal fluid (2, 13). sKlotho elicits multiple biologic effects on target cells, including antagonism of Wnt signaling to suppress cellular senescence and tissue fibrosis, and inhibition of retinoic acid–inducible gene I (Rig1)-mediated inflammation (14, 15). Moreover, sKlotho regulates signaling by multiple growth factors: sKlotho inhibits growth factor-driven PI3K/Akt signaling, contributing to extension of life span and cardioprotection in mice, and inhibition of tumor cell proliferation in culture (2, 16, 17). sKlotho also inhibits TGF-β1 signaling and suppresses kidney fibrosis and cancer metastasis in mice (18). Besides choroid plexus epithelial cells, α-Klotho expression was recently found in other brain regions, including hippocampus, and important roles of sKlotho in synaptic function and cognition in animals have been reported (7, 19). Thus, deficiency of sKlotho in animals also contributes to aging phenotypes. How sKlotho elicits pleiotropic cellular effects is poorly understood.
sKlotho has been reported to exhibit weak glucuronidase activity (20), but its physiologic role as glucuronidase is unclear. We reported that sKlotho functions as a sialidase that targets α2-6-sialyllactose in the N-glycans of glycoproteins (21). This action of sKlotho regulates cell-surface retention of target proteins. More recently, we reported that sKlotho also targets α2-3-sialyllactose of glycolipid gangliosides highly enriched in lipid rafts and affects multiple cellular effects mediated by lipid rafts (22). Lipid rafts are small heterogeneous, dynamic cholesterol- and sphingolipid-rich membrane microdomains that serve as platforms for multiple cellular processes such as signal transduction and membrane trafficking (23). We showed that sKlotho binds to α2-3-sialyllactose of gangliosides GM1 and GM3 that are highly enriched in lipid rafts and inhibits raft-dependent PI3K signaling and PI3K-dependent exocytosis of transient receptor potential canonical type isoform 6 (TRPC6) channels, thus down-regulating TRPC6 (22). In this study, we sought to elucidate the structural basis of sKlotho for α2-3-sialyllactose recognition.
MATERIALS AND METHODS
Modeling KL1 and KL2 domains
KL1 and KL2 domains have a sequence identity of 33% between them (Supplemental Table S1). To obtain structures sharing maximal sequence identity with the KL1 and KL2 domains, a Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information, Bethesda, MD, USA; https://blast.ncbi.nlm.nih.gov/Blast.cgi) search was performed against sequences in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB) (24, 25). The Klotho-related protein (KLrP; PDB entry 2e9l, 1.6-Å resolution) was found to have 40% sequence identity with residues 60 to 506 in the KL1 domain (26), whereas β-glucosidase (PDB entry 3vii, 1.0-Å resolution) shared a sequence identity of 33% with residues 517 to 962 in the KL2 domain (27). On the basis of these 2 templates, the homology modeling program Modeler (28, 29) was used to produce 20 structures per domain; the lowest DOPE-scoring model was chosen for further evaluation.
Preparation of α2-3-sialyllactose ligand for docking
A search of the PDB archive yielded 6 structures that contained α2-3-sialyllactose: 1s0i (1.60 Å), 1z4x (2.50 Å), 4c1w (1.84 Å), 4wvw, (1.47 Å), 4y20, (2.20 Å), and 4yz5 (2.27 Å). The α2-3-sialyllactose was taken from 1s0i, and hydrogen atoms were added using the Chimera program (30–35). Its geometry was then optimized at the RHF/6-31G* level using Gaussian09 (36), and the charges were derived at the MP2/6-31G* level using the NBO 5.9 program (37).
Preparation of receptor for docking
The receptor was prepared using the Dock Prep module of Chimera and the Amber FF14SB force field (38). Spheres representing potential atoms that can contact the receptor surface were mapped onto the protein surface using the Sphgen from the DOCK6 suite of programs (39–42). Grid-based electrostatic and van der Waals (vdW) potentials were then calculated for a box containing the largest sphere cluster with an additional 15 Å in each direction using the grid program (43, 44).
Docking protocol
The DOCK6 program was used to dock α2-3-sialyllactose as a flexible ligand into the KL1 binding site described as spheres via an anchor-and-grow methodology. For each configuration, an anchor of 5 atoms from one of the ligand rings was aligned with 5 spheres to locate the initial docked structure of that ring; the rest of the ligand was then grown out from this anchor. Each docked configuration was scored using electrostatic- and vdW-grid-based potentials and optimized using simplex energy minimization. The 500 best-scoring conformations were clustered using a 1.0-Å root-mean-square deviation (RMSD) cutoff over all atoms such that in a given cluster, the RMSD of the backbone atoms of any 2 structures differed by ≤1 Å. The resulting top-scoring clusters were analyzed for hydrogen bonds and vdW contacts using the ViewDock module from Chimera. A Klotho residue was deemed to be in vdW contact with the ligand if one of the atoms was within 0.4 Å of the sum of the vdW radii of the residue and ligand atoms. Hydrogen bonds were defined by the reference values given in Mills and Dean (45), but with the criteria relaxed by 0.4 Å and 20°.
Docking calibrations
Before using the above docking protocol to determine the complex structure of α2-3-sialyllactose bound to Klotho, we first assessed whether it could reproduce the known X-ray structures of KLrP bound to glucose (PDB entry 2e9l) as well as α2-3-sialyllactose bound to Trypanosoma cruzi trans-sialidase (PDB entry 1s0i). The conservation score for each residue was obtained from the Consurf server (46).
Docking of glucose to KLrP
The largest sphere cluster for the KLrP [PDB entry 2e9l (26)] consisted of 70 spheres. Docking glucose into this cluster generated 83,700 unique starting orientations, which were minimized and clustered using 1-Å backbone RMSD cutoff yielding 74 distinct docked conformations whose RMSDs differed by >1 Å. Among the docked conformations, the top-scoring one was closest to the native structure with a 2-Å RMSD from the glucose-bound KLrP X-ray structure (Supplemental Fig. S1A). Furthermore, all but one contact residue found in the 2e9l crystal structure were found in vdW contact with glucose in the top-docked structure (Table 1). This shows that the docking protocol could yield a nativelike structure of glucose bound to KLrP.
TABLE 1.
Residues in contact with glucose in X-ray structure (PDB entry 2e9l) compared to those in top-scoring docked configuration of glucose to KLrP
| X-ray contact residue | Docked structure #1 contact residue | Corresponding residue in KL1 and Consurf value |
|---|---|---|
| Q17 | Q17 | Q75 (9) |
| H120 | H120 | H193 (9) |
| F121 | F121 | W194 (8) |
| N164 | N164 | D238 (9) |
| E165 | E165 | N239 (9) |
| Y309 | Y309 | G372 (8) |
| W345 | W345 | — |
| E373 | E373 | E414 (9) |
| W417 | W417 | W458 (8) |
| E424 | E424 | E465 (9) |
| W425 | W425 | W466 (9) |
| F433 | — | R474 (9) |
Consurf values (in parentheses) calculated by the Consurf server (46).
Docking of α2-3-sialyllactose with PDB 1s0i
The above docking protocol was also used to dock α2-3-sialyllactose bound to T. cruzi trans-sialidase consisting of 632 residues (31). This yielded 103,352 orientations, which were minimized and sorted into 58 clusters at 1-Å backbone RMSD cutoff. As for docking of glucose to KLrP, the docking protocol could also yield a native-like structure of α2-3-sialyllactose-bound trans-sialidase; the top-scoring one was again close to the 1s0i X-ray structure with a RMSD of 2.2 Å (Supplemental Fig. S1B), but the fourth and fifth top-scoring conformations yielded a slightly smaller RMSD of 1.8 Å. Nine of the 13 residues found within hydrogen bonding or vdW contact with α2-3-sialyllactose in the 1s0i crystal structure were found in the top-docked structure (Table 2).
TABLE 2.
Comparison between residues in hydrogen-bonding or vdW contact with α2-3-sialyllactose in X-ray structure of 1s0i and top-ranked docked structure
| 1s0i X-ray structure | Top-ranked docked structure |
|---|---|
| Arg35** | Arg35** |
| Leu36 | — |
| Arg53* | — |
| Asp96* | Asp96 |
| Tyr119 | Tyr119 |
| Trp120 | Trp120 |
| Thr121 | — |
| Leu176 | — |
| Gln195 | |
| Ser229 | |
| Glu230* | |
| Arg245** | Arg245* |
| Asp247 | |
| Arg311 | |
| Trp312 | Trp312 |
| Arg314**** | Arg314*** |
| Tyr342 | Tyr342 |
| Glu362 | |
| Tyr364 | Tyr364 |
Each asterisk denotes a hydrogen bond with α2-3-sialyllactose.
Electrophysiological recordings of TRPC6 currents
TRPC6 currents were recorded from human embryonic kidney 293 (HEK293 cells expressing recombinant TRPC6 with or without ectodomain of Klotho proteins, as previously described (16, 22). For testing recombinant KL1, cells expressing TRPC6 were treated with or without KL1 (300 pM) in serum-containing medium overnight. Currents were recorded by voltage clamp in ruptured whole-cell mode with an Axopatch 200B patch-clamp amplifier and Pulse software (Axon Instruments, Foster City, CA, USA). Cells were held at 0 mV membrane potential and stimulated with repetitive ascending ramp pulse from –100 to +100 mV for 400 ms every 10 s. The pipette and bath solution for recording HEK293 cells contained (mM) 140 CsCl, 1 MgCl2 1.5 CaCl2, 2 ATP-Mg, 5 EGTA, 10 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.2 with CsOH) and 140 NaCl, 0.5 EGTA, 10 HEPES, 10 glucose, and 10 mannitol (pH 7.4 with NaOH), respectively. The resistance of electrodes containing pipette solution was 1.5 ∼3 MΩ. TRPC6 currents were activated by bath application of oleoyl-acetyl-glycerol (OAG; 100 μM), low-pass filtered at 2 kHz and sampled every 0.1 ms (10 kHz). At the end of experiments, the bath solution was replaced by a solution containing nonpermeant cation N-methyl-d-glucamine to assess TRPC6-mediated inward currents. Data acquisition and analysis were performed by the pClamp9.2 program (Axon Instruments) and GraphPad Prism 3.0 software (GraphPad Software, La Jolla, CA, USA).
Production and characterization of recombinant human KL1
Codon-optimized cDNA encoding amino acids of the alternatively spliced human KL1 (UniProtKB identifier Q9UEF7-2) and a C-terminal His-tag were used for production of recombinant KL1. Protein was expressed in HEK293 cells, and conditioned media containing the secreted protein with cleaved signal sequence (aa 1–33) was collected and purified by nickel column chromatography. Purified KL1 was kept in 20 mM 4-morpholineethanesulfonic acid, 20% glycerol, 500 mM NaCl, 1 mM ethylenediaminetetraacetic acid, pH 6, and stored in aliquots at −70°C until use. Native and denaturing SDS-PAGE confirmed expected molecular size, indicating a purity of >95% and a lack of higher-molecular-weight aggregates. Monomer predominance (>95%) was confirmed by size exclusion chromatography. Endotoxin determined by limulus amebocyte lysate assay was <0.00025 EU/μg.
Binding of KL1 domain to α2-3-sialyllactose
Binding was performed by biolayer interferometry performed using the Octet Red384 system with ForteBio Octet analysis software (ForteBio, Menlo Park, CA, USA) as previously described for sKlotho (22). Briefly, streptavidin sensors were uncoated in measurement buffer for 15 min, then loaded with 150 nM biotindiol, biotinylated lactose, or biotinylated α2-3-sialyllactose for 4 min, followed by incubation with fresh Klotho solution at 30°C for 400 s and dissociation for 500 s. Biotindiol served as reference. The data were processed by subtraction of the signal for biotindiol, alignment to association, interstep correction to dissociation, and Savitzky-Golay filtering to result in the given sensorgram. The respective values for kon, koff, and Kd were determined by analyzing association and dissociation in a 1:1 model, full local fitting.
Analysis of effect of KL1 domain on cardiac hypertrophy in mice
All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda, MD, USA) using protocols approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee. To induce cardiac hypertrophy, mice (129/S1SvImJ strain, male, ∼25 kg) were administrated isoproterenol (ISO; 3 mg/kg subcutaneously) daily for 10 d. Recombinant KL1 domain (10 μg/kg body weight) was administered via intraperitoneal injection to mice beginning at day 1. At the end of d 10, mice were humanely killed and hearts isolated. After measurement of heart weight, a portion of the hearts was snap-frozen in liquid N2 and saved for RNA isolation and analysis of brain natriuretic peptide (BNP) transcript by real-time PCR.
RESULTS
Common features of protein recognition of α2-3-sialyllactose
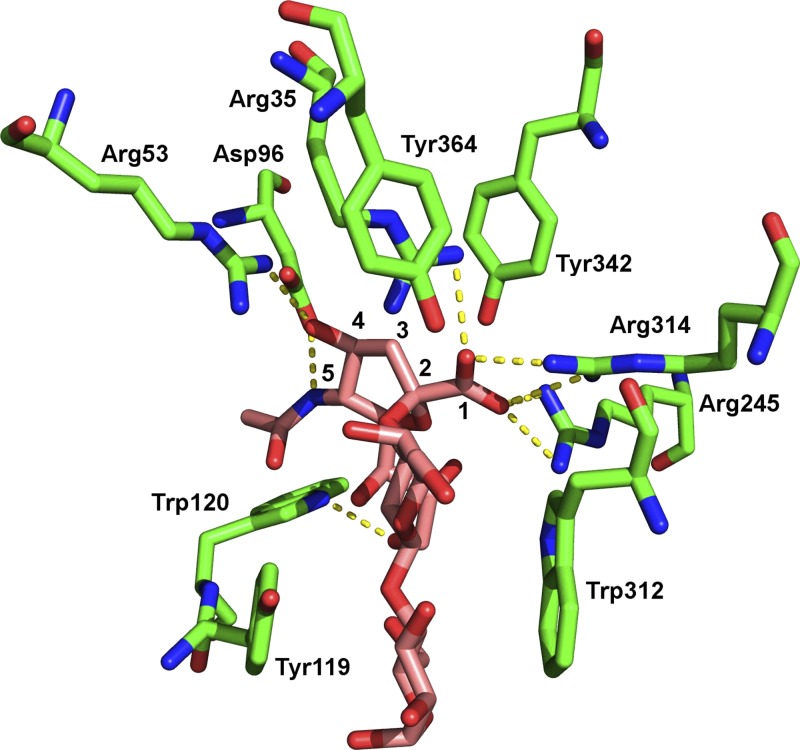
To help determine how α2-3-sialyllactose would bind to sKlotho, all X-ray/nuclear magnetic resonance structures containing α2-3-sialyllactose (ligand ID SLT) bound to proteins were retrieved from the PDB (24). This yielded 6 protein structures containing α2-3-sialyllactose, but one of them did not possess any interactions from the sialic group and so was discarded. The remaining 5 protein structures—T. cruzi trans-sialidase (PDB id: 1s0i), parainfluenza virus 5 hemagglutinin–neuraminidase (1z4x), Streptococcus pneumoniae NanA sialidase (4c1w), chicken galectin-8 N-terminal domain (4wvw), and S. pneumoniae NanC sialidase (4yz5)—showed the following common features shared by all the proteins in binding α2-3-sialyllactose: 2 or more hydrogen bonds (salt bridges) from the sialic acid carboxylate (carbon atom #1 in Fig. 1) oxygen atoms to Arg (Table 3 and Fig. 1). In addition, for 3 proteins (1s0i, 4c1w, and 4yz5), the sialic acid amide N (attached to carbon atom #5) and the adjacent hydroxyl O (attached to carbon #4) each formed a hydrogen bond to an acidic Asp/Glu residue (Table 3). Hence, the PDB structures show that ≥1 Arg and an acidic residue (Asp/Glu) involved in binding the sialic acid moiety of α2-3-sialyllactose.
Figure 1.
Protein recognition of α2-3-sialyllactose. Side chains of several amino acids of Trypanosoma cruzi trans-sialidase (PDB 1s0i) are shown in complex with α2-3-sialyllactose. Backbone carbon atoms of trans-sialidase and α2-3-sialyllactose are in green and purple, respectively. Nitrogen atoms are in blue, oxygen atoms in red. Carbon atoms 1 through 5 of sialic acid are numbered; Scheme 2 provides complete carbon numbering for 1 through 9. Hydrogen bonds are shown by yellow dashed lines.
TABLE 3.
Hydrogen bonds made by sialic acid in protein structures containing α2-3-sialyllactose with protein–sialic acid interactions
| PDB code | Resoln (Å) | Protein name | 2-3-sialic acid atoms | |||
|---|---|---|---|---|---|---|
| O(COO–) | O(COO–) | OH | NH | |||
| 1s0i | 1.60 | Trypanosoma cruzi trans-sialidase | Arg35 | Arg245 | Arg53 | Asp96 |
| Arg314 | Arg314 | Asp96 | ||||
| 1z4x | 2.50 | Parainfluenza hemagglutinin-neuraminidase | Arg163 | Arg405 | ||
| Arg495 | Arg495 | |||||
| 4c1w | 1.84 | Streptococcus pneumoniae NanA neuraminidase | Arg197 | Arg274 | Glu195 | Glu195 |
| Arg274 | ||||||
| 4wvw | 1.47 | Galectin-8 N-terminal domain | Arg58 | Gln46 | ||
| Trp85 | Arg58 | |||||
| 4yz5 | 2.27 | S. pneumoniae NanC neuraminidase | Arg237 | Arg161 | Glu159 | Glu159 |
| Arg237 | ||||||
Model structure of KL1
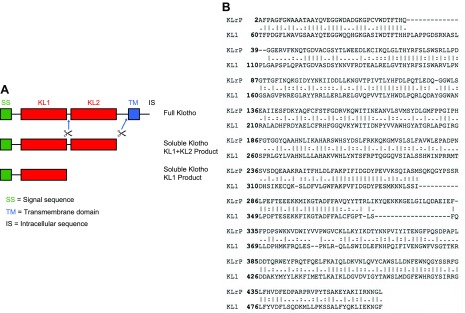
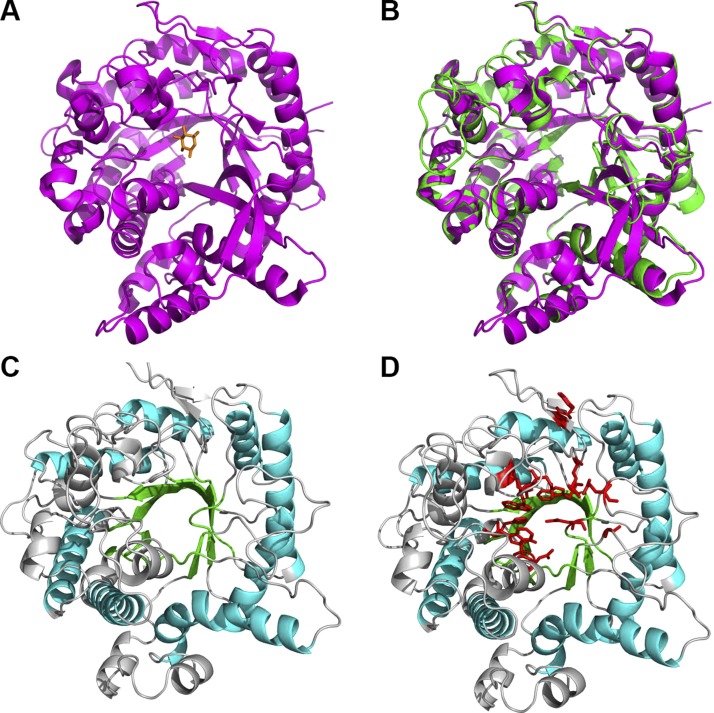
Next, we searched the PDB for structures homologous to sKlotho and found that the KLrP shares the highest sequence identity (∼40%) with the KL1 domain of Klotho ectodomain, whose sequence alignment with KLrP is shown in Fig. 2. KLrP is a cytosolic glycosyl hydrolase that hydrolyzes the glycosidic bond of its natural substrate β-glucosylceramide (26). On the basis of the 1.6-Å structure of glucose-bound KLrP (PDB entry 2e9l) shown in Fig. 3A, a model structure of the Klotho KL1 domain was generated using homology modeling. This KL1 model structure closely resembled the KLrP structure with a root-mean-square deviation (RMSD) of 0.6 Å when the backbone atoms were superimposed (Fig. 3B). The modeled KL1 domain is tightly packed, with a central core formed by 8 parallel β strands that are stabilized by 8 external α helices (Fig. 3C). Although the parallel β strands appear to define a cavity, the side chains of several hydrophobic residues extend inward to form a densely packed core. The sugar-binding site, delineated by those KL1 residues that are sequence aligned with the sugar-binding KLrP residues, is in fact outside this central core, as in the case of KLrP (Fig. 3D). This sugar-binding site enables the docking of α2-3-sialyllactose to the KL1 model structure.
Figure 2.
A) Domain structure of Klotho. Full-length Klotho is type 1 membrane protein with large ectodomain containing 2 related domains named KL1 and KL2. entire ectodomain can be cleaved and functions as extracellular endocrine or paracrine hormone. KL1-KL2 ectodomain domain may be further cleaved to become 2 separate KL1 and KL2 domains. Separate KL2 domain is not shown. B) Amino acid alignment of KLrP and KL1 domain of sKlotho illustrating homology.
Figure 3.
A) Structure of glucose-bound KLrP. B) Model of KL1 domain overlaid with glucose-bound KLrP showing modeled KL1 domain is structurally similar to KLrP domain. C) Model structure of sKlotho KL1 domain showing central parallel core of β strands (green) stabilized by α helixes (cyan); other β sheets and α helices and loops in light gray. D) Model structure of KL1 domain with residues in KLrP that bind to glucose mapped to corresponding residues in Klotho KL1 domain (red).
Model structure of α2-3-sialyllactose bound to KL1
The 5 structures containing α2-3-sialyllactose in Table 3 could not be used as templates to generate a α2-3-sialyllactose/KL1 complex structure using homology modeling, as the proteins in these structures have low sequence homology with KL1 or KL2 (Supplemental Table S1). These 5 proteins also exhibit insignificant sequence identity with each other (Supplemental Table S1); nevertheless, they share common features in binding α2-3-sialyllactose (Table 3 and Fig. 1). Hence, we docked α2-3-sialyllactose to the modeled sKlotho KL1 domain structure, but before this procedure, we validated the docking protocol used by showing that it could reproduce a near-native structure of glucose bound to KLrP (PDB 2e9l) as well as α2-3-sialyllactose bound to T. cruzi trans-sialidase (PDB 1s0i). For both validation cases, the RMSD of the ligand atoms in the top-ranked docked conformation from those in the respective X-ray structure was ∼2 Å, and most of the protein-ligand vdW contacts in the crystal structure were preserved.
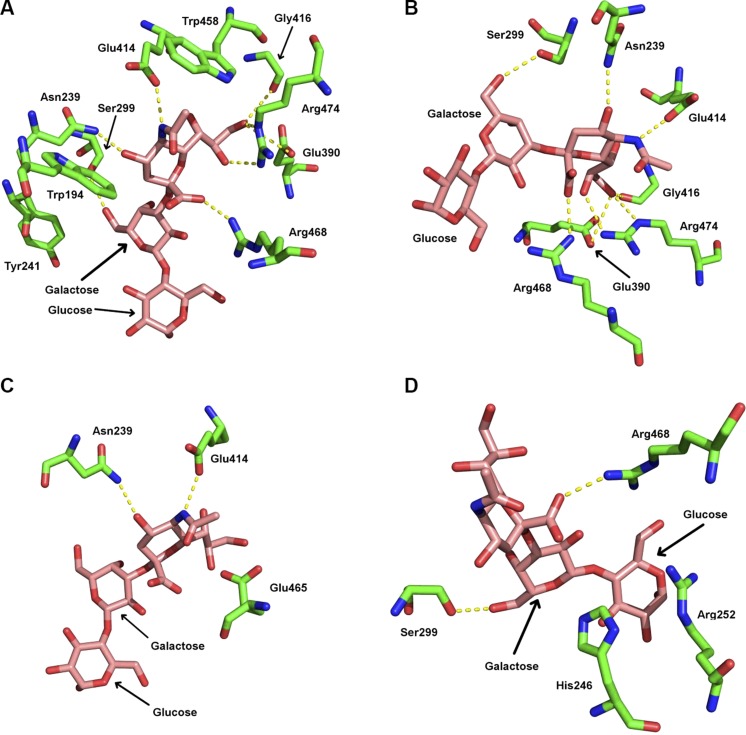
The docking protocol was then used to generate 236,229 initial orientations of α2-3-sialyllactose bound to the Klotho KL1 domain. After minimization, many of these initial orientations converged to the same structure. The resulting 139 configurations were clustered using a 1.0 Å backbone RMSD cutoff into 81 distinct clusters. The top-scoring 5 clusters were analyzed for residues in contact with α2-3-sialyllactose. These residues are listed in Table 4 along with their Consurf conservation scores (46), where 1 indicates a variable residue and 9 a highly conserved residue. In the #1 docked orientation (Table 4, orientation 1), the α2-3-sialyllactose deviated from that in the 1s0i X-ray structure by a RMSD of 1.4 Å, and its binding pocket is similar that in the 1s0i structure: both binding sites contain multiple arginine and acidic residues as well as tyrosine and tryptophan residues, which are also seen in the X-ray structures of other α2-3-sialyllactose-bound proteins (Figs. 1 and 4A). Thus, the #1 docked orientation contains many elements of binding seen in the X-ray structures of α2-3-sialyllactose-bound proteins.
TABLE 4.
KL1 residues that are in contact with α2-3-sialyllactose for best-scoring 5 clusters
| KL1 residues that bind α2-3-sialyllactose in model complex |
Consurf value | KLrP residue | ||||
|---|---|---|---|---|---|---|
| Orientation 1 | Orientation 2 | Orientation 3 | Orientation 4 | Orientation 5 | ||
| Trp194 | Trp194 | Trp194 | Trp194 | Trp194 | 8 | Phe121 |
| Asn239 | Asn239 | Asn239 | Asn239 | Asn239 | 9 | Glu165 |
| Tyr241 | Tyr241 | Tyr241 | Tyr241 | 1 | ||
| Val242 | Val242 | Val242 | Val242 | Val242 | 4 | |
| His246 | His246 | His246 | 1 | |||
| Thr250 | 1 | |||||
| Arg252 | Arg252 | Arg252 | 1 | |||
| Ser299 | Ser299 | Ser299 | Ser299 | Ser299 | 5 | |
| His301 | His301 | 5 | ||||
| Val323 | Val323 | Val323 | Val323 | Val323 | 1 | |
| Gly372 | Gly372 | Gly372 | Gly372 | Gly372 | 8 | Tyr309 |
| Gln378 | Gln378 | Gln378 | 1 | |||
| Leu389 | Leu389 | Leu389 | Leu389 | 1 | ||
| Glu390 | Glu390 | Glu390 | Glu390 | Glu390 | 7 | |
| Glu414 | Glu414 | Glu414 | Glu414 | Glu414 | 9 | Glu373 |
| Gly416 | Gly416 | Gly416 | Gly416 | Gly416 | 9 | |
| Trp458 | Trp458 | Trp458 | Trp458 | Trp458 | 8 | Trp417 |
| Glu465 | Glu465 | Glu465 | Glu465 | Glu465 | 9 | Glu424 |
| Trp466 | 9 | Trp425 | ||||
| Arg468 | Arg468 | Arg468 | Arg468 | Arg468 | 1 | |
| Arg474 | Arg474 | Arg474 | Arg474 | Arg474 | 9 | Phe433 |
Consurf values calculated by the Consurf server (46). KLrP residues were found to bind glucose in the 2e9l X-ray structure.
Figure 4.
α2-3-Sialyllactose bound to modeled sKlotho KL1 domain. A) Binding site contains multiple charged and hydrophobic residues seen in X-ray structures of α2-3-sialyllactose-bound proteins in Fig. 1. B) Hydrogen bonds (dashed lines) formed by α2-3-sialyllactose with KL1 domain from #1 docked conformation. C) Hydrogen bonds formed by highly conserved KL1 residues with α2-3-sialyllactose. D) Hydrogen bonds formed by variable KL1 residues with α2-3-sialyllactose.
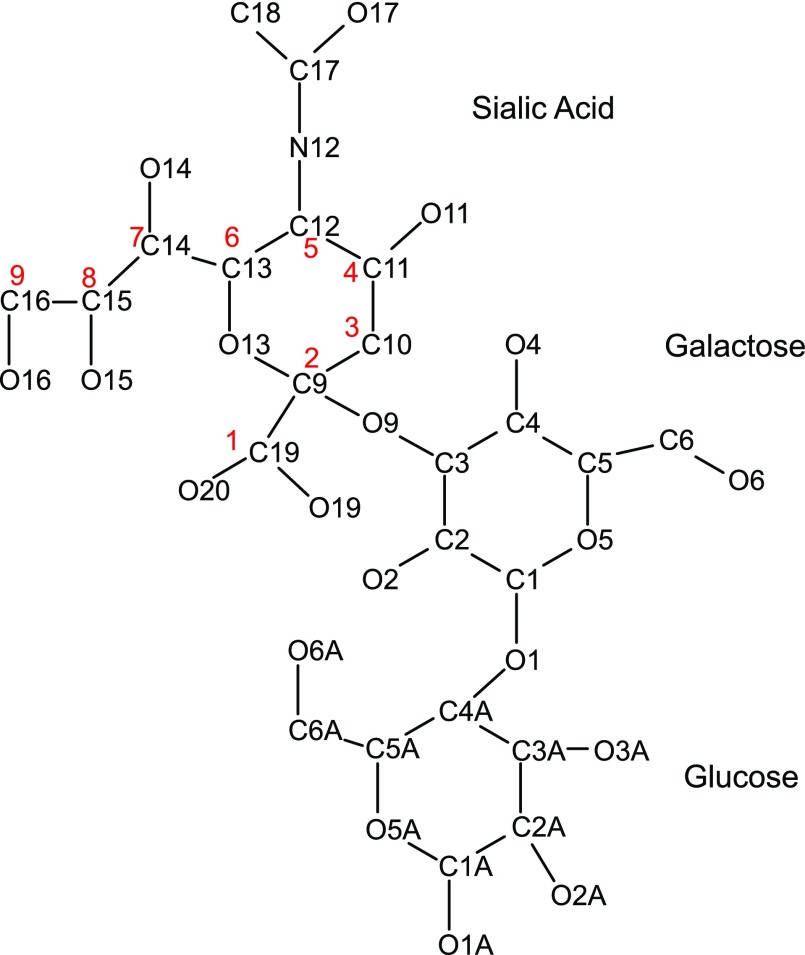
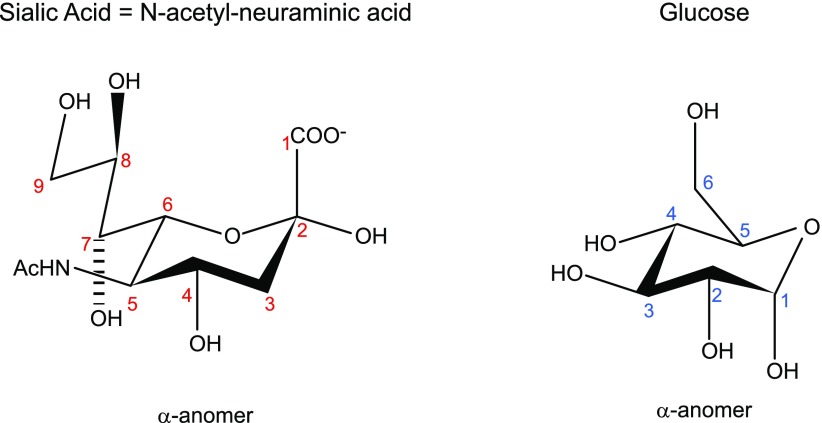
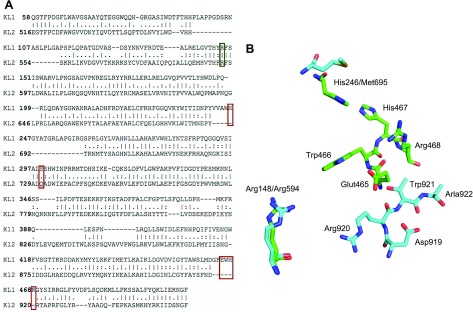
For the modeled KL1 domain, the α2-3-sialyllactose forms hydrogen bonds with 7 KL1 residues: the sialic acid carboxylate O19/O20 and amide N12 form hydrogen bonds with Arg468 (Fig. 4A, B, D) and Glu414 side chains (not shown), respectively. Its hydroxyl O16H group donates a hydrogen bond to the Gly416 carbonyl O and/or the Glu390 carboxylate oxygen as well as accepts a hydrogen bond from the Arg474 imino group, whereas its hydroxyl O11 and O15 atoms accept hydrogen bonds from the Asn239 amino and Arg474 imino groups, respectively. In addition, the galactose hydroxyl O6 group forms a hydrogen bond with the Ser299 side chain. The galactose also forms vdW contacts with Tyr241, whereas the glucose makes vdW contacts with the Leu389, Thr250, Arg252, and Arg468. Scheme 1 provides the numbering used for α2-3-sialyllactose.
Scheme 1.
PDB numbering scheme used for 2-3-sialyllactose. Corresponding carbon atom numbering 1–9 of sialic acid shown in Fig. 1 and scheme are in red.
The #1 docked orientation also contains nearly all the equivalent residues from KLrP that are involved in binding glucose in the X-ray structure of glucose-bound KLrP (PDB 2e9l). In particular, the charged residues that bind glucose in KLrP (Glu165, Glu373, and Glu424) are also found to bind α2-3-sialyllactose in KL1—Asn239, Glu414, and Glu465 (Fig. 4C and Table 4). These residues are highly conserved (Consurf score of 9) because Glu165 and Glu373 of KLrP serve as the acid–base catalyst and nucleophile, respectively, in the KLrP-catalyzed hydrolysis of the glucose–galactose glycosidic bond (26).
Given that sialic acid (9-carbon sugar acid) differs from glucose (6-carbon sugar), we focused on those variable residues (Consurf score of ≤5) interacting with sialic acid groups that are absent in glucose, notably the extra carboxylate group (carbon #1), acetamide (attached to carbon #5), and CH(OH)CH2(OH) (carbons #7–9) (Scheme 2). Among residues that interact with these sialic acid groups, only Arg468, which forms a salt bridge with the sialic acid carboxylate, is variable (Consurf score of 1), whereas Glu414 and Arg474, which interact with the sialic acid acetamide and CH(OH)CH2(OH), respectively, are highly conserved (Consurf score of 9). Given the ligand for sKlotho is a trisaccharide, we also considered those variable residues (Consurf score of ≤5) that interact with glucose and galactose in addition to sialic acid: His246, Arg252, and Ser299 (Fig. 4D). These variable residues, as well as some of the conserved residues in the sugar-binding site (Trp194, Trp458, Glu465, Trp466), were then experimentally verified using site-directed mutagenesis.
Scheme 2.
Chemical structure of sialic acid vs. glucose.
Experimental verification of α2-3-sialyllactose binding sKlotho residues
Coexpression of the plasmid encoding sKlotho inhibits diacylglycerol (DAG)-stimulated TRPC6 channel function by ∼50% compared to the control vector (Fig. 5A) (16, 22). DAG increases TRPC6 channel activity by activation of channel gating as well as by stimulation of TRPC6 exocytosis (16, 22). sKlotho inhibits DAG-activated TRPC6 channel activity partially because it inhibits DAG stimulation of channel exocytosis but does not affect channel gating. We used the regulation of TRPC6 by sKlotho as an experimental system to test the functional significance of the amino acid residues implicated in binding α2-3-sialyllactose.
Figure 5.
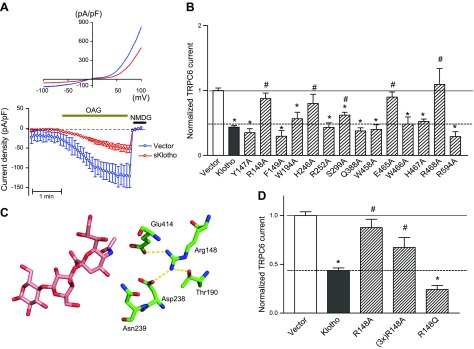
A) sKlotho partially inhibits TRPC6 channel. Cultured HEK cells were cotransfected with plasmid encoding for TRPC6 and with or without sKlotho. Whole-cell recording of OAG-stimulated TRPC6 currents was performed 48 h later. OAG is membrane-permeant DAG analog. Top, current–voltage relationship curve of TRPC6 current revealing characteristic doubly-rectifying current. Bottom, time course of inward TRPC6 current density (current at −100 mV divided by capacity). At end of recording, N-methyl-d-glucamine was added to inhibit inward current in order to verify TRPC6-specific current. B) Comparison of activity of mutated sKlotho proteins compared to wild-type sKlotho activity. HEK cells were cotransfected with plasmid encoding for TRPC6 and for wild-type (Klotho) or mutant sKlotho (labeled for amino acid mutated, such as Y149A) or control empty vector (vector). Current density for vector transfected is 1. Current density for wild-type and mutant sKlotho is normalized to vector group; n = 6–15 each. *P < 0.01 vs. vector; #P < 0.05 vs. wild-type sKlotho. C) Illustration of hydrogen-bonding network formed by Arg148, showing why mutation of Arg148 can cause loss of binding. D) Effect of 3-fold excess of R148A mutant and R148Q mutant on TRPC6. Experimental paradigm as in A and B.
We systemically mutated the predicted α2-3-sialyllactose-binding residues (Trp194, His246, Arg252, Ser299, Trp458, Glu465, Trp466, and Arg468) as well as control residues outside the predicted binding site (Tyr147, Arg148, Phe149, Gln388, His467, and Arg594) to Ala and compared the effect of wild-type vs. mutant sKlotho on TRPC6 channel function. As shown in Fig. 5B, alanine substitution of Arg148 (R148A), His246 (H246A), Ser299 (S299A), Glu465 (E465A), and Arg468 (R468A) each resulted in a significant loss of inhibition of TRPC6 channel function relative to the wild-type sKlotho (normalized TRPC6 current; i.e., the current density normalized to vector group; P < 0.05 vs. wild-type sKlotho). The results supported the notion that His246, Ser 299, Glu465, and Arg468 among the predicted α2-3-sialyllactose-binding residues are important. Notably, among the predicted binding residues, His246, Ser 299, and Arg468 are the most variable compared to KLrP, with a Consurf value of 1 (Table 4). In contrast, sKlotho mutants carrying alanine substitution of the residues not expected to contact the α2-3-sialyllactose in the modeled complex retained the ability to inhibit TRPC6 as well as wild-type sKlotho with the exception of the R148A mutant.
The experimental result of the R148A mutant may be rationalized by the elaborate network of hydrogen bonds formed by the Arg148 side chain with Thr190, Asp238, and Glu414 (Fig. 5C). One of the Arg148 amino groups forms a salt bridge with the Glu414 carboxylate in the model complex. The other Arg148 amino group forms hydrogen bonds with both the Thr190 and Asp238 side chains. Loss of this elaborate network of hydrogen bonds to Thr190, Asp238, and Glu414 upon mutation of Arg148 to Ala could lead to protein conformational changes resulting in a loss of α2-3-sialyllactose binding—thus the observed loss of sKlotho function. To test this notion, we mutated Arg148 to glutamine, which potentially can form hydrogen bonding interactions in place of Arg. Additionally, we transfected cells with 3-fold excess of R148A plasmid DNA to test whether the observed loss of function of R148A mutant may be due to a reduced level of protein expression. As shown in Fig. 5D, transfection of R148A mutant sKlotho DNA at 3-fold excess failed to cause inhibition of TRPC6, as observed for wild-type sKlotho. In support of an Arg148 role in forming hydrogen bonds, the R148Q mutant functioned nearly as well as wild-type sKlotho. Overall, our results are consistent with the notion that, while Arg148 is not in direct contact with α2-3-sialyllactose in the model complex, loss of the hydrogen-bonding network mediated by Arg148 may impact structure important for α2-3-sialyllactose recognition.
KL1 domain exhibits the key binding and biologic activities of sKlotho
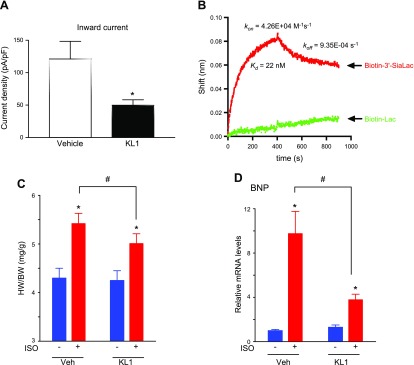
To support the notion that these residues in KL1 domain are important for binding α2-3-sialyllactose, we next examined the ability of recombinant KL1 domain to regulate TRPC6 channel activity. As shown, relative to vehicle, extracellular application of purified recombinant KL1 caused ∼60% inhibition of TRPC6 current (Fig. 6A). Direct binding assay using biolayer interferometry showed that KL1 domain binds α2-3-sialyllactose immobilized in a biosensor with Kd ∼22 nM (Fig. 6B). This Kd value is similar to the ∼10 nM reported for sKlotho (22), and therefore, like sKlotho, KL1 is capable of binding α2-3-sialyllactose-containing gangliosides. Binding to sialogangliosides and down-regulation of PI3K-dependent TRPC6 channel function in lipid rafts is believed to be the underlying mechanism for cardioprotection by sKlotho (16, 22). Hence, we tested the effect of KL1 in cardioprotection in mice using the ISO-induced cardiac hypertrophy model. As shown, ISO induced cardiac hypertrophy in mice, as was evident by an increase in the heart weight/body weight ratio (Fig. 6C) and in BNP gene expression (Fig. 6D). Administration of KL1 partially ameliorated ISO-induced cardiac hypertrophy. These findings are similar to results from previous studies with sKlotho (22) and thus indicate that like sKlotho, KL1 can mitigate pathologic cardiac remodeling and protect against stress-induced cardiac hypertrophy in mice.
Figure 6.
A) Whole-cell recording of OAG-stimulated TRPC6 currents expressed in HEK cells with KL1 domain (300 nM) or vehicle. Experimental paradigm is as Fig. 5A, B. Means ± sem of multiple recordings are shown (n = 6 each). *P < 0.01 KL1 vs. vehicle. B) Binding of KL1 domain to α2-3-sialyllactose analyzed by biolayer interferometry. Spectral shifts of reflected light due to changes in biosensor thickness (Δλ; nm) were measured after 170 nM KL1 domain binding to streptavidin sensors loaded with 150 nM biotin-labeled α2-3-sialyllactose or lactose against biotindiol reference. Similar results were observed in 2 experiments. C, D) Heart weight/body weight ratio (C) and BNP gene expression (D) of mice with or without ISO to induce cardiac hypertrophy, and after intraperitoneal administration of KL1 or vehicle. *P < 0.01 +ISO vs. −ISO; #P < 0.05 between indicated groups.
DISCUSSION
The mechanism of action for pleiotropic effects of sKlotho is poorly understood. Recently, we reported that sKlotho binds α2-3-sialyllactose of gangliosides in lipid rafts to affect multiple raft-dependent cellular effects (22). Here, we provide a mechanism for understanding how sKlotho binds lipid rafts and regulates their functions. By combining theory with experiment, we have delineated the critical residues in sKlotho involved in α2-3-sialyllactose recognition. We investigated the structural basis of sKlotho for recognition of α2-3-sialyllactose by first searching the PDB for homologous structures and identified glucose-bound KLrP to be most homologous to the KL1 domain. It is important to note that KLrP binds glucose whereas sKlotho binds α2-3-sialyllactose. Docking experiments using the KL1 model structure identified > 10 potential amino acids for binding α2-3-sialyllactose. Functional experiments revealed that, among the potential binding residues, Arg148, His246, and the 465EWHR468 motif are essential for binding α2-3-sialyllactose by sKlotho. Interestingly, these residues are not found in their entirety in the KLrP. The equivalent residues in the KLrP are Arg75, Met172, and the 424EWNQ427 (Fig. 2B), giving a plausible reason why KLrP and sKlotho bind different ligands: glucose and α2-3-sialyllactose, respectively.
sKlotho has 2 large domains, KL1 and KL2. Our results indicate that the KL1 domain is important for binding α2-3-sialyllactose. Computational and functional studies show that the KL1 domain contains all essential amino acid residues important for binding α2-3-sialyllactose, and this is supported by biolayer interferometry studies using recombinant human KL1. Furthermore, these results suggest that the KL2 domain cannot compensate for loss of binding activity of the KL1 domain in the context of sKlotho: mutations of the predicted residues in the KL1 domain result in loss of binding to α2-3-sialyllactose, as evidenced by loss of TRPC6 regulation. The role of the KL2 domain remains unclear. Our results indicate the likelihood that the KL2 domain alone (independently of KL1) cannot bind α2-3-sialyllactose in the same way the KL1 domain does and regulate TRPC6 channel function. This is probably because the KL2 domain lacks residues that are equivalent to His246, Ser299, and the 465EWHR468 motif when the KL1 and KL2 domains are both sequence and structurally aligned (Fig. 7A, B). Interestingly, although the KL1 Arg148 is conserved with Arg594 being the equivalent residue in the KL2 domain, its hydrogen-bonding partners, Thr190, Asp238, and Glu414 (Fig. 5C), are not conserved, as the equivalent KL2 residues are Ala615, Asn688, and Ser872, respectively (Fig. 7A). sKlotho is reported to form dimers (6) and KL2 domain may play a role in dimer formation, enhancing the in vivo function of KL1. Future studies will address the role of the KL2 domain in the binding and regulation of raft-dependent function.
Figure 7.
A) Amino acid alignment of KL1 and KL2 domains of sKlotho. Arg148 of KL1 is conserved in KL2 domain (green box). His246, Ser299, and motif 465GluTrpHisArg468 of KL1 domain are not conserved in KL2 (red box). B) Structural differences between KL1 and KL2 domain of sKlotho for residues implicated in binding to α2-3-sialyllactose (Arg148, His246, and motif 465GluTrpHisArg468). KL1 residues are green. Closest matching residues from KL2 domain (turquoise) for same KL1 residues show considerable local differences in structure, giving plausible reason why KL2 domain may be unable to bind α2-3-sialyllactose.
Our findings have important implications for the role of sKlotho in physiology and disease. Chronic kidney disease is a major public health problem affecting ∼13% of the population worldwide (47). The main cause of death for chronic kidney disease patients is cardiac morbidity. sKlotho is mainly produced in the kidney (1). Recent studies have shown that reduced blood levels of sKlotho in patients with chronic kidney disease are an important risk factor of cardiac hypertrophy (48). sKlotho down-regulates TRPC6 channel function by inhibiting PI3K-dependent exocytosis of TRPC6 channels. Here, we show that sKlotho and KL1 exert cardioprotection by targeting α2-3-sialyllactose-containing gangliosides in lipid rafts to inhibit TRPC6-mediated calcium entry and calcium signaling in cardiomyocytes (16, 22). Our results provide insights into how sKlotho binds α2-3-sialyllactose and regulate lipid raft function, and support the notion that ganglioside-enriched lipid rafts are receptors for sKlotho. Thus, sKlotho targets sialyllactose in glycoproteins and glycolipids (21, 22). Targeting sialic acids may be a general mechanism for the pleiotropic actions of sKlotho. Finally, there has been considerable interest in the potential applications of sKlotho and analogs for therapeutics in clinical medicine. KL1, which is about half the molecular size of sKlotho protein, may be a better molecule for production and/or for therapeutic application. Additional structure function analysis based on the modeled structure of Klotho-α2-3-sialyllactose interaction may provide further insight into the feasibility of designing mimetic structures of the α2-3-sialyllactose binding site and providing guidance for the design of simpler Klotho therapeutics.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported, in part, by the Academia Sinica and Ministry of Science and Technology (MOST) of Taiwan (Grant 98-2113-M-001-011), the U.S. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (Grants DK109887 and DK100605), the Welch Foundation (Grant I-1686), and by a sponsored research agreement between University of Texax Southwestern and Boehringer Ingelheim Pharmaceuticals Inc. (BIPI). BIPI provided purified recombinant KL1 domain protein. N.N. is supported by a postdoctoral fellowship from the German Academic Exchange Service (DAAD). The authors thank M. Diamond and J. Vaquer-Alicea (both from the University of Texas Southwestern Medical Center, Dallas, TX, USA) for provision of the Octet.
Glossary
- BNP
brain natriuretic peptide
- DAG
diacylglycerol
- FGF
fibroblast growth factor
- HEK
human embryonic kidney
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- ISO
isoproterenol
- KLrP
Klotho-related protein
- OAG
oleoyl-acetyl-glycerol
- PDB
Protein Data Bank
- RMSD
root-mean-square deviation
- sKlotho
soluble Klotho
- TRPC6
transient receptor potential canonical type isoform 6
- vdW
van der Waals
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
J. D. Wright, S.-W. An, J. Xie, J. Yoon, and N. Nischan designed the experiment, performed research, and analyzed data; J. J. Kohler, C. Lim, and C.-L. Huang analyzed data and supervised research; N. Oliver contributed critical reagents, designed the experiment, and analyzed data; and all authors participated in writing the article and approved the final article.
REFERENCES
- 1.Kuro-o M., Matsumura Y., Aizawa H., Kawaguchi H., Suga T., Utsugi T., Ohyama Y., Kurabayashi M., Kaname T., Kume E., Iwasaki H., Iida A., Shiraki-Iida T., Nishikawa S., Nagai R., Nabeshima Y. I. (1997) Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390, 45–51 [DOI] [PubMed] [Google Scholar]
- 2.Kurosu H., Yamamoto M., Clark J. D., Pastor J. V., Nandi A., Gurnani P., McGuinness O. P., Chikuda H., Yamaguchi M., Kawaguchi H., Shimomura I., Takayama Y., Herz J., Kahn C. R., Rosenblatt K. P., Kuro-o M. (2005) Suppression of aging in mice by the hormone Klotho. Science 309, 1829–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arking D. E., Krebsova A., Macek M. Sr., Macek M. Jr., Arking A., Mian I. S., Fried L., Hamosh A., Dey S., McIntosh I., Dietz H. C. (2002) Association of human aging with a functional variant of klotho. Proc. Natl. Acad. Sci. USA 99, 856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arking D. E., Atzmon G., Arking A., Barzilai N., Dietz H. C. (2005) Association between a functional variant of the KLOTHO gene and high-density lipoprotein cholesterol, blood pressure, stroke, and longevity. Circ. Res. 96, 412–418 [DOI] [PubMed] [Google Scholar]
- 5.Ogata N., Matsumura Y., Shiraki M., Kawano K., Koshizuka Y., Hosoi T., Nakamura K., Kuro-O M., Kawaguchi H. (2002) Association of klotho gene polymorphism with bone density and spondylosis of the lumbar spine in postmenopausal women. Bone 31, 37–42 [DOI] [PubMed] [Google Scholar]
- 6.Tucker Zhou T. B., King G. D., Chen C., Abraham C. R. (2013) Biochemical and functional characterization of the klotho-VS polymorphism implicated in aging and disease risk. J. Biol. Chem. 288, 36302–36311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubal D. B., Yokoyama J. S., Zhu L., Broestl L., Worden K., Wang D., Sturm V. E., Kim D., Klein E., Yu G.-Q., Ho K., Eilertson K. E., Yu L., Kuro-o M., De Jager P. L., Coppola G., Small G. W., Bennett D. A., Kramer J. H., Abraham C. R., Miller B. L., Mucke L. (2014) Life extension factor klotho enhances cognition. Cell Rep. 7, 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurosu H., Ogawa Y., Miyoshi M., Yamamoto M., Nandi A., Rosenblatt K. P., Baum M. G., Schiavi S., Hu M.-C., Moe O. W., Kuro-o M. (2006) Regulation of fibroblast growth factor-23 signaling by klotho. J. Biol. Chem. 281, 6120–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urakawa I., Yamazaki Y., Shimada T., Iijima K., Hasegawa H., Okawa K., Fujita T., Fukumoto S., Yamashita T. (2006) Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 444, 770–774 17086194 [Google Scholar]
- 10.White K. E., Evans W. E., O’Riordan J. L. H., Speer M. C., Econs M. J., Lorenz-Depiereux B., Grabowski M., Meitinger T., Strom T. M.; ADHR Consortium (2000) Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat. Genet. 26, 345–348 [DOI] [PubMed] [Google Scholar]
- 11.Morishita K., Shirai A., Kubota M., Katakura Y., Nabeshima Y., Takeshige K., Kamiya T. (2001) The progression of aging in klotho mutant mice can be modified by dietary phosphorus and zinc. J. Nutr. 131, 3182–3188 [DOI] [PubMed] [Google Scholar]
- 12.Ito S., Fujimori T., Hayashizaki Y., Nabeshima Y. (2002) Identification of a novel mouse membrane-bound family 1 glycosidase-like protein, which carries an atypical active site structure. Biochim. Biophys. Acta 1576, 341–345 [DOI] [PubMed] [Google Scholar]
- 13.Imura A., Iwano A., Tohyama O., Tsuji Y., Nozaki K., Hashimoto N., Fujimori T., Nabeshima Y. (2004) Secreted Klotho protein in sera and CSF: implication for post-translational cleavage in release of Klotho protein from cell membrane. FEBS Lett. 565, 143–147 [DOI] [PubMed] [Google Scholar]
- 14.Liu H., Fergusson M. M., Castilho R. M., Liu J., Cao L., Chen J., Malide D., Rovira I. I., Schimel D., Kuo C. J., Gutkind J. S., Hwang P. M., Finkel T. (2007) Augmented Wnt signaling in a mammalian model of accelerated aging. Science 317, 803–806 [DOI] [PubMed] [Google Scholar]
- 15.Liu F., Wu S., Ren H., Gu J. (2011) Klotho suppresses RIG-I-mediated senescence-associated inflammation. Nat. Cell Biol. 13, 254–262 [DOI] [PubMed] [Google Scholar]
- 16.Xie J., Cha S.-K., An S.-W., Kuro-O M., Birnbaumer L., Huang C.-L. (2012) Cardioprotection by Klotho through downregulation of TRPC6 channels in the mouse heart. Nat. Commun. 3, 1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf I., Levanon-Cohen S., Bose S., Ligumsky H., Sredni B., Kanety H., Kuro-o M., Karlan B., Kaufman B., Koeffler H. P., Rubinek T. (2008) Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene 27, 7094–7105 [DOI] [PubMed] [Google Scholar]
- 18.Doi S., Zou Y., Togao O., Pastor J. V., John G. B., Wang L., Shiizaki K., Gotschall R., Schiavi S., Yorioka N., Takahashi M., Boothman D. A., Kuro-o M. (2011) Klotho inhibits transforming growth factor-β1 (TGF-β1) signaling and suppresses renal fibrosis and cancer metastasis in mice. J. Biol. Chem. 286, 8655–8665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubal D. B., Zhu L., Sanchez P. E., Worden K., Broestl L., Johnson E., Ho K., Yu G.-Q., Kim D., Betourne A., Kuro-O M., Masliah E., Abraham C. R., Mucke L. (2015) Life extension factor klotho prevents mortality and enhances cognition in hAPP transgenic mice. J. Neurosci. 35, 2358–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tohyama O., Imura A., Iwano A., Freund J.-N., Henrissat B., Fujimori T., Nabeshima Y. (2004) Klotho is a novel β-glucuronidase capable of hydrolyzing steroid β-glucuronides. J. Biol. Chem. 279, 9777–9784 [DOI] [PubMed] [Google Scholar]
- 21.Cha S.-K., Ortega B., Kurosu H., Rosenblatt K. P., Kuro-O M., Huang C.-L. (2008) Removal of sialic acid involving Klotho causes cell-surface retention of TRPV5 channel via binding to galectin-1. Proc. Natl. Acad. Sci. USA 105, 9805–9810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalton G., An S.-W., Al-Juboori S. I., Nischan N., Yoon J., Dobrinskikh E., Hilgemann D. W., Xie J., Luby-Phelps K., Kohler J. J., Birnbaumer L., Huang C.-L. (2017) Soluble klotho binds monosialoganglioside to regulate membrane microdomains and growth factor signaling. Proc. Natl. Acad. Sci. USA 4, 752–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simons K., Sampaio J. L. (2011) Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3, a004697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman H. M., Battistuz T., Bhat T. N., Bluhm W. F., Bourne P. E., Burkhardt K., Feng Z., Gilliland G. L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., Thanki N., Weissig H., Westbrook J. D., Zardecki C. (2002) The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 58, 899–907 [DOI] [PubMed] [Google Scholar]
- 25.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 26.Hayashi Y., Okino N., Kakuta Y., Shikanai T., Tani M., Narimatsu H., Ito M. (2007) Klotho-related protein is a novel cytosolic neutral β-glycosylceramidase. J. Biol. Chem. 282, 30889–30900 [DOI] [PubMed] [Google Scholar]
- 27.Jeng W.-Y., Wang N.-C., Lin C.-T., Chang W.-J., Liu C.-I., Wang A. H.-J. (2012) High-resolution structures of Neotermes koshunensis β-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates. Acta Crystallogr. D Biol. Crystallogr. 68, 829–838 [DOI] [PubMed] [Google Scholar]
- 28.Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 29.Shen M. Y., Sali A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 31.Amaya M. F., Watts A. G., Damager I., Wehenkel A., Nguyen T., Buschiazzo A., Paris G., Frasch A. C., Withers S. G., Alzari P. M. (2004) Structural insights into the catalytic mechanism of Trypanosoma cruzi trans-sialidase. Structure 12, 775–784 [DOI] [PubMed] [Google Scholar]
- 32.Yuan P., Thompson T. B., Wurzburg B. A., Paterson R. G., Lamb R. A., Jardetzky T. S. (2005) Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13, 803–815 [DOI] [PubMed] [Google Scholar]
- 33.Connaris H., Govorkova E. A., Ligertwood Y., Dutia B. M., Yang L., Tauber S., Taylor M. A., Alias N., Hagan R., Nash A. A., Webster R. G., Taylor G. L. (2014) Prevention of influenza by targeting host receptors using engineered proteins. Proc. Natl. Acad. Sci. USA 111, 6401–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruiz F. M., Gilles U., Lindner I., André S., Romero A., Reusch D., Gabius H.-J. (2015) Combining crystallography and hydrogen-deuterium exchange to study galectin–ligand complexes. Chemistry 21, 13558–13568 [DOI] [PubMed] [Google Scholar]
- 35.Owen C. D., Lukacik P., Potter J. A., Sleator O., Taylor G. L., Walsh M. A. (2015) Streptococcus pneumoniae NanC: structural insights into the specificity and mechanism of a sialidase that produces a sialidase inhibitor. J. Biol. Chem. 290, 27736–27748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A. Jr., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam J. M., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas O., Foresman J. B., Ortiz J. V., Cioslowski J., Fox D. J. (2009) Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford, CT, USA [Google Scholar]
- 37.Reed A., Weinstock R., Weinhold F. (1985) Natural population analysis. J. Chem. Phys. 83, 735–746 [Google Scholar]
- 38.Maier J. A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K. E., Simmerling C. (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuntz I. D., Blaney J. M., Oatley S. J., Langridge R., Ferrin T. E. (1982) A geometric approach to macromolecule–ligand interactions. J. Mol. Biol. 161, 269–288 [DOI] [PubMed] [Google Scholar]
- 40.Richards F. M. (1977) Areas, volumes, packing and protein structure. Annu. Rev. Biophys. Bioeng. 6, 151–176 [DOI] [PubMed] [Google Scholar]
- 41.Connolly M. L. (1983) Analytical molecular surface calculation. J. Appl. Cryst. 16, 548–558 [Google Scholar]
- 42.Allen W. J., Balius T. E., Mukherjee S., Brozell S. R., Moustakas D. T., Lang P. T., Case D. A., Kuntz I. D., Rizzo R. C. (2015) DOCK 6: impact of new features and current docking performance. J. Comput. Chem. 36, 1132–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoichet B. K., Bodian D. L., Kuntz I. D. (1992) Molecular docking using shape descriptors. J. Comput. Chem. 13, 380–397 [Google Scholar]
- 44.Meng E. C., Shoichet B. K., Kuntz I. D. (1992) Automated docking with grid-based energy evaluation. J. Comput. Chem. 13, 505–524 [Google Scholar]
- 45.Mills J. E. J., Dean P. M. (1996) Three-dimensional hydrogen-bond geometry and probability information from a crystal survey. J. Comput. Aided Mol. Des. 10, 607–622 [DOI] [PubMed] [Google Scholar]
- 46.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44(W1), W344–W350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coresh J., Selvin E., Stevens L. A., Manzi J., Kusek J. W., Eggers P., Van Lente F., Levey A. S. (2007) Prevalence of chronic kidney disease in the United States. JAMA 298, 2038–2047 [DOI] [PubMed] [Google Scholar]
- 48.Xie J., Yoon J., An S.-W., Kuro-o M., Huang C.-L. (2015) Soluble Klotho protects against uremic cardiomyopathy independently of fibroblast growth factor 23 and phosphate. J. Am. Soc. Nephrol. 26, 1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.