Abstract
The on-site and real-time detection of metal ions is important for environmental monitoring and for understanding the impact of metal ions on human health. However, developing sensors selective for a wide range of metal ions that can work in the complex matrices of untreated samples and cells presents significant challenges. To meet these challenges, DNAzymes, an emerging class of metal ion-dependent enzymes selective for almost any metal ion, have been functionalized with fluorophores, nanoparticles and other imaging agents and incorporated into sensors for the detection of metal ions in environmental samples and for imaging the metal ions in living cells. Herein, we highlight the recent developments of DNAzyme-based fluorescent, colorimetric, SERS, electrochemical and electrochemiluminscent sensors for metal ions for these applications.
Introduction
Metal ions are major targets for environmental sensing, because the presence, distribution and speciation of metal ions in the environment – such as in ground water, lakes, rivers and oceans – as well as in living cells can have a significant impact on human health. A primary example is the elevated levels of lead (Pb) in the drinking water in cities such as Flint, Michigan, due to aged water pipes and improper water treatment. Fast, accurate detection and quantification of metal ion concentrations in different locations in the environment is important in preventing heavy metal contamination. At the same time, it is also imperative to investigate the effects of distribution and speciation of these metal ions in living cells to determine their potential effects on biological species such as plants, animals and humans.
While metal analysis has been established using techniques such as atomic absorption spectroscopy, atomic emission spectroscopy (AES), inductively coupled plasma (ICP)-AES and ICP-mass spectroscopy, these methods of instrumental analysis are limited to laboratory settings, are expensive to operate and cannot provide real-time information. To overcome these limitations, the field of portable detection has been growing over the past decade, which has been facilitated in part by the application of advances in biotechnology. In particular, a new class of sensors based on DNAzymes have been used to impart metal ion selectivity into otherwise non-selective sensing modalities [1-5].
DNAzymes are single-stranded (ss) DNA sequences which fold into complex tertiary structures and are able to catalyze a number of reactions, including cleavage of the phosphodiester backbone at a ribonucleotide or deoxyribonucleotide site [6]. Metal ions have been shown to play a critical role in the catalytic process and are constitutively required for the catalytic activity of most known DNAzymes. DNAzymes are discovered by an iterative screening and amplification process known as in vitro selection (Fig. 1A) [6,7]. In vitro selection starts with approximately 1015 different DNA sequences and, based on the selection conditions, can produce DNAzymes which are highly specific for metal ion cofactors in a range of concentrations. Thus far, DNAzymes have been selected that are specific for Pb2+ [6]-[8], Zn2+ [9,10], Mg2+ [8,11], Cu2+ [12], Ca2+ [13], UO22+ [14], Na+ [15], Hg2+ [16], Cd2+ [17], Cr3+ [18], Ln3+ [19-21], Ce3+ [22], and Ag+ [23].
Figure 1.
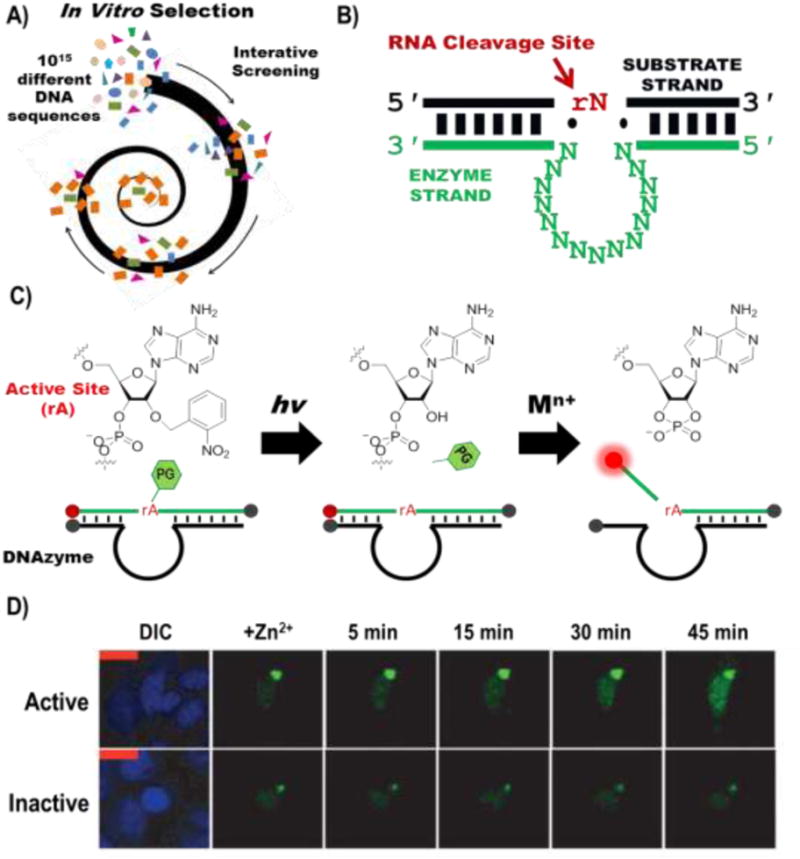
DNAzymes are selected via an iterative combinatorial selection strategy called in vitro selection (A). The resulting DNAzyme (B) has binding arms which hybridize according to Watson-Crick base pairing, indicated by the black and green bars, which are highly programmable, an enzymatic region, shown as repeated N, where N is A, C, G or T, and an RNA cleavage site indicated in red at rN, where N can be A, C, G, T or U. (C) By functionalizing the DNAzyme with fluorophores (red) and quenchers (grey), it can be turned into a catalytic beacon, which has a turn-on fluorescent signal in the presence of a selective monovalent, divalent, or trivalent metal-ion cofactor. The catalytic beacon can be caged using the photolabile nitrobenzyl group on the 2′-OH. An example of the efficacy of the photocaged catalytic beacon is shown in D (figure modified from ref. 49)
DNAzymes typically have three main components in their secondary structures: two binding arms, an active site, and an enzymatic region (Fig. 1B). The ability to select a DNAzyme with metal ion specific activity without prior chemical knowledge of the DNAzyme structure, and then to subsequently modify DNAzyme binding arms and other non-essential nucleotides with minimal to no effect on selectivity and sensitivity has made DNAzymes ideal metal-selective components for new metal ion sensing technologies. These attributes in conjunction with the relatively low-cost synthesis, chemical stability, and ease of DNA functionalization with a plethora of chemical modifications available, has made DNAzymes easy to attach to nanomaterials and other molecular signaling groups such as fluorophores and electrochemical agents. Due to this ease of conjugating different sensing agents with different DNAzyme sensors in a modular fashion, these sensors can readily be adapted for multiplexed sensing, allowing for simultaneous imaging of multiple metal ions within the same system. Additionally, the modular structure of DNAzymes presents a unique advantage because it allows them to be used interchangeably to detect different metal ions using the same sensing strategy. If a new sensing platform is developed for detection of a certain metal ion using DNAzymes, it can often be easily adapted to sense various other metal ions simply by exchanging the DNAzyme with another. Because of this advantage, while this review highlights DNAzyme-based sensors developed for specific metal ions, the same sensing technique can theoretically be applied to detect any of the previously listed metal ions by simply exchanging the DNAzyme. By adapting DNAzymes for different sensing platforms, metal ions have been selectively detected using colorimetric [24], fluorescent [25], electrochemical [26,27], electrochemiluminescent, chemiluminescent [28], luminescent [29], SERS, glucose-meter [30-32], PCR [33], nanopore [34,35] and cantilever-based methods [36,37]. This article seeks to review recent progress, focusing largely on new developments within the past year, of DNAzyme-based metal ion sensing, with an emphasis on fluorescent, colorimetric, SERS, electrochemical, and electrochemiluminescent detection methods.
Fluorescent Metal Ion Sensors for Environmental Monitoring and Cellular Imaging
One of the earliest and most established DNAzyme-based sensors is the fluorescent sensor based on the novel design of the catalytic beacon, which typically places a fluorophore-quencher pair on adjacent ends of the enzyme and substrate strands, respectively (Fig 1C) [14,38]. Upon metal ion-dependent cleavage of the substrate strand at a specific ribonucleotide (e.g. rA) or deoxyribonucleotide site, the fluorophore will dissociate to generate a turn-on fluorescent signal, due to a change in the melting temperature after the cleavage reaction. The metal ion concentration can then be determined based on the kinetic rate of fluorescence increase. Since the invention of the catalytic beacon, it has been modified to decrease the background and has utilized various different fluorophores. The ability to change fluorophores and the separation between the metal ion binding site and the fluorophore give a distinct advantage to DNAzyme-based fluorescent metal ion sensors. These fluorescence sensors typically have a limit of detection (LOD) in the low nanomolar to picomolar range and have been successfully applied to the detection of metal ion contaminants, including UO22+, Pb2+, Hg2+, Cu2+, Ag+, Cr3+, and Ce3+, in tap [39,40], ground [18], pond [41,42], waste, river and lake water [23,43-45], as shown in Table 1. As a result, fluorescent sensors based on DNAzymes are commercially available and have been used to monitor drinking water systems in public schools [46]. Recent work has focused on signal amplification by protein enzymes like exonuclease [42] and alkaline phosphatase [47], or by enzyme-free amplification methods which are solely DNA-based, such as hybridization chain reaction (HCR) [39].
Table 1. Recent metal ion sensors developed using DNAzymes, the type of DNAzyme and the samples in which sensors were tested.
| Detection Mode | Analyte | LOD | DNAzyme Name | Sample | Ref. | ||||
|---|---|---|---|---|---|---|---|---|---|
| Fluorescence-B Detectionased | |||||||||
| UO22+ | 2.4 pM | 39E DNAzyme | Mine and Pond water | [42] | |||||
| UO22+, Pb2+ | 0.6 nM, 3.9 nM | 39E and 8-17 DNAzymes | Cellular | [51] | |||||
| Pb2+ | 1.7 nM | 8-17 DNAzyme | River, Lake, and Waste water | [45] | |||||
| Pb2+ | 2 nM | GR-5 L-DNAzyme | River water + salt and buffer | [53] | |||||
| Hg2+ | None Given | UV1C DNAzyme | Tap Water | [40] | |||||
| Cu2+ | 500 pM | CA3 variant DNAzyme | Tap | [39] | |||||
| Cu2+ | 1.6 nM | PsCu10 DNAzyme | Lake water | [44] | |||||
| Cd2+ | 1.6 nM | BN-Cd16 DNAzyme | Rice extract | [17] | |||||
| Ag+ | 24.9 nM | Ag10c DNAzyme | Lake water | [23] | |||||
| Cr3+ | 70 nM | Ce13d DNAzymes | Groundwater | [18] | |||||
| Zn2+ | None Given | 8-17 DNAzymes | Cellular | [58] | |||||
| Electrochemiluminescent Detection | |||||||||
| Pb2+ | 6.4 pM | 8-17 DNAzyme | Buffer | [59] | |||||
| Pb2+ | ∼50 pM | 8-17 DNAzyme | Mineral, Drinking, and Snow water | [60] | |||||
| Cu2+ | 1 nM | CA3 variant DNAzyme | Human hair extract buffer | [61] | |||||
| Ag+ | 2 pM | HRP-mimicking DNAzyme | River water | [62] | |||||
| Electrochemical Detection | |||||||||
| UO22+ | 2 pM | 39E DNAzyme | River water | [63] | |||||
| UO22+ | 20 pM | 39E DNAzyme | River water | [64] | |||||
| Pb2+ | 0.034 pM | 8-17 DNAzyme | Tap and lake water | [65] | |||||
| Pb2+ | 0.005 nM | 8-17 DNAzyme | Lake water | [66] | |||||
| Pb2+ | 37 pM | 8-17 DNAzyme | Buffer | [67] | |||||
| Pb2+ | 43 pM | 8-17 DNAzyme | Tap water (5%) | [68] | |||||
| Pb2+ | 0.25 nM | 8-17cis DNAzyme | Lake water sample | [69] | |||||
| Pb2+ | 500 pM | HRP-mimicking DNAzyme | Buffer | [70] | |||||
| Hg2+ | 5.8 nM | HRP-mimicking DNAzyme | Rap and Waste water | [71] | |||||
| Cu2+ | 0.1 aM | CA3 variant DNAzyme | River water | [72] | |||||
| Colorimetric Detection | |||||||||
| UO22+ | 37 nM | 39E DNAzyme | Filtered (0.25 μM) Lake water | [73] | |||||
| Pb2+ | 12 pM | 8-17 DNAzyme | Tap and River water, and Landfill leachate | [74] | |||||
| Pb2+ | 20 pM | 8-17 DNAzyme | River water | [75] | |||||
| Ce3+ | 20 nM | Multiple DNAzymes | Lake water | [76] | |||||
| Surface Enhanced Raman Scattering Detection | |||||||||
| UO22+ | 1 pM | 39E DNAzyme | Sea, River, and Pond water | [77] | |||||
| Pb2+ | 8.9 pM | 8-17 DNAzyme | Lake, Tap, and Industrial waste water | [78] | |||||
While the high selectivity and sensitivity of the catalytic beacon has made it widely applicable to environmental sensing, a key advantage for its application in cells is the ability to easily choose the desired fluorophore, which allows it to be used with other cellular dyes and sensors without worry of overlapping fluorescence and even allows for detection of multiple metal ions simultaneously. Therefore, in 2013, the Lu Group reported the first application of metal ion sensing in cells using a catalytic beacon [48]. In this study the catalytic beacon was delivered into cells via functionalization to gold nanoparticles, and used phosphorothioate modifications on the 3′- and 5′-ends to prevent degradation by exonucleases. Subsequent studies have used cationic liposomes [49], MnO2 nanosheets [50], DNA nanostructures [51] and cell penetrating peptides [49,52] to deliver DNAzymes to cells. To increase biostability, other groups have used non-natural L-DNA, which has similar reactivity to the D-DNA enantiomer but cannot be degraded by native nucleases [53,54]. Perhaps the most significant advancement in cellular imaging was the introduction of a photocaging group, to allow for light-controlled activation of DNAzymes; in this way, nonspecific cleavage of DNAzymes during their delivery into cells can be prevented and metal ion imaging can be controlled both temporally and spatially using light. Photoactivation of DNAzymes by modifying the active site or backbone, and subsequent light activation was demonstrated as early as 2004, and earlier yet for ribozymes in 1998 [55-57]; however, not until 2014 was a photocaged DNAzyme used to detect the presence of metal ions in cells [49]. The photocaging process requires synthesis of a photolabile nitrobenzyl group on the 2′ -OH of the phosphoroamidite which will be then become the RNA active site. Though this provides for reliable photocaging, the synthesis of the photocaged phosphoroamidite is complex, thus in 2016 Yu Xiang's lab simplified the photocaging process to a simple post-synthetic modification of a non-bridging phosphorothioate with bromo-4′ -hydroxyacetophenone on three nucleobases essential for DNAzyme activity [58]. Furthermore, Xiang et al. was able to detect both UO22+ and Pb2+ simultaneously in cells using DNAzyme-DNA nanostructure complexes, using another photocaging method [51]. All of these studies have enhanced our ability to probe labile metal ion distributions in cells; however, further work needs to be performed in order to use DNAzymes to accurately quantify metal ion concentrations in the cellular environment.
Colorimetric sensors
While fluorescent sensors provide quantitative information about metal ions, the need for a fluorimeter and costs associated with it may deter the application of fluorescent sensors for on-site and real-time environmental monitoring. The visible spectrum (300 nm – 700 nm) offers a myriad of opportunities for the development of sensors where the presence of an analyte can be observed by the naked-eye, allowing for fast and simple detection of analytes without the need of sophisticated instrumentation [79]. Over the past decade, there have been significant advancements in the field of colorimetric-based biosensors. One interesting emerging trend in the past decade was the extensive use of colorimetric gold and silver nanomaterials for the detection of metal ions [80,81].
In the past several years, several systems utilizing DNAzymes for colorimetric metal ion sensing have been reported. Most of these sensor designs take advantage of recent progress in the field of nanotechnology to couple activity of the DNAzyme with a colorimetric output. Yun et al. [75] designed a sensor which uses a DNAzyme to cleave a DNA hairpin with low stability (Fig. 2A). In the presence of Pb2+, the cleaved hairpin dissociates, generating short DNA stands that adsorb on the surface of gold nanoparticles, thus stabilizing the nanoparticles and yielding a bright red solution. In the absence of lead, the hairpin remains uncleaved and so is unable to stabilize the nanoparticles. The gold nanoparticles aggregate without the stabilizing single-stranded DNA ligands, which shifts the surface plasmon resonance, resulting in a purple solution. This detection scheme has a reported LOD of 20 pM [75]. Similar nanoparticle-based colorimetric sensing strategies have been demonstrated in hydrogel systems. In these systems, DNAzymes were embedded with gold nanoparticles within a hydrogel which is cross-linked with the substrate strand. In the presence of the target metal ion, the DNAzymes cleaves the substrate strands which destabilize the hydrogel. The subsequent release of the embedded gold nanoparticles resulting in a similar change in color, based on detection of the desired metal ion. This hydrogel-DNAzyme-based colorimetric biosensor was demonstrated effectively for sensing lead [82], lanthanide [76] and uranyl ions [73] (Fig. 2B) with 20 and 37 nM LOD values, respectively.
Figure 2.
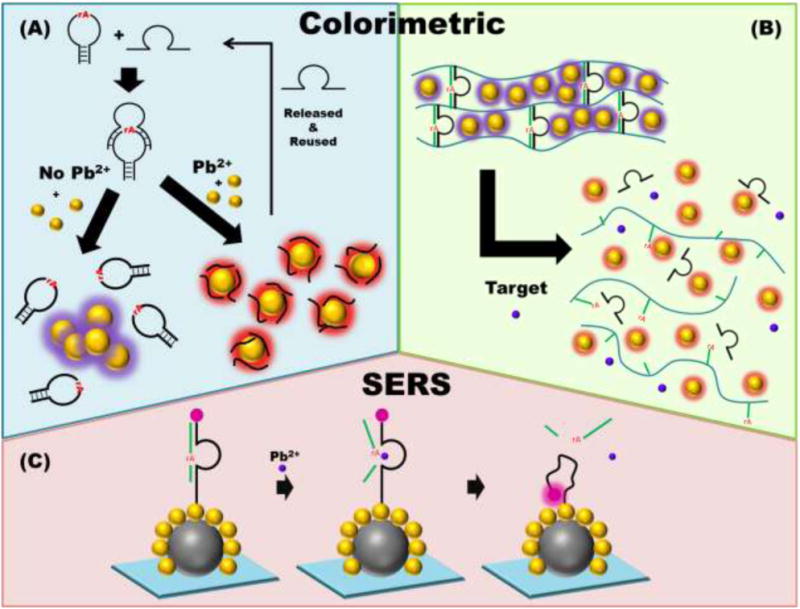
A schematic representation of DNAzyme-based colorimetric and SERS detection methods using nanoparticles. In (A), the presence of the target analyte (Pb2+) cleaves the DNA hairpin, and as a result the released ssDNA is able to adsorb on the gold nanoparticle surface, stabilizing the nanoparticles and preventing aggregation, resulting in a red solution. Without analyte-induced cleavage by the DNAzyme, no free singled-stranded fragments are available to stabilize the nanoparticles, leading to aggregation of the nanoparticles and a purple colored solution. In section (B), nanoparticles are enclosed within a hydrogel polymer with DNAzyme cross linkers. The presence of target analyte cleaves the DNAzyme, destabilizing the hydrogel system and releasing the gold nanoparticles to allow a recovery of absorbance unique to gold nanoparticles. In the bottom section, gold satellite coated silver core nanoparticles were synthesized. Using gold-thiol chemistry, DNAzymes were functionalized on the surface of the nanoparticles where the Cy5 fluorescence dye is far from the surface of the probe. The presence of target would cleave the substrate strand, resulting in the deformation of the enzyme strand to increase the proximity of the Cy5 dye and the nanoparticle.
Other approaches to colorimetric sensing involving DNAzymes not covered by this review use redox chemistry to induce a color change in certain redox active compounds, using the redox-active horseradish peroxidase (HRP) mimicking-DNAzyme [79].
SERS sensors
Surface-enhanced Raman scattering (SERS) is the enhanced Raman signal of a molecule in close proximity to or direct contact with a roughened metal surface or nanoparticle. This analytical technique is capable of producing a Raman enhancement factor of 1011 as compared to that of single molecule fluorescence detection methods [83,84]. Therefore it is not surprising that SERS-based DNAzyme metal ion sensors have been utilized to develop sensitive metal ion sensors for lead and uranyl ions [85-89]. Inspired by a previously reported highly reproducible SERS system [90], Shi et al. [78] reported using the 8-17 DNAzyme to detect Pb2+ with a SERS gold-silver nanoparticle satellite core structure system on a silicon chip (Fig. 2C). This system yielded a LOD of 8.9 pM with high selectivity and was able to be recycled over multiple rounds. In another study, the 8-17 DNAzyme was functionalized on the surface of gold nanoparticles in the presence of Raman-active Victoria blue B dye to detect Pb2+ with a LOD of 10 fM [91]. In a different system, the 39E DNAzyme was used to detect UO22+ using a plasmonic nanowire interstice sensor to reach a LOD of 1 pM with high selectivity [77]. The use of DNAzymes in SERS applications has seen its translation into field of environmental monitoring for Pb2+[78,85], UO22+ [77,87] and Hg2+ metal ions [90].
Electrochemical Sensors
While fluorescent, colorimetric and SERS sensors are powerful methods to detect metal ions, they are often vulnerable to high background noise from environmental samples. Therefore, DNAzyme-based electrochemical detection of metal ions has been a popular field since its conception in 2007, because of its relative imperviousness to signal interference [92]. The field has expanded significantly since the original concept of a DNAzyme-based electrochemical sensor, which used the cleavage and release of a substrate of the 8-17 DNAzyme in the presence of Pb2+ to increase the flexibility of a thiol-DNA-methylene blue (MB) tethered enzyme and thus decrease the distance between the electrode and MB [26,69]. Much of the progress in this field has dealt with amplifying the signal either via the introduction of enzymes, changing the redox active group or through use of hybridization chain reaction (HCR), for which this review will focus.
In order to overcome the inherent limitation of one signal output per single DNAzyme reaction, several differing methods of HCR have been applied. One of the most sensitive of these methods employed the highly selective UO22+-dependent DNAzyme, 39E [14]. Yun et al. was able to achieve an LOD of 2 pM (Fig. 3A) by using a non-functionalized 39E DNAzyme which, when cleaved, would release an initiator sequence to open hairpin 2 (H2). Hairpin 1 (H1) would subsequently bind to H2, which is functionalized on the electrode through a thiol modification, resulting in release of the initiator sequence. Methylene blue (MB), a minor groove binder, would then bind the dsDNA attached to the electrode to generate an electrochemical signal [63]. A similar method was employed again using the 39E DNAzyme, which allows the H1/H2 complex to form upon cleavage by the tethered DNAzyme, revealing the substrate HCR initiator on the electrode. The initiator opens H2, which subsequently opens H1 to form a nicked dsDNA polymer chain, which is again filled with minor groove binder MB to give an electrochemical signal, with an LOD of 20 pM (Fig. 3B) [64]. It should be noted that a further study uses the formation of a hairpin polymer for the electrochemical detection of Pb2+ by tethering the enzyme sequence of a DNAzyme to a magnetic bead, and using the cleavage of the substrate and exposure of the enzyme sequence to initiate a hairpin chain assembly on the magnetic beads with a ferrocene-tagged hairpin, generating an electrochemical signal, with an LOD of 37 pM [67].
Figure 3.
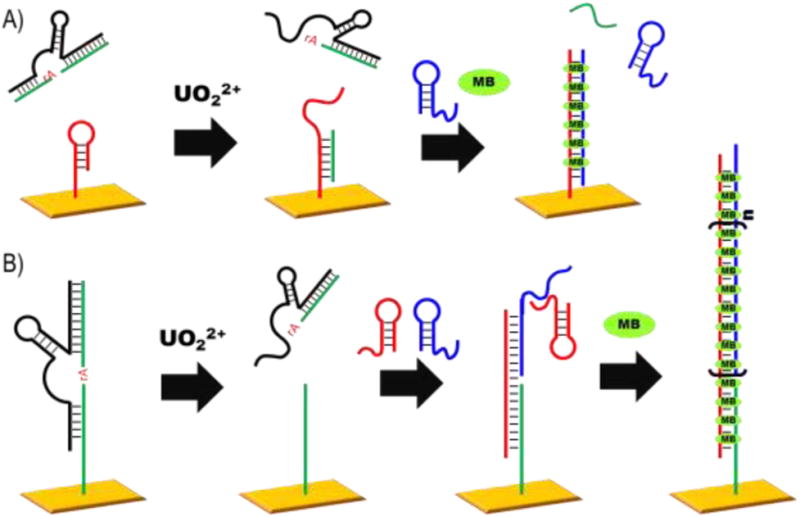
Electrochemical signals enhanced by HCR, with the substrate acting as an initiator for H2 (shown in red) to open and hybridize with H1 (in blue), which kicks-off the substrate initiator so it can open another H2 (A). The formation of a nicked dsDNA polymer chain initiated by cleavage of the DNAzyme tethered to the electrode (B).
Electrochemiluminescence
Even though electrochemical sensors are quite useful in detecting metal ions in complex environmental samples with minimal interference, these sensors, often do not have enough sensitivity for environmental detection of metal ions without resorting to the signal amplification methods described in the previous section. A further method of enhancing the sensitivity of these metal ion sensors is with electrochemiluminescence (ECL), which excites a luminophore via a high-energy electron transfer reaction. This technique has recently gained popularity in analytical detection because of its relatively simple detection equipment, coupled with the reactivity-based luminescence signal output, leading to low background and high sensitivity ranging from low picomolar to low nanomolar analyte concentrations (Table 1). In 2009, Zhu et al. developed a lead sensor by functionalizing an Au-electrode with a thiol-modified 8-17 DNAzymes, which can cleave the substrate sequence in the presence of Pb2+. The substrate is labeled with the luminophore Ru(bpy)3-NHS, so the subsequent release of the cleaved substrate generates a turn-off signal (Fig. 4A) [93]. The sensitivity imparted by this technique meant that the LOD was 11 pM. Using a different approach, Guangpeng Liu et al. was able to detect Ag+ with an LOD of 2 pM, by using DNAzyme nanowires composed of horseradish peroxidase-mimicking DNAzyme, also called the hemin/G-quadruplex DNAzyme, to accelerate the luminescence of luminol [62]. In the presence of Ag+, the C-Ag+-C interaction promoted the release of the DNAzyme nanowires, which inhibits peroxidase activity and decreases overall luminescence.
Figure 4.
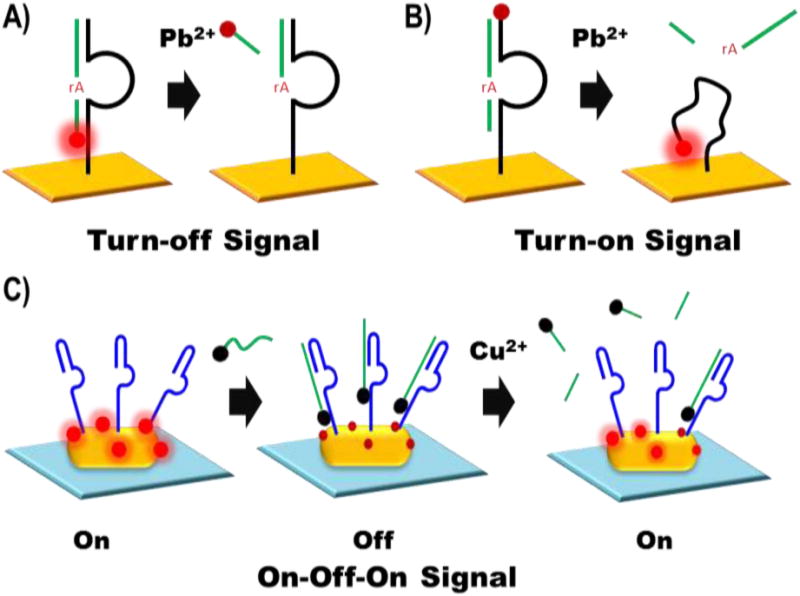
Schematics of the turn-off (A) turn-on (B) and “On-Off-On” signal (C) DNAzyme sensor constructs. The turn-off sensor (A) uses a DNAzyme enzyme sequence (black) tethered to an electrode (gold surface) to cleave a DNAzyme substrate sequence (green) at the RNA active site (indicated by rA in red). The cleavage reaction will release the Ru(bpy)3-NHS tag (shown in bright red when “on” and dark red when “off”) into solution turning “off” the luminescence signal. The turn-on sensor uses release of the DNAzyme substrate to allow the Ru(bpy)3-NHS labeled enzyme sequence to bend towards the electrode and generate a turn-on signal. The “On-Off-On” sensor (C) initially has a gold nanorod (yellow oval) coated in 3,4,9,10-perylenetetracarboxylic acid (indicated in bright red when excited and dark red when quenched) and the enzyme sequence of the Cu2+-dependent DNAzyme (dark blue). The introduction of a substrate strand (green) attached to a ferrocene quencher (black circle) turns off the signal and the subsequent introduction of Cu2+ cause the cleavage and subsequent release of the quencher to regenerate a turn-on signal.
To further improve the sensitivity, decrease background and mitigate the risk of false positives, Zhang and co-workers were able to report a turn-on DNAzyme-based ECL sensor for Pb2+ with an LOD of 1.4 pM (Fig. 4B) [94]. They were able to accomplish this feat by tagging the DNAzyme enzyme sequence with Ru(bpy)3-NHS and annealing the enzyme and substrate sequence to form the rigid hybridized structure which keeps the luminophore further from the electrode. Upon cleavage and release of the substrate, the ssDNA of the enzyme is able to bend and move the luminophore closer to the electrode, increasing electron transfer to ruthenium and thereby increasing the luminescence signal. Wu and co-workers built upon this design to create another turn-on sensor with an LOD of 6.4 pM Pb2+ by attaching the enzyme sequence to the electrode via a 4-amino benzoic acid linker, and using the substrate strand to block hybridization of free Ru(bpy)3-NHS-labeled hairpin sequences, which are complementary to the enzyme [59]. In the presence of Pb2+, the substrate will cleave and be replaced by the Ru(bpy)3-NHS-labeled hairpin, placing the luminophore next to the electrode. However, even with the significantly improved sensitivity and decreased background signal of the turn-on ECL sensors, further improvements to sensitivity and selectivity have been shown with the “on-off-on” signaling method. A DNAzyme-based “on-off-on” ECL sensor for Cu2+ with picomolar sensitivity, and with selectivity for 1 nM copper over 1 μM levels of competing metal ions has thus been developed (Fig. 4C) [61]. This sensor used a 3,4,9,10-perylenetetracarboxylic acid (PTCA)/ aniline mixture to generate a turn-on signal. The presence of ferrocene, tagged to the substrate sequence would quench the PTCA luminescence signal, generating a “turn-off” signal proportional to the amount of fully formed enzyme-substrate complex. The “on” signal is then regenerated by cleavage and dissociation of the ferrocence-tagged substrate in the presence of the analyte. Overall, the highly sensitive ECL sensor platform has shown great promise developing turn-on, turn-on and “on-off-on” sensors.
Conclusions and Perspectives
In this mini-review, we have summarized recent advances in DNAzyme-based metal ion sensing, focusing on fluorescent, colorimetric, SERS, electrochemical and electrochemiluminescent sensors. Of these techniques, fluorescent, colorimetric, electrochemical and ECL sensors have been able to successfully detect metal ions in environmental samples and fluorescent sensors have been developed for the detection of metal ions within living cells. Improvements on DNAzyme-based metal ion sensors have successfully lowered the limit of detection for many of these techniques by changing the method of signal output, whether by converting systems with turn-off signals is to turn-on outputs, or by incorporating methods of signal amplification such as HCR. Additionally, DNAzyme fluorescent sensors have been modified to allow temporal control through the introduction of a photo-caged switch, which can enable time-dependent detection of native metal ion concentrations. Furthermore, initial studies have demonstrated the simultaneous monitoring of two different metal ions in cells. The generality of many of these techniques means that they can typically be used for multiple detection methods and across different sample conditions, including both environmental and cellular samples. There can be room for significant growth in detection of metal ions in cells by applying techniques like HCR to amplify fluorescent signals, and further work must be done to accurately quantify metal ions in a cellular environment using catalytic beacon sensors. The field of DNAzyme-based environmental sensors can also be improved by generating robust portable sensors for all sensing modalities, and for multiplex detection of many metal ions simultaneously. Overall, the field of DNAzyme-based metal ion sensing is continuing to develop with promising prospects for future cellular and portable detection technologies.
Highlights.
DNAzymes are an emerging class of metal ions sensors with high selectivity
Recent progress has been made in using DNAzymes for environmental detection of metal ions with high sensitivity
Cellular imaging of metal ions in living cells using DNAzymes with high temporal control has also bfigeen reported
Acknowledgments
We wish to thank Lu group members who contributed to the work described in this review, Mr. Ryan Lake for proof-reading, and the U.S. National Institutes of Health (R21MH110975 and R41MH111337) for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Zhang XB, Kong RM, Lu Y. Metal ion sensors based on DNAzymes and related DNA molecules. Annu Rev Anal Chem. 2011;4:105–128. doi: 10.1146/annurev.anchem.111808.073617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JH, Wang Z, Lu Y. DNAzyme-based sensing for metal ions in ocean platform. In: Tiquia-Arashiro SM, editor. Molecular biological technologies for ocean sensing. Humana Press; Totowa, NJ: 2012. pp. 103–116. [Google Scholar]
- 3.Xiang Y, Lu Y. DNA as sensors and imaging agents for metal ions. Inorg Chem. 2014;53:1925–1942. doi: 10.1021/ic4019103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhan S, Wu Y, Wang L, Zhan X, Zhou P. A mini-review on functional nucleic acids-based heavy metal ion detection. Biosens Bioelectron. 2016;86:353–368. doi: 10.1016/j.bios.2016.06.075. [DOI] [PubMed] [Google Scholar]
- 5.Saidur MR, Aziz ARA, Basirun WJ. Recent advances in DNA-based electrochemical biosensors for heavy metal ion detection: A review. Biosens Bioelectron. 2017;90:125–139. doi: 10.1016/j.bios.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 6.Breaker RR, Joyce GF. A DNA enzyme that cleaves rna. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 7.Ihms HE, Lu Y. In vitro selection of metal ion-selective DNAzymes. In: Hartig JS, editor. Ribozymes: Methods and protocols. Humana Press; Totowa, NJ: 2012. pp. 297–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF. RNA cleavage by a DNA enzyme with extended chemical functionality. J Am Chem Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Zheng W, Kwon AH, Lu Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breaker RR, Joyce GF. A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem Biol. 1995;2:655–660. doi: 10.1016/1074-5521(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 12.Carmi N, Shultz LA, Breaker RR. In vitro selection of self-cleaving DNAs. Chem Biol. 1996;3:1039–1046. doi: 10.1016/s1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]
- 13.Zhou W, Zhang Y, Ding J, Liu J. In vitro selection in serum: RNA-cleaving DNAzymes for measuring Ca2+ and Mg2+ ACS Sensors. 2016;1:600–606. [Google Scholar]
- 14.Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc Natl Acad Sci. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Torabi SF, Wu P, McGhee CE, Chen L, Hwang K, Zheng N, Cheng J, Lu Y. In vitro selection of a sodium-specific DNAzyme and its application in intracellular sensing. Proc Natl Acad Sci. 2015;112:5903–5908. doi: 10.1073/pnas.1420361112. A DNAzyme with high selectivity for Na+ over other mono-, di- and tri-valent metal ions have been obtained, demonstrating that the in vitro selection can be a poweful method to obtain selective agents for metal ions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollenstein M, Hipolito C, Lam C, Dietrich D, Perrin DM. A highly selective DNAzyme sensor for mercuric ions. Angew Chem, Int Ed. 2008;47:4346–4350. doi: 10.1002/anie.200800960. [DOI] [PubMed] [Google Scholar]
- 17.Huang PJJ, Liu J. Rational evolution of Cd2+-specific DNAzymes with phosphorothioate modified cleavage junction and Cd2+ sensing. Nucleic Acids Res. 2015;43:6125–6133. doi: 10.1093/nar/gkv519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou W, Vazin M, Yu T, Ding J, Liu J. In vitro selection of chromium-dependent DNAzymes for sensing chromium(III) and chromium(VI) Chem Eur J. 2016;22:9835–9840. doi: 10.1002/chem.201601426. [DOI] [PubMed] [Google Scholar]
- 19.Huang PJ, Vazin M, Liu J. In vitro selection of a new lanthanide-dependent DNAzyme for ratiometric sensing lanthanides. Anal Chem. 2014;86:9993–9999. doi: 10.1021/ac5029962. [DOI] [PubMed] [Google Scholar]
- 20.Huang PJ, Lin J, Cao J, Vazin M, Liu J. Ultrasensitive DNAzyme beacon for lanthanides and metal speciation. Anal Chem. 2014;86:1816–1821. doi: 10.1021/ac403762s. [DOI] [PubMed] [Google Scholar]
- 21.Huang PJ, Vazin M, Matuszek Z, Liu J. A new heavy lanthanide-dependent DNAzyme displaying strong metal cooperativity and unrescuable phosphorothioate effect. Nucleic Acids Res. 2015;43:461–469. doi: 10.1093/nar/gku1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W, Ding J, Liu J. An efficient lanthanide-dependent DNAzyme cleaving 2′-5′-linked RNA. Chembiochem. 2016;17:890–894. doi: 10.1002/cbic.201500690. [DOI] [PubMed] [Google Scholar]
- 23.Saran R, Liu J. A silver DNAzyme. Anal Chem. 2016;88:4014–4020. doi: 10.1021/acs.analchem.6b00327. [DOI] [PubMed] [Google Scholar]
- 24.Priyadarshini E, Pradhan N. Gold nanoparticles as efficient sensors in colorimetric detection of toxic metal ions: A review. Sens Actuators B. 2017;238:888–902. [Google Scholar]
- 25.Zhang J, Cheng F, Li J, Zhu JJ, Lu Y. Fluorescent nanoprobes for sensing and imaging of metal ions: Recent advances and future perspectives. Nano Today. 2016;11:309–329. doi: 10.1016/j.nantod.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui L, Wu J, Ju H. Electrochemical sensing of heavy metal ions with inorganic, organic and bio-materials. Biosens and Bioelectron. 2015;63:276–286. doi: 10.1016/j.bios.2014.07.052. [DOI] [PubMed] [Google Scholar]
- 27.Cui H, Xiong X, Gao B, Chen Z, Luo Y, He F, Deng S, Chen L. A novel impedimetric biosensor for detection of lead (II) with low-cost interdigitated electrodes made on PCB. Electroanalysis. 2016;28:2000–2006. [Google Scholar]
- 28.Wu Q, Shen H, Shen H, Sun Y, Song L. Study on sensing strategy and performance of a microfluidic chemiluminescence aptazyme sensor. Talanta. 2016;150:531–538. doi: 10.1016/j.talanta.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 29.Kleinke K, Saran R, Liu J. Label-free Ag+ detection by enhancing DNA sensitized Tb3+ luminescence. Sensors. 2016;16:1370. doi: 10.3390/s16091370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang Y, Lu Y. An invasive DNA approach toward a general method for portable quantification of metal ions using a personal glucose meter. Chem Commun. 2013;49:585–587. doi: 10.1039/c2cc37156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan T, Zhang J, Lu Y. Transforming the blood glucose meter into a general healthcare meter for in vitro diagnostics in mobile health. Biotechnol Adv. 2016;34:331–341. doi: 10.1016/j.biotechadv.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ming J, Fan W, Jiang TF, Wang YH, Lv ZH. Portable and sensitive detection of Copper(II) ion based on personal glucose meters and a ligation DNAzyme releasing strategy. Sens Actuators B. 2017;240:1091–1098. [Google Scholar]
- 33.Zhu P, Shang Y, Tian W, Huang K, Luo Y, Xu W. Ultra-sensitive and absolute quantitative detection of Cu2+ based on DNAzyme and digital PCR in water and drink samples. Food Chem. 2017;221:1770–1777. doi: 10.1016/j.foodchem.2016.10.106. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, Zhang L, Dong D, Liu Y, Li J. A label-free DNAzyme-based nanopore biosensor for highly sensitive and selective lead ion detection. Anal Methods. 2016;8:7040–7046. [Google Scholar]
- 35.Liu N, Hou R, Gao P, Lou X, Xia F. Sensitive Zn2+ sensor based on biofunctionalized nanopores via combination of DNAzyme and DNA supersandwich structures. Analyst. 2016;141:3626–3629. doi: 10.1039/c6an00171h. [DOI] [PubMed] [Google Scholar]
- 36.Peng RP, Xing LB, Wang XJ, Wu Cj, Chen B, Ji HF, Wu LZ, Tung CH. Detection of Pb2+ in aqueous solution by using a DNA-modified microcantilever. Anal Sci. 2016;32:1065–1069. doi: 10.2116/analsci.32.1065. [DOI] [PubMed] [Google Scholar]
- 37.Du Y, Dong S. Nucleic acid biosensors: Recent advances and perspectives. Anal Chem. 2017;89:189–215. doi: 10.1021/acs.analchem.6b04190. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Lu Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J Am Chem Soc. 2000;122:10466–10467. [Google Scholar]
- 39.Chen Y, Chen L, Ou Y, Wang Z, Fu F, Guo L. DNAzyme-based biosensor for Cu2+ ion by combining hybridization chain reaction with fluorescence resonance energy transfer technique. Talanta. 2016;155:245–249. doi: 10.1016/j.talanta.2016.04.057. [DOI] [PubMed] [Google Scholar]
- 40.Ma L, Liu H, Wu G, Sun N, Meng L, Li Y, Liu Z, Diao A. A dual-channel detection of mercuric ions using a label free G-quadruplex-based DNAzyme molecule. Analyst. 2016;141:3997–4000. doi: 10.1039/c6an00795c. [DOI] [PubMed] [Google Scholar]
- 41.Lin YW, Liu CW, Chang HT. Fluorescence detection of mercury(II) and lead(II) ions using aptamer/reporter conjugates. Talanta. 2011;84:324–329. doi: 10.1016/j.talanta.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Cao JX, Wang YS, Xue JH, Huang YQ, Li MH, Chen SH, Zhou B, Tang X, Wang XF, Zhu YF. Exonuclease III-assisted substrate fragment recycling amplification strategy for ultrasensitive detection of uranyl by a multipurpose DNAzyme. RSC Adv. 2016;6:108662–108667. [Google Scholar]
- 43.Wei H, Wang L, Xu X, Zhu J, Jiang W. A T-Hg2+-T metallo-base pair-mediated dual amplification fluorescent strategy for the selective and sensitive detection of Hg2+ RSC Adv. 2016;6:70984–70989. [Google Scholar]
- 44.Huang PJJ, Liu J. An ultrasensitive light-up Cu2+ biosensor using a new DNAzyme cleaving a phosphorothioate-modified substrate. Anal Chem. 2016;88:3341–3347. doi: 10.1021/acs.analchem.5b04904. [DOI] [PubMed] [Google Scholar]
- 45.Wang XY, Niu CG, Guo LJ, Hu LY, Wu SQ, Zeng GM, Li F. A fluorescence sensor for lead (II) ions determination based on label-free gold nanoparticles (GNPs)-DNAzyme using time-gated mode in aqueous solution. J Fluoresc. 2016:1–7. doi: 10.1007/s10895-016-1993-y. [DOI] [PubMed] [Google Scholar]
- 46.Chaloux E, Altmann E. Lead in Water: St. Paul Schools Delayed Fixes. [accessed Jan 10, 2017]; http://www.kaaltv.com/article/stories/S4265242.shtml.
- 47.Zhao M, Guo Y, Wang L, Luo F, Lin C, Lin Z, Chen G. A sensitive fluorescence biosensor for alkaline phosphatase activity based on the Cu(II)-dependent DNAzyme. Anal Chim Acta. 2016;948:98–103. doi: 10.1016/j.aca.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 48.Wu P, Hwang K, Lan T, Lu Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J Am Chem Soc. 2013;135:5254–5257. doi: 10.1021/ja400150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Hwang K, Wu P, Kim T, Lei L, Tian S, Wang Y, Lu Y. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew Chem Int Ed. 2014;53:13798–13802. doi: 10.1002/anie.201408333. The development of photo-caging of the DNAzymes made it possible for imaging metal ions in cells with spatial and temperal control without the DNAzyme being activated prematuraly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan H, Zhao Z, Yan G, Zhang X, Yang C, Meng H, Chen Z, Liu H, Tan W. A smart DNAzyme-MnO2 nanosystem for efficient gene silencing. Angew Chem Int Ed. 2015;54:4801–4805. doi: 10.1002/anie.201411417. [DOI] [PubMed] [Google Scholar]
- 51•.Zhou W, Liang W, Li D, Yuan R, Xiang Y. Dual-color encoded DNAzyme nanostructures for multiplexed detection of intracellular metal ions in living cells. Biosens Bioelectron. 2016;85:573–579. doi: 10.1016/j.bios.2016.05.058. In this article Zhou et al. are able to simultaneously imaging of two different metal ions in living cells. [DOI] [PubMed] [Google Scholar]
- 52.Xing Z, Gao S, Han H, Zhang J, Chen X, Yang Y, Li Q. Hydrophobic N-acetyl-l-leucine grafted polyethylenimine as an efficient carrier for DNAzyme delivery. J Controlled Release. 2015;213:e146–e147. doi: 10.1016/j.jconrel.2015.05.248. [DOI] [PubMed] [Google Scholar]
- 53.Liang H, Xie S, Cui L, Wu C, Zhang X. Designing a biostable L-DNAzyme for lead(II) ion detection in practical samples. Anal Methods. 2016;8:7260–7264. doi: 10.1039/C6AY01791F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui L, Peng R, Fu T, Zhang X, Wu C, Chen H, Liang H, Yang CJ, Tan W. Biostable L-DNAzyme for sensing of metal ions in biological systems. Anal Chem. 2016;88:1850–1855. doi: 10.1021/acs.analchem.5b04170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaulk SG, MacMillan AM. Caged RNA: Photo-control of a ribozyme reaction. Nucleic Acids Res. 1998;26:3173–3178. doi: 10.1093/nar/26.13.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Sen D. Light-regulated catalysis by an RNA-cleaving deoxyribozyme. J Mol Biol. 2004;341:887–892. doi: 10.1016/j.jmb.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 57.Keiper S, Vyle JS. Reversible photocontrol of deoxyribozyme-catalyzed RNA cleavage under multiple-turnover conditions. Angew Chem, Int Ed. 2006;45:3306–3309. doi: 10.1002/anie.200600164. [DOI] [PubMed] [Google Scholar]
- 58••.Wang X, Feng M, Xiao L, Tong A, Xiang Y. Postsynthetic modification of DNA phosphodiester backbone for photocaged DNAzyme. ACS Chem Biol. 2016;11:444–451. doi: 10.1021/acschembio.5b00867. Yu Xiang et al. create a simple, post-synthetic method to modify DNAzymes with a photocage group, to allow for temporal control of DNAzyme reactivity. [DOI] [PubMed] [Google Scholar]
- 59.Wu Yf, Cai Zm, Wu Gh, Rong Mc, Jiang Yq, Yang CyJ, Chen X. A novel signal-on DNAzyme-based electrochemiluminescence, sensor for Pb2+ Sens Actuators B. 2014;191:60–66. [Google Scholar]
- 60.Zhang Y, Zhang L, Kong Q, Ge S, Yan M, Yu J. Electrochemiluminescence of graphitic carbon nitride and its application in ultrasensitive detection of lead(II) ions. Analytical and Bioanalytical Chemistry. 2016;408:7181–7191. doi: 10.1007/s00216-016-9718-2. [DOI] [PubMed] [Google Scholar]
- 61.Lei YM, Zhao M, Wang A, Yu YQ, Chai YQ, Yuan R, Zhuo Y. Electrochemiluminescence of supramolecular nanorods and their application in the “on-off-on” detection of copper ions. Chem Eur J. 2016;22:8207–8214. doi: 10.1002/chem.201504995. [DOI] [PubMed] [Google Scholar]
- 62.Liu G, Yuan Y, Wang J. Hemin/G-quadruplex DNAzyme nanowires amplified luminol electrochemiluminescence system and its application in sensing silver ions. RSC Adv. 2016;6:37221–37225. [Google Scholar]
- 63••.Yun W, Cai D, Jiang J, Wang X, Liao J, Zhang P, Sang G. An ultrasensitive electrochemical biosensor for uranyl detection based on DNAzyme and target-catalyzed hairpin assembly. Microchimica Acta. 2016;183:1425–1432. Using DNAzymes and the hybridization chain reaction, the authors were able to develop a highly sensitive electrochemical sensor with a limit of detection of 2 pM. [Google Scholar]
- 64•.Yun W, Jiang J, Cai D, Wang X, Sang G, Liao J, Lu T, Yan K. Ultrasensitive electrochemical detection of UO22+ based on DNAzyme and isothermal enzyme-free amplification. RSC Adv. 2016;6:3960–3966. By using HCR to form a nicked dsDNA polymer chain, the authors created an electrochemical sensor with a limit of detection of 20 pM. [Google Scholar]
- 65.Xue S, Jing P, Xu W. Hemin on graphene nanosheets functionalized with flower-like MnO2 and hollow AuPd for the electrochemical sensing lead ion based on the specific DNAzyme. Biosens Bioelectron. 2016;86:958–965. doi: 10.1016/j.bios.2016.07.111. [DOI] [PubMed] [Google Scholar]
- 66.Ge S, Wu K, Zhang Y, Yan M, Yu J. Paper-based biosensor relying on flower-like reduced graphene guided enzymatically deposition of polyaniline for Pb2+ detection. Biosens Bioelectron. 2016;80:215–221. doi: 10.1016/j.bios.2016.01.072. [DOI] [PubMed] [Google Scholar]
- 67.Zhuang J, Fu L, Xu M, Zhou Q, Chen G, Tang D. DNAzyme-based magneto-controlled electronic switch for picomolar detection of lead (II) coupling with DNA-based hybridization chain reaction. Biosens Bioelectron. 2013;45:52–57. doi: 10.1016/j.bios.2013.01.039. [DOI] [PubMed] [Google Scholar]
- 68.Liu S, Wei W, Sun X, Wang L. Ultrasensitive electrochemical DNAzyme sensor for lead ion based on cleavage-induced template-independent polymerization and alkaline phosphatase amplification. Biosens Bioelectron. 2016;83:33–38. doi: 10.1016/j.bios.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y, Xiao S, Li H, Liu H, Pang P, Wang H, Wu Z, Yang W. A Pb2+-ion electrochemical biosensor based on single-stranded DNAzyme catalytic beacon. Sens Actuators B. 2016;222:1083–1089. [Google Scholar]
- 70.Li F, Yang L, Chen M, Qian Y, Tang B. A novel and versatile sensing platform based on HRP-mimicking DNAzyme-catalyzed template-guided deposition of polyaniline. Biosens Bioelectron. 2013;41:903–906. doi: 10.1016/j.bios.2012.09.048. [DOI] [PubMed] [Google Scholar]
- 71.Li G, Li Z, You X, Chen J, Tang S. A novel label-free and sensitive electrochemical biosensor for Hg2+ based on ligase-mediated formation of DNAzyme. Talanta. 2016;161:138–142. doi: 10.1016/j.talanta.2016.08.042. [DOI] [PubMed] [Google Scholar]
- 72.Tian R, Chen X, Liu D, Yao C. A sensitive biosensor for determination of Cu2+ by one-step electrodeposition. Electroanalysis. 2016;28:1617–1624. [Google Scholar]
- 73.Huang Y, Fang L, Zhu Z, Ma Y, Zhou L, Chen X, Xu D, Yang C. Design and synthesis of target-responsive hydrogel for portable visual quantitative detection of uranium with a microfluidic distance-based readout device. Biosens Bioelectron. 2016;85:496–502. doi: 10.1016/j.bios.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 74.Zhang C, Lai C, Zeng G, Huang D, Tang L, Yang C, Zhou Y, Qin L, Cheng M. Nanoporous au-based chronocoulometric aptasensor for amplified detection of Pb2+ using dnazyme modified with Au nanoparticles. Biosens Bioelectron. 2016;81:61–67. doi: 10.1016/j.bios.2016.02.053. [DOI] [PubMed] [Google Scholar]
- 75•.Yun W, Cai D, Jiang J, Zhao P, Huang Y, Sang G. Enzyme-free and label-free ultra-sensitive colorimetric detection of Pb2+ using molecular beacon and DNAzyme based amplification strategy. Biosens Bioelectron. 2016;80:187–193. doi: 10.1016/j.bios.2016.01.053. Yun et al. presents a simple colorimetric sensor based on gold nanoparticle stabilization by ssDNA, without the need for DNAzyme conjugation to nanoparticle surfaces. [DOI] [PubMed] [Google Scholar]
- 76.Huang Y, Wu X, Tian T, Zhu Z, Lin H, Yang C. Target-responsive DNAzyme hydrogel for portable colorimetric detection of lanthanide(III) ions. Sci China Chem. 2016:1–6. [Google Scholar]
- 77.Gwak R, Kim H, Yoo SM, Lee SY, Lee GJ, Lee MK, Rhee CK, Kang T, Kim B. Precisely determining ultralow level UO22+ in natural water with plasmonic nanowire interstice sensor. Sci Rep. 2016;6:19646. doi: 10.1038/srep19646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi Y, Wang H, Jiang X, Sun B, Song B, Su Y, He Y. Ultrasensitive, specific, recyclable, and reproducible detection of lead ions in real systems through a polyadenine-assisted, surface-enhanced Raman scattering silicon chip. Anal Chem. 2016;88:3723–3729. doi: 10.1021/acs.analchem.5b04551. [DOI] [PubMed] [Google Scholar]
- 79.Tang G, Zhao C, Gao J, Tan H. Colorimetric detection of hydrogen sulfide based on terbium-G-quadruplex-hemin DNAzyme. Sens Actuators B. 2016;237:795–801. [Google Scholar]
- 80.Liu J, Lu Y. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J Am Chem Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 81.Lee JH, Wang Z, Liu J, Lu Y. Highly sensitive and selective colorimetric sensors for uranyl (uo22+): Development and comparison of labeled and label-free dnazyme-gold nanoparticle systems. J Am Chem Soc. 2008;130:14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang Y, Ma Y, Chen Y, Wu X, Fang L, Zhu Z, Yang CJ. Target-responsive DNAzyme cross-linked hydrogel for visual quantitative detection of lead. Anal Chem. 2014;86:11434–11439. doi: 10.1021/ac503540q. [DOI] [PubMed] [Google Scholar]
- 83.Stiles PL, Dieringer JA, Shah NC, Van Duyne RP. Surface-enhanced Raman spectroscopy. Annu Rev Anal Chem. 2008;1:601–626. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 84.Blackie EJ, Ru ECL, Etchegoin PG. Single-molecule surface-enhanced Raman spectroscopy of nonresonant molecules. J Am Chem Soc. 2009;131:14466–14472. doi: 10.1021/ja905319w. [DOI] [PubMed] [Google Scholar]
- 85.Wang Y, Irudayaraj J. A SERS DNAzyme biosensor for lead ion detection. Chem Commun. 2011;47:4394–4396. doi: 10.1039/c0cc04140h. [DOI] [PubMed] [Google Scholar]
- 86.Ye S, Guo Y, Xiao J, Zhang S. A sensitive sers assay of L-histidine via a DNAzyme-activated target recycling cascade amplification strategy. Chem Commun. 2013;49:3643–3645. doi: 10.1039/c3cc41619d. [DOI] [PubMed] [Google Scholar]
- 87.Jiang Z, Yao D, Wen G, Li T, Chen B, Liang A. A label-free nanogold DNAzyme-cleaved surface-enhanced resonance Raman scattering method for trace UO22+ using rhodamine 6G as probe. Plasmonics. 2013;8:803–810. [Google Scholar]
- 88.Fu C, Xu W, Wang H, Ding H, Liang L, Cong M, Xu S. DNAzyme-based plasmonic nanomachine for ultrasensitive selective surface-enhanced Raman scattering detection of lead ions via a particle-on-a-film hot spot construction. Anal Chem. 2014;86:11494–11497. doi: 10.1021/ac5038736. [DOI] [PubMed] [Google Scholar]
- 89.Gao F, Du L, Tang D, Lu Y, Zhang Y, Zhang L. A cascade signal amplification strategy for surface enhanced Raman spectroscopy detection of thrombin based on DNAzyme assistant DNA recycling and rolling circle amplification. Biosens Bioelectron. 2015;66:423–430. doi: 10.1016/j.bios.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 90.Sun B, Jiang X, Wang H, Song B, Zhu Y, Wang H, Su Y, He Y. Surface-enhancement Raman scattering sensing strategy for discriminating trace mercuric ion (II) from real water samples in sensitive, specific, recyclable, and reproducible manners. Anal Chem. 2015;87:1250–1256. doi: 10.1021/ac503939d. [DOI] [PubMed] [Google Scholar]
- 91.Ye L, Wen G, Ouyang H, Liu Q, Liang A, Jiang Z. A novel and highly sensitive nanocatalytic surface plasmon resonance-scattering analytical platform for detection of trace Pb ions. Sci Rep. 2016;6:24150. doi: 10.1038/srep24150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiao Y, Rowe AA, Plaxco KW. Electrochemical detection of parts-per-billion lead via an electrode-bound DNAzyme assembly. J Am Chem Soc. 2007;129:262–263. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]
- 93.Zhu X, Lin Z, Chen L, Qiu B, Chen G. A sensitive and specific electrochemiluminescentl sensor for lead based on DNAzyme. Chem Commun. 2009:6050–6052. doi: 10.1039/b911191c. [DOI] [PubMed] [Google Scholar]
- 94••.Ma F, Sun B, Qi H, Zhang H, Gao Q, Zhang C. A signal-on electrogenerated chemiluminescent biosensor for lead ion based on DNAzyme. Anal Chim Acta. 2011;683:234–241. doi: 10.1016/j.aca.2010.10.030. A turn-on electrochemiluminescent DNAzyme-base sensor is reported. [DOI] [PubMed] [Google Scholar]


