Abstract
Veliparib (ABT-888) is a novel oral poly-ADP-ribose polymerase (PARP) inhibitor that is being developed for the treatment of hematologic malignancies and solid tumors. Although the pharmacokinetics of veliparib has been studied in combination with cytotoxic agents, limited information exists regarding the pharmacokinetics of chronically-dosed, single-agent veliparib, in patients with either BRCA 1/2–mutated cancer or PARP sensitive tumors. The objectives of the current analysis were to characterize the population pharmacokinetics of veliparib and its primary, active metabolite, M8, and to evaluate the relationship between veliparib and M8 concentrations and poly-ADP-ribose (PAR) level observed in peripheral blood mononuclear cells (PBMC). Seventy-one subjects contributed with veliparib plasma concentrations, M8 plasma concentrations, and PAR levels in PBMC. Veliparib and M8 concentrations were modeled simultaneously using a population PK approach. A two-compartment model with delayed first-order absorption and the elimination parameterized as renal (CLR/F) and non-renal clearance (CLNR/F) adequately described veliparib pharmacokinetics. The pharmacokinetics of the M8 metabolite was described with a two-compartment model. Creatinine clearance and lean body mass were identified as significant predictors of veliparib CLR/F and central volume of distribution, respectively. For a typical subject (LBM, 48 kg; CLCR, 95 mL/min), total clearance (CLR/F +CLNR/F), central and peripheral volume of distribution for veliparib were estimated as 17.3 L/h, 98.7 L and 48.3 L, respectively. At least 50% inhibition of PAR levels in PBMCs was observed at dose levels ranging from 50 to 500 mg.
Keywords: Veliparib, PARP inhibitor, BRCA, parent-metabolite modelling, population pharmacokinetics
Introduction
Poly-ADP-ribose polymerase (PARP) is a nuclear enzyme that signals DNA damage and contributes to the maintenance of genomic integrity1. After single- and double-stranded DNA breaks, the catalytic domains of PARP are immediately activated to regulate DNA damage repair through the base excision repair (BER) pathway2,3. Elevated levels of PARP in cancer cells compared to normal cells are recognized as one of the mechanisms by which tumor cells avoid apoptosis caused by DNA damaging agents4. Thus, inhibition of DNA repair by small-molecule PARP inhibitors potentiates DNA damage caused by cytotoxic chemotherapies and radiation therapy5.
Veliparib (ABT-888) is a novel oral PARP inhibitor that is being developed for the treatment of hematologic malignancies and solid tumors primarily as combination therapy. However, patients with BRCA 1/2-mutated cancer or PARP sensitive tumors could potentially derive clinical benefit from monotherapy with a PARP inhibitor7,8, as these tumors have compromised ability to repair double-stranded DNA breaks, resulting in an accumulation of DNA strand breaks, which are lethal in these cells.
The pharmacokinetics (PK) of veliparib has been characterized in cancer patients8–10. Renal excretion is the primary route of veliparib elimination, with approximately 70% of the administered oral drug excreted as unchanged parent drug via urine in patients10. In addition to renal excretion, veliparib is metabolized in the liver by at least four cytochrome P450 (CYP450) isoenzymes including CYP2D6, 1A2, 2C19, and 3A411, with CYP2D6 playing the key role in the formation of M8, the primary, active metabolite in humans12. PARP inhibition in peripheral blood mononuclear cells (PBMCs) or peripheral blood lymphocytes (PBLs), measured by poly-ADP-ribose (PAR) formation, has been used as a pharmacodynamic biomarker in the development of PARP inhibitors13–16. Significant inhibition of PAR levels in PBMC was observed in a phase 0 study in patients receiving a single dose of veliparib9.
In light of strong rationale that veliparib can be developed as a monotherapy, a clinical study was conducted to evaluate the safety, tolerability, and PK of single-agent veliparib in patients with either BRCA 1/2-mutated cancer or PARP sensitive tumors. The objectives of this analysis were to (1) characterize the population PK of veliparib and M8 and to assess the potential impact of intrinsic factors on the PK parameters; (2) explore the exposure response relationship for PAR in PBMC.
Methods
Subjects, Study Design, and Treatment
Data was obtained from a phase 1, multicenter, randomized, open-label, dose-escalation study (ClinicalTrials.gov Identifier: NCT00892736) evaluating the safety, tolerability, and PK of chronically-dosed, single-agent veliparib, in patients with either BRCA 1/2-mutated cancer or PARP sensitive tumors. The study protocol was approved by the Institutional Review Boards of the study sites, and written informed consent was obtained from each participant prior to enrollment. The effects of veliparib treatment on the level of PARP inhibition in PBMCs were also determined.
Patients were treated at 1 of 9 different dose levels of oral veliparib without regards to meals at 50/50, 100/50, 100/100, 150/100, 150/150, 200/200, 300/300, 400/400 and 500/500 mg (am/pm dosing in mg) twice daily (b.i.d). Only the morning dose was given on day 1, and b.i.d dosing started on day 2 for at least one cycle (28 days). About 6 patients (range 5–16), including at least 1 patient with a known BRCA germline mutation, were treated at each dose level.
Sampling and Assay for Veliparib and M8 Serum Concentration
Peripheral venous blood samples (4 mL each) were collected on days 1 and 15 of cycle 1 at the following time points: before morning dose, 0.5, 1, 1.5, 2, 3, 4, 6, 8, and 24 h post morning dose administration. For dose level 200/200 mg and above, an additional single PK blood draw was obtained just before the 6th dose on day 4. The plasma concentrations of veliparib and M8 were analyzed using a validated liquid chromatography-mass spectrometric (LC-MS) assay17.
Sampling and assay for PAR levels in PBMCs were obtained on consented patients on days 1 and 28 of cycle 1 before the morning dose and at 2, 4, 8, and 24 h post morning dose. An additional blood draw for PBMC, collected during cycle 4, was only available for a small proportion of patients. PAR concentration was determined by enzyme-linked immunosorbent assay (ELISA)9.
Population Pharmacokinetic Modeling
Software and Estimation Methods
Veliparib and M8 plasma concentration-time data were fitted simultaneously using Phoenix® NLME software (version 1.4, build 7.0.0.2535, Certara USA, Inc; Princeton, NJ, USA), and the first-order conditional estimation–extended least squares method. Data preparation and visualization were performed using R software (version 3.1.1, R Foundation for Statistical Computing). The molecular weights of veliparib and M8 are similar with a ratio of M8 to veliparib of 1.05711. Hence, no corrections for the concentration units were performed and ng/mL was used for analysis purposes.
Base Model
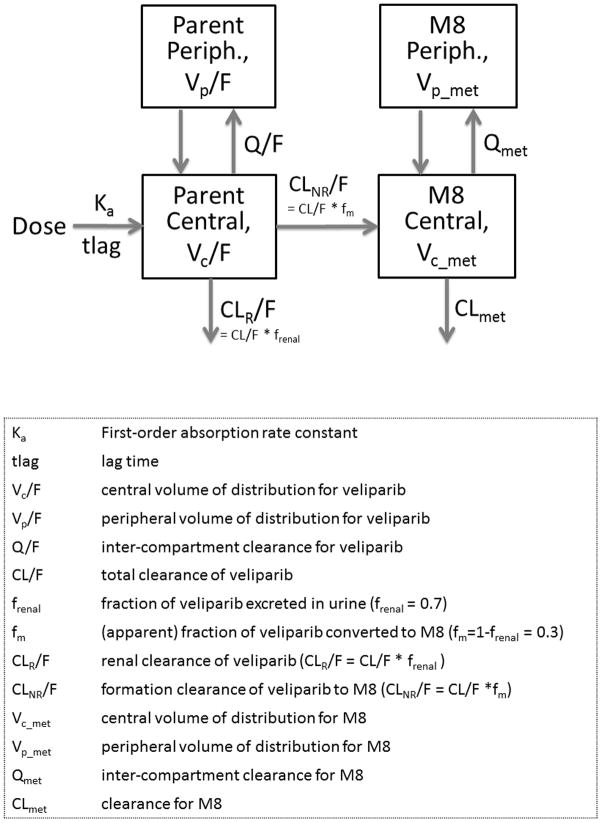
Graphical analysis was conducted to assess trends in the data and one- and two-compartment models were investigated to describe the concentration-time profile of veliparib and M8. Different absorption models (zero- or first-order absorption with or without lag time) were explored. Total clearance for veliparib (CL) was parameterized as renal (CLR) and non-renal (CLNR) clearance, the former was assumed to represent renal excretion and the latter referring to the conversion to metabolites. Using current data, the fraction of veliparib metabolized to M8 (fm) was not identifiable; however, from the literature, it is known that an average of 70% oral veliparib is renally excreted (frenal=70%)9. Thus, veliparib not cleared by renal excretion was assumed to be metabolized into M8 (fm= 1−frenal =30%).
The between-subject variability (BSV) was incorporated using an exponential error structure, as PK parameters are assumed to be log normally distributed:
| (1) |
Where θi is the estimated parameter value (post hoc value) for individual i, θTV is the population mean parameter value, ηi is the between-subject random effects for individual i and is assumed to follow normal distribution with a mean of zero and a variance of ω2. The structure of random effect correlations were explored after incorporating covariates into the base model.
Residual variability was tested as additive, proportional and a combination of the two. Correlation was considered between residual error terms for veliparib and M8 because their quantitation by LC-MS utilized a single, shared internal standard.
After incorporating covariates and correlation between random effects, between-occasion variability (BOV) was also tested, defined for parameter θ in an individual i at occasion j, as shown in Equation 2.
| (2) |
Where θij is the estimated parameter value for the individual i at occasion j, and κij is BOV on parameter θ for individual i at occasion j, and is assumed to follow a normal distribution with a mean of zero and a variance of π2. The BOV was considered because pharmacokinetic sampling was conducted on two occasions for each participant, on study day 1 or day 4 of cycle 1, and on study day 15 of cycle 1.
Base model was selected based on likelihood ratio test and standard goodness of fit plots, including observed concentrations (DV) versus population-predicted concentrations (PRED), DV versus individual-predicted concentrations (IPRED), conditional weighted residuals (CWRES) versus PRED, and CWRES versus time after dose plots.
Covariate Model Development
Once the base model was established, the influence of covariates on the PK parameters was explored. Covariate selection was based on physiological relevance, visual inspection of between-subject random effects–covariate relationships and likelihood ratio test. In the forward addition and backward elimination process, decrease or increase of 6.63 units in the objective function value (OFV) for inclusion or exclusion of one parameter was considered statistically significant (df=1; α=0.01). The covariates that were screened included age, total body weight (WT), lean body mass (LBM)18, body surface area (BSA), creatinine clearance (CLCR), sex, and liver function (ALT, AST and total bilirubin). CLCR and BSA were estimated using Cockcroft–Gault and Mosteller formulas19,20, respectively. Continuous covariates (age, WT, LBM, BSA, liver function and CLCR) and categorical covariate (sex) were investigated as shown in Equation 3 and 4, respectively.
| (3) |
In Equation 3, covi is the value of covariate in individual i, covmed is the median value in the population, and θ cov is the slope factor describing the effect of centered covariate on parameter θ as a power relationship.
| (4) |
In Equation 4, covi is the value of covariate in individual i, which was coded as 0 for females and 1 for males.
Population Model Qualification
The final model was qualified with a visual predictive check (VPC), quantitative predictive check (QPC)21 and non-parametric bootstrap analysis. The final model was evaluated using VPC by comparing the observed 5th, 50th and 95th percentiles of concentration time profiles with the simulated percentiles obtained from 500 replicates. QPC was performed by graphically overlaying the observed 50th percentiles of Cmax and AUC derived from non-compartmental analysis (NCA) on the distribution of the 50th percentiles derived from 500 final model simulated replicates. AUC calculation was performed by Phoenix WinNonlin®, following its AUC methodology22. The predictability of the final model was deemed acceptable if the observed median Cmax and AUC were in the middle of the distribution of model derived percentiles. Similarly, QPC comparing the observed 25th and 75th percentile with corresponding distribution of simulated percentiles was also performed. Additionally, the robustness of the final model parameter estimates was checked by comparing the model predicted estimates with the bootstrap estimates (n=250).
Evaluations of PAR levels in PBMCs
Baseline and post-veliparib administration PAR levels were measured in PBMCs as indicated previously. PARP inhibition in PBMCs was expressed as a percentage of PARP activity after veliparib administration in subjects. Quintile plots23 were made to explore the veliparib concentration-response relationship for PAR in PBMC, where the concentrations are divided into five equal bins, and mean PAR response in each bin, as well as the 95% confidence interval, are plotted against the mean veliparib concentration in the corresponding bin. As PK and PBMC samples were not collected simultaneously, model predicted individual veliparib and M8 plasma concentrations at PBMC sampling points were used as pharmacokinetic endpoint and PARP inhibition was defined as percentage of pre-dose baseline PAR levels.
Results
A population PK model was developed using veliparib (n= 1214) and M8 (n=656) concentrations obtained from 67 patients with BRCA 1/2–mutated cancer or PARP sensitive tumor types (Supplemental Figure S1). A total of 295 PAR concentrations from 41 patients were available for pharmacodynamic (PD) analysis. The demographic and clinical characteristics of the patient population included in the PK/PD analysis are summarized in Table 1. For a few patients (n=20), the estimated CLCR with the Cockcroft-Gault formula19 exceeded 120 mL/min. Their CLCR was assumed to be 120 mL/min as a physiologically reasonable upper limit of CLCR in adults.
Table 1.
Summary of patient characteristics.
| Characteristic | DOSE LEVEL | Total (n=71) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| 50/50 mg (n = 9) | 100/50 mg (n = 6) | 100/100 mg (n = 5) | 150/100 mg (n = 7) | 150/150 mg (n = 6) | 200/200 mg (n = 6) | 300/300 mg (n = 8) | 400/400 mg (n = 16) | 500/500 mg (n = 8) | ||
| Demographics | ||||||||||
| Age, years, mean (range) | 49.2 (28.0 – 64.0) | 51.0 (39.0 – 61.0) | 59.8 (49.0 – 84.0) | 56.1 (47.0 – 64.0) | 50.0 (37.0 – 62.0) | 58.2 (50.0 – 64.0) | 50.9 (33.0 – 66.0) | 56.2 (40.0 – 74.0) | 50.5 (37.0 – 65.0) | 53.5 (28.0 – 84.0) |
| Body weight, kg, mean (range) | 70.4 (45.0 – 98.2) | 81.6 (59.0 – 102) | 75.9 (61.4 – 92.1) | 68.9 (46.3 – 92.0) | 64.2 (51.8 – 84.0) | 87.3 (64.5 – 109) | 90.1 (58.3 – 113) | 73.4 (51.3 – 119) | 66.2 (47.0 – 92.2) | 74.9 (45.0 – 119) |
| Lean body mass, kg, mean (range) | 47.5 (36.9 – 62.6) | 49.2 (39.8 – 53.5) | 47.5 (43.1 – 54.4) | 45.3 (37.3 – 51.2) | 45.3 (42.9 – 48.4) | 52.4 (43.2 – 61.3) | 52.4 (44.2 – 58.3) | 47.4 (36.6 – 63.7) | 42.6 (38.6 – 49.8) | 47.6 (36.6 – 63.7) |
| Height, cm, mean (range) | 167 (160 – 177) | 163 (157 – 175) | 163 (155 – 174) | 163 (155 – 170) | 167 (160 – 172) | 167 (161 – 173) | 165 (160 – 179) | 165 (154 – 175) | 159 (152 – 165) | 164 (152 – 179) |
| Body mass index, kg/m2, mean (range) | 25.2 (17.6 – 31.9) | 30.8 (20.0 – 40.9) | 28.7 (20.3 – 35.8) | 25.8 (18.1 – 35.5) | 23.3 (18.0 – 32.8) | 31.1 (24.8 – 36.4) | 33.3 (20.7 – 44.1) | 26.9 (19.4 – 40.6) | 26.5 (17.8 – 37.2) | 27.8 (17.6 – 44.1) |
| Sex, n (%) | ||||||||||
| Female | 8 (89%) | 6 (100%) | 5 (100%) | 7 (100%) | 6 (100%) | 6 (100%) | 8 (100%) | 15 (94%) | 8 (100%) | 69 (97%) |
| Male | 1 (11%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (6%) | 0 (0%) | 2 (3%) |
| Clinical | ||||||||||
| Serum Creatinine, mg/dL, mean (range) | 0.77 (0.65–1.15) | 0.85 (0.56–1.00) | 0.80 (0.50–1.32) | 0.85 (0.57–1.10) | 0.69 (0.47–0.90) | 1.00 (0.60–1.37) | 0.79 (0.62–1.37) | 0.71 (0.40–1.30) | 0.73 (0.50–0.80) | 0.78 (0.4–1.37) |
| CLCR, mL/min*, mean (range) | 96.8 (71.3 – 120) | 95.4 (61.8 – 120) | 92.8 (66.7 – 120) | 89.6 (48.5 – 120) | 98.4 (74.5 – 120) | 85.4 (51.6 – 120) | 108 (72.0 – 120) | 95.4 (48.9 – 120) | 90.7 (65.9 – 120) | 95.2 (48.5 – 120) |
| Albumin (g/L), mean (range) | 3.51 (2.00 – 4.30) | 3.95 (3.60 – 4.30) | 3.72 (3.10 – 4.40) | 3.87 (2.60 – 4.50) | 3.80 (3.20 – 4.40) | 3.73 (2.70 – 4.20) | 3.88 (3.20 – 4.40) | 3.67 (2.60 – 4.30) | 3.93 (3.60 – 4.20) | 3.77 (2.00 – 4.50) |
| ALT (U/L), mean (range) | 28.2 (7.00 – 73.0) | 25.5 (8.00 – 55.0) | 17.8 (15.0 – 23.0) | 40.4 (13.0 – 124) | 27.8 (10.0 – 45.0) | 27.5 (14.0 – 42.0) | 20.4 (13.0 – 39.0) | 32.2 (13.0 – 135) | 27.9 (17.0 – 43.0) | 28.3 (7.00 – 135) |
| AST (U/L), mean (range) | 44.4 (21.0 – 117) | 31.3 (18.0 – 49.0) | 28.2 (18.0 – 53.0) | 44.0 (21.0 – 81.0) | 34.8 (20.0 – 55.0) | 29.3 (22.0 – 52.0) | 23.4 (11.0 – 32.0) | 33.0 (13.0 – 76.0) | 32.4 (21.0 – 54.0) | 33.7 (11.0 – 117) |
| Total bilirubin (mg/dL), mean (range) | 0.39 (0.10 – 0.70) | 0.40 (0.30 – 0.70) | 0.56 (0.30 – 0.90) | 0.49 (0.30 – 0.90) | 0.50 (0.30 – 0.90) | 0.67 (0.40 – 0.80) | 0.47 (0.20 – 0.80) | 0.49 (0.20 – 1.00) | 0.64 (0.20 – 1.10) | 0.51 (0.10 – 1.10) |
| ECOG performance Status, n (%) | ||||||||||
| 0 | 4 (44%) | 2 (33%) | 3 (60%) | 4 (57%) | 3 (50%) | 3 (50%) | 5 (63%) | 12 (75%) | 7 (88%) | 43 (61%) |
| 1 | 4 (44%) | 2 (33%) | 1 (20%) | 3 (43%) | 3 (50%) | 3 (50%) | 2 (25%) | 3 (19%) | 1 (13%) | 22 (31%) |
| 2 | 1 (11%) | 2 (33%) | 1 (20%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (13%) | 1 (6%) | 0 (0%) | 6 (8%) |
Notes: CLCR was estimated using Cockcroft–Gault formula19. Estimated CLCR values higher than 120 were assigned a value of 120 mL/min as a reasonable upper limit for the analysis.
67 of the 71 patients had pharmacokinetic samples for veliparib, and 38 out of the 67 had M8 pharmacokinetic samples collected.
41 of the 71 patients had PAR levels in PBMC collected. 4 patients did not have pharmacokinetic results but had PAR levels in PBMC.
Dose level was depicted as am/pm dose in mg.
The population PK model was developed for veliparib and its metabolite, M8, simultaneously. A two-compartment model with first order absorption and lag time adequately described the concentration time profile of veliparib. In comparison with a one-compartment model, a two-compartment model decreased the OFV by 173 units (df=4, P<0.001, Supplemental Table S1) and improved the goodness of fit plots, thus was considered as the structural model. The M8 PK was adequately described with a two-compartment model. Figure 1 shows the schematic of simultaneous drug-metabolite model used to describe the PK of veliparib and M8. Between-subject variability was estimated on Ka, tlag, Vc/F, Vp/F, CLR/F, CLNR/F, Vc_met, CLmet and Vp_met, while that on Q and Qmet was not estimated in the final model. Correlation between random effects of CLNR/F, CLR/F and Vc/F decreased the OFV by 64 units but worsened the parameter precision and increased the condition number to 108. Hence, it was dropped from further analysis. Proportional plus additive residual error model best accounted for the unexplained variability of the observed concentrations for veliparib and M8. The expected correlation between residual errors of parent and metabolite, due to sampling from the same matrix, was accounted using a fixed effect correlation term (Table 2).
Figure 1.
Overview of final population pharmacokinetic model for veliparib and its major metabolite M8.
Table 2.
Final parameter estimates and the bootstrapped parameter estimates(95% CI) from 250 replicates.
| Parameter | Estimate | %RSE | Bootstrap estimate | ||
|---|---|---|---|---|---|
| Median | 2.5% CI | 97.5% CI | |||
| Fixed effects | |||||
| Ka (hr−1) | 2.02 | 16.5 | 2.02 | 1.59 | 2.93 |
| tlag (h) | 0.272 | 9.53 | 0.270 | 0.241 | 0.318 |
| Vc/F (L) | 99.2 | 6.91 | 99.1 | 89.5 | 119.6 |
| Vp/F (L) | 47.8 | 10.2 | 47.5 | 31.2 | 56.7 |
| Q/F (L·h−1) | 17.9 | 18.1 | 17.9 | 11.4 | 23.6 |
| CL/F (L·h−1) | 17.3 | 3.85 | 17.3 | 15.9 | 19.1 |
| frenal | 0.7 (fix) | - | - | - | - |
| fm | 0.3 (fix) | - | - | - | - |
| Vc_met (L) | 23.6 | 16.6 | 23.5 | 16.3 | 33.8 |
| Vp_met (L) | 51.4 | 15.4 | 51.3 | 33.0 | 70.2 |
| Qmet (L·h−1) | 29.0 | 18.2 | 29.0 | 21.0 | 43.6 |
| CLmet (L·h−1) | 22.8 | 7.74 | 22.7 | 19.2 | 28.8 |
| LBM effect on Vc/F, power_LBM | 1.21 | 20.8 | 1.21 | 0.72 | 1.56 |
| CLCR effect on CLrenal/F, power_CLcr | 0.903 | 20.1 | 0.901 | 0.525 | 1.33 |
| Between-subject variability | |||||
| ω for Ka (CV%) | 78.1 % | 33.7 | 78.1% | 64.0% | 114.4% |
| ω for tlag(CV%) | 45.7 % | 22.3 | 45.6% | 24.6% | 45.9% |
| ω Vc/F (CV%) | 26.7 % | 29.4 | 26.7% | 18.7% | 35.3% |
| ω Vp/F (CV%) | 38.3 % | 47.6 | 38.3% | 29.0% | 47.3% |
| ω CLR/F (L·h−1) | 34.7 % | 29.5 | 34.7% | 27.2% | 45.2% |
| ω CLNR/F (CV%) | 47.1 % | 27.9 | 47.1% | 34.6% | 57.4% |
| ω Vc_met (CV%) | 61.9 % | 35.5 | 62.0% | 47.2% | 76.7% |
| ω Vp_met (CV%) | 66.3 % | 31.7 | 66.3% | 42.4% | 85.4% |
| ω CLmet (CV%) | 32.8 % | 22.2 | 32.8% | 21.6% | 39.0% |
| PK Residual error | |||||
| Proportional error for veliparib (%CV), CMultStdev | 25.1% | - | 26.0% | 21.4% | 30.5% |
| Proportional error for M8 (%CV), MultStdev | 100% (fix) | - | - | - | - |
| Ratio between veliparib and M8 Proportional error, Ratio | 0.808 | - | 0.806 | 0.639 | 0.926 |
| Additive error for veliparib (ng/mL) | 0.607 | - | 0.183 | 0 | 0.607 |
| Additive error for M8 (ng/mL) | 3.37 | - | 3.37 | 1.60 | 5.60 |
| OFV = −3608, Subjects = 67, Concentration observations = 1214 for Veliparib and M8 concentration observations = 656 | |||||
- typical , where mL/min was the unit of CLCR.
- typical CLNR /F=CL/F · fm,
- typical , where kg was the unit of LBM.
The proportional residual error for M8 is expanded as: CMultStdev·Ratio·MultStdev, where MultStdev is the coefficient variation for proportional error for M8 and the value (%CV) was fixed as 100%.
%RSE was calculated from bootstrap results.
Clinically relevant covariates that depicted definitive trend with the random effects of the PK parameters were evaluated using forward addition and backward elimination. LBM and CLCR on Vc/F and CLR/F, respectively, were found to be significant covariates (ΔOFV = −23 and ΔOFV = −11, df=1, P<0.01, Supplemental Table S1). The coefficient for the effect of LBM on Vc/F and CLCR on CLR/F were estimated to be 1.21 and 0.903, close to a linear relationship. Covariates explained 17% and 16% of the variability on CLR/F and Vc/F, respectively, relative to the base model. Supplemental Figure S2 shows that the inclusion of covariates in the final model accounted for the trend observed between the random effects of veliparib PK parameters and LBM and CLCR in the base model. Adding BOV to the model did not result in significant decrease in OFV and hence was not included. Table 2 shows the parameter estimates for the final model. The final equations for the typical values of CLR/F, CLNR/F and Vc/F were as follows:
| (5) |
where mL/min is the unit of CLCR.
| (6) |
| (7) |
where kg is the unit of LBM.
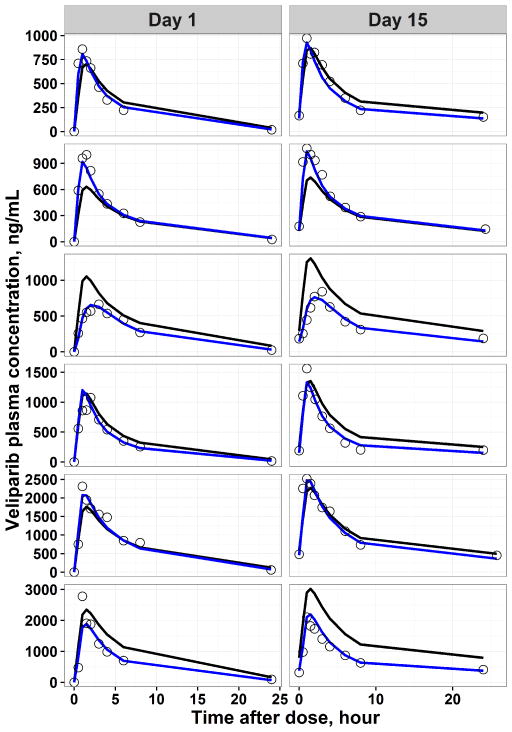
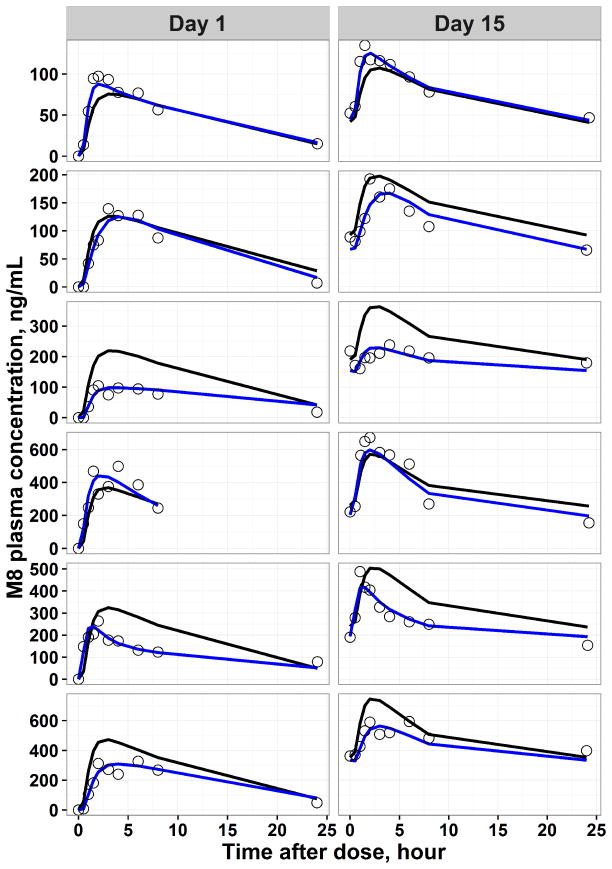
Final model predicted PK profile in representative individuals and the goodness of fit plots are shown in Figure 2–Figure 3 and Supplemental Figure S3, respectively. The individual predicted concentrations for veliparib and M8 are in good agreement with the observed concentrations. In order to evaluate the precision of estimated PK parameters, a non-parametric bootstrap analysis was performed. The population estimates of the final model showed close agreement with the median obtained from the bootstrap replicates and were within the 2.5th to 97.5th percentile obtained from bootstrap (Table 2), indicating model stability.
Figure 2.
Representative individual pharmacokinetic observations (black dots), population predicted (black line) and individual predicted (blue line) pharmacokinetic profiles derived from the final parent-metabolite model for veliparib. The horizontal axis represented time after the morning dose on respective day.
Figure 3.
Representative individual pharmacokinetic observations (black dots), population predicted (black line) and individual predicted (blue line) pharmacokinetic profiles derived from the final parent-metabolite model for M8. The horizontal axis represented time after the morning dose on respective day.
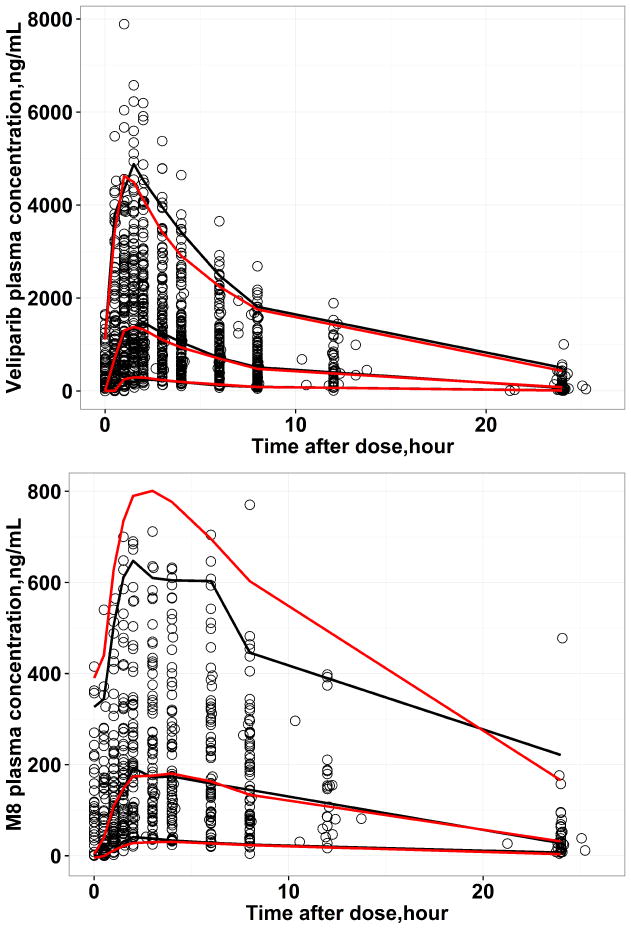
Figure 4 shows the VPC of the final model. There is a substantial overlap between the 5th, 50th and the 95th percentiles of the observed and the simulated data indicating the simulated data from the final model can reproduce the observed data. However, for M8, there were minor over predictions at the 95th percentile of the observed data. Similar results were observed for QPC, that the observed Cmax and AUC were at the center of predicted distribution of the 25th, 50th and 75th percentile of Cmax and AUC from 500 replicated datasets for both veliparib and M8, indicating the final model could predict the central tendency as well as the variability of the observed data adequately (Figure 5 & Supplemental Figure S3–10).
Figure 4.
Visual predictive check (VPC) for 500 replicated datasets for veliparib (top) and M8 (bottom), respectively. The black lines represent the 5%, 50% and 95% observation intervals and the red lines represent the 5%, 50% and 95% prediction intervals.
Figure 5.
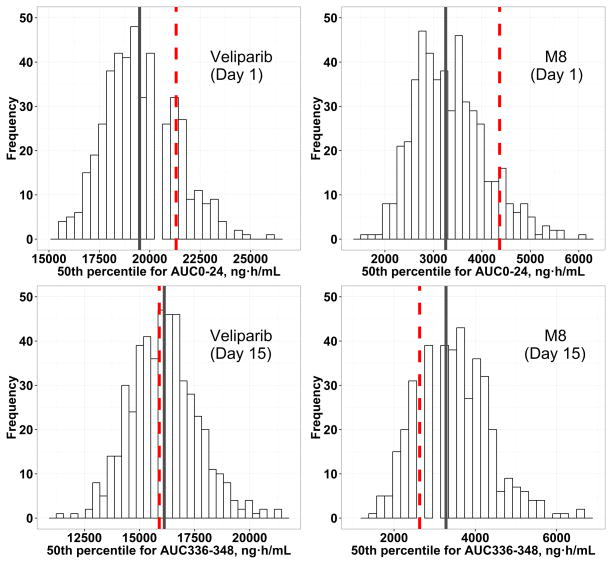
Quantitative predictive check (QPC) for 500 replicated datasets for veliparib and M8. The histograms represent the distribution of the 50th percentile of AUC on day 1 or day 15 from 500 replicated datasets after administration of the 400/400 mg dose. The red comparison lines represent the observed median AUC on day 1 or day 15 from the original dataset, while the grey comparison lines represent the median values of simulated AUC on day 1 or day 15.
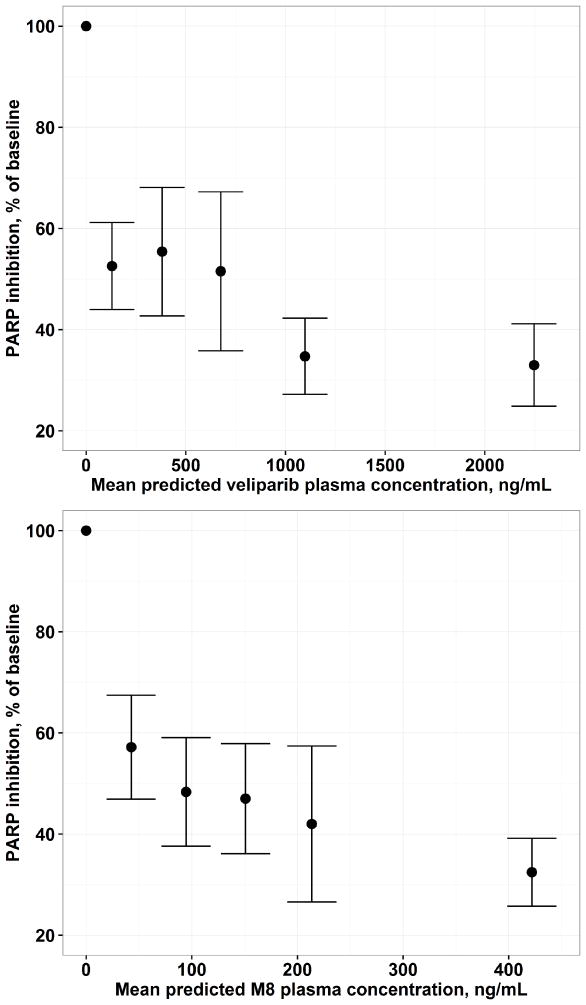
Figure 6 depicts the concentration-response relationship for PAR in PBMC. Inhibition of PAR by ~ 50%, as compared with the pre-dose baseline, was observed in patients treated at the 50/50 mg dose level, and the inhibition of PARP gradually increased to ~ 70% when the dose increased to 500/500 mg.
Figure 6.
Exposure-response relationship for PAR in PBMC based on individual predicted veliparib (top) and M8 (bottom) plasma concentrations from the final pharmacokinetic model. The vertical axis represented PAR activity as percentage of baseline.
Discussion
Veliparib is a potent oral PARP inhibitor, and it has been predominantly studied in combination with cytotoxic agents and radiation therapy for the treatment of a wide range of malignancies18,19,12,20. However, limited information exists regarding the PK and PD of chronically-dosed, single-agent veliparib, in patients with either BRCA 1/2 –mutated cancer or PARP sensitive tumor types. Based on data from a phase 1 clinical trial, we developed a simultaneous population PK model for veliparib, as well as its primary, active metabolite M8. An effect of increasing CLR/F and Vc/F with increasing CLCR and LBM, respectively, were identified to be the most significant covariates. The PD analysis used PAR as the PD biomarker13–15 and inhibition of PAR level was observed in PBMCs.
Individual model predictions of representative parent-metabolite PK profiles (Figure 2–Figure 3), goodness-of-fit plots (Supplemental Figure S2) and visual/quantitative predictive check (Figure 4–Figure 5 and Supplemental Figure S3–10) suggested an adequate overall fit of the final population PK model to the data. All structural parameters were precisely estimated with relative standard errors (%RSE) within 20%. With parameters standardized to 48 kg in LBM and 95 mL/min in CLCR, the final model presented herein predicts values of 17.3 L/h and 147 L for CL/F and Vd/F (=Vc/F+ Vp/F), respectively. The estimated parameters from the population PK model are in good agreement with those previously reported based on NCA (18 L/h and 145 L, respectively)26 and on population PK analysis by Salem et al (20.9 L/h and 173 L, respectively) or by Li et al (13.3 L/h and 126 L, respectively) using a one-compartment model10,27. In our case, the superiority of a two-compartment model was well indicated by decrease of OFV and improvement of goodness of fit plots. Dose proportionality in dose level from 50 to 500 mg was supported herein, and no induction or suppression of its own clearance is associated with chronic dosing of veliparib. Negligible accumulation was observed within the dose range tested using the NCA approach (manuscript in preparation).
Given that about 70% of veliparib was recovered in the urine as unchanged in cancer patients receiving 50 mg single oral dose9, it is not surprising that renal function, measured with estimated creatinine clearance, was identified as the predictor on the CLR/F, which explained 17% of the between subject variability. A similar finding has recently been reported by Salem et al. from population PK analysis of 325 patients with solid tumors10, in which creatinine clearance was modelled with the exponent of 0.48 on oral clearance. The predominant role of renal function on veliparib clearance was supported by Li et al by a physiologically based pharmacokinetic (PBPK) model integrated with a mechanistic kidney module27. According to our covariate model, patients with mild (~25% decrease in CLCR) and moderate (~50% decrease in CLCR) renal function are associated with ~10% and ~20% increase in veliparib exposure, respectively. Whether patients with renal impairment need dose adjustment will rely on the tolerability profile of veliparib with therapeutic dose in targeted patient populations. An ongoing phase 1 study in cancer patients with renal dysfunction will shed light on the impact of varying degree of renal dysfunction (NCT01366144). After adding CLCR on CLR/F, none of the other patient demographic measures, such as body weight, LBM or age, were identified as significant covariates due to possible collinearity with CLCR.
Instead of total body weight, LBM was found to be a significant covariate and was considered in explaining veliparib PK variability on Vc/F, similar to that reported previously10. Given the hydrophilic nature of veliparib28, our finding supported the results of studies for hydrophilic drugs, that a better correlation between LBM and volume of distribution than body weight has been found. LBM has been proposed as a better predictor of drug dosage, especially in obese patients29. Nearly 60% of the patients included in our study were either overweight or obese (BMI of 25 or larger). Therefore, the use of LBM over body weight on Vc/F could be attributed to the more accurate description of LBM for the difference in body composition in our patient population.
To date, no report with respect to the volume of distribution of M8 or the fraction of veliparib converted to M8 has been published. We assumed that veliparib not cleared by renal excretion was metabolized to M8. An average of 70% of veliparib oral doses was excreted as the unchanged parent drug in the urine in cancer patients9, thus, the remaining 30% is assumed to be converted to M8 by CYP2D6 metabolism. Furthermore, on the basis of results from an in vitro study that CYP2D6 accounts for ~72% of overall veliparib metabolism11, we expected that the “true” fraction converted to M8 might be 22%, i.e. 30% (percent of non-renal elimination of veliparib ) * 72%.
CYP2D6 polymorphism was not determined for the current study. Based on simulations from a PBPK model, CYP2D6 poor metabolizers (PM) exhibited ~ 20% higher exposure and the ultra-extensive metabolizers (UM) had ~20% lower exposure in comparison with the extensive metabolizers27. Given that CYP2D6 metabolism contributes to ~22% of veliparib total clearance, changes in CYP2D6 activity caused by polymorphism is likely to have insignificant influence on veliparib PK. The impact of CYP2D6 deficiency, when collectively evaluated with other factors (i.e., LBM and CLCR ), has the potential to be clinically meaningful and needs further evaluation.
PARP inhibitors are believed to have adequate activity as monotherapy in tumors with defects in homologous recombination such as those with BRCA mutation or PARP sensitive cancer types16. PARP inhibition in surrogate samples, including PBMCs or PBLs, reflects the inhibition of PAR formation in tumor tissues, thus has been widely used in the development of PARP inhibitors13–16. In the present analysis, with increase in veliparib and M8 concentrations, a trend in decrease in PAR levels in PBMC was observed. We attempted to quantitate the exposure-PAR relationship with the concentrations of veliparib as driver for PAR reduction using an Emax model, assuming a direct effect between exposures and PAR reduction. A similar structure model was reported by Wang et al to describe the inhibitory effect of rucaparib, a novel PARP inhibitor, on PAR formation15. However, an extremely low IC50 (30 ng/mL) was estimated with low precision. The doses studied were higher such that PAR inhibition was approaching plateau even with the lowest dose. The estimated IC50 was higher than the lower limit of quantification for veliparib (10 ng/mL), but limited data was available in the range of 10–30 ng/mL. Therefore, only exploratory exposure-PAR results are presented. The concentration of the parent was used as driver since it has been reported that M8 is approximately 5-fold less potent as compared to veliparib30. Furthermore, the concentrations of veliparib and M8 are correlated. Exposure-PAR in PBMC relationship needs to be further characterized, and ideally when applied with cytotoxic agents which would be expected to increase the level of PARP activity susceptible to inhibition by veliparib. The exposure-PAR relationship depicts the mechanism of action of veliparib. However, the relationship between level of PAR inhibition and efficacy is unknown as shown in the case of approved PARP inhibitors such as rucaparib and olaparib15,30.
Conclusions
In conclusion, we characterized the PK of veliparib and M8, the primary, active metabolite, using data collected in patients with BRCA 1/2-mutated cancer or PARP sensitive tumors, and evaluated the effect of patient demographics and clinical factors on veliparib PK parameters. CLCR and LBM were identified as significant predictors of veliparib CLR/F and Vc/F, respectively. The exploratory PK/PD analysis demonstrated that veliparib inhibits PAR levels in PBMC supporting the intended mechanism of action.
Supplementary Material
Acknowledgments
Grant UM1-CA186690 (NCI-CTEP) and support from AbbVie. This project used the UPCI Cancer Pharmacokinetics and Pharmacodynamics Facility (CPPF) and was supported in part by award P30-CA47904. Work was supported by the National Cancer Institute [contract number HHSN261200800001E].
Footnotes
ACCP fellows: Jogarao Gobburu is a fellow of the American College of Clinical pharmacology (ACCP)
Disclosures:
JN: None
CS: None
SM: None
SK: None
SP: Research support and consulting from AbbVie.
BK: None
JJ: None
EC: None
MG: None
VI: None
JG: None
JB: Research support from AbbVie.
References
- 1.Jubin T, Kadam A, Jariwala M, et al. The PARP family: insights into functional aspects of poly (ADP-ribose) polymerase-1 in cell growth and survival. Cell Prolif. 2016;49(4):421–437. doi: 10.1111/cpr.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee J–, Ledermann JA, Kohn EC. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann Oncol. 2014;25(1):32–40. doi: 10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean E, Middleton MR, Pwint T, et al. Phase I study to assess the safety and tolerability of olaparib in combination with bevacizumab in patients with advanced solid tumours. Br J Cancer. 2012;106(3):468–474. doi: 10.1038/bjc.2011.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Murcia JM, Niedergang C, Trucco C, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94(14):7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandhu SK, Yap TA, de Bono JS. Poly(ADP-ribose) polymerase inhibitors in cancer treatment: A clinical perspective. Eur J Cancer. 2010;46(1):9–20. doi: 10.1016/j.ejca.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 6.van der Noll R, Marchetti S, Steeghs N, et al. Long-term safety and anti-tumour activity of olaparib monotherapy after combination with carboplatin and paclitaxel in patients with advanced breast, ovarian or fallopian tube cancer. Br J Cancer. 2015;113(3):396–402. doi: 10.1038/bjc.2015.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner N, Tutt A, Ashworth A. Hallmarks of “BRCAness” in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 8.Mizugaki H, Yamamoto N, Nokihara H, et al. A phase 1 study evaluating the pharmacokinetics and preliminary efficacy of veliparib (ABT-888) in combination with carboplatin/paclitaxel in Japanese subjects with non-small cell lung cancer (NSCLC) Cancer Chemother Pharmacol. 2015;76(5):1063–1072. doi: 10.1007/s00280-015-2876-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kummar S, Kinders R, Gutierrez ME, et al. Phase 0 Clinical Trial of the Poly (ADP-Ribose) Polymerase Inhibitor ABT-888 in Patients With Advanced Malignancies. J Clin Oncol. 2009;27(16):2705–2711. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salem AH, Giranda VL, Mostafa NM. Population pharmacokinetic modeling of veliparib (ABT-888) in patients with non-hematologic malignancies. Clin Pharmacokinet. 2014;53(5):479–488. doi: 10.1007/s40262-013-0130-1. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Delzer J, Voorman R, de Morais SM, Lao Y. Disposition and drug-drug interaction potential of veliparib (ABT-888), a novel and potent inhibitor of poly(ADP-ribose) polymerase. Drug Metab Dispos Biol Fate Chem. 2011;39(7):1161–1169. doi: 10.1124/dmd.110.037820. [DOI] [PubMed] [Google Scholar]
- 12.Wiegand R, Wu J, Sha X, LoRusso P, Li J. Simultaneous determination of ABT-888, a poly (ADP-ribose) polymerase inhibitor, and its metabolite in human plasma by liquid chromatography/tandem mass spectrometry. J Chromatogr B. 2010;878(3–4):333–339. doi: 10.1016/j.jchromb.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plummer R, Jones C, Middleton M, et al. Phase I Study of the Poly(ADP-Ribose) Polymerase Inhibitor, AG014699, in Combination with Temozolomide in Patients with Advanced Solid Tumors. Am Assoc Cancer Res. 2008;14(23):7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kummar S, Kinders R, Gutierrez ME, et al. Phase 0 Clinical Trial of the Poly (ADP-Ribose) Polymerase Inhibitor ABT-888 in Patients With Advanced Malignancies. J Clin Oncol. 2009;27(16):2705–2711. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang DD, Li C, Sun W, et al. PARP activity in peripheral blood lymphocytes as a predictive biomarker for PARP inhibition in tumor tissues – A population pharmacokinetic/pharmacodynamic analysis of rucaparib. Clin Pharmacol Drug Dev. 2015;4(2):89–98. doi: 10.1002/cpdd.176. [DOI] [PubMed] [Google Scholar]
- 16.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 17.Parise RA, Shawaqfeh M, Egorin MJ, Beumer JH. Liquid chromatography-mass spectrometric assay for the quantitation in human plasma of ABT-888, an orally available, small molecule inhibitor of poly(ADP-ribose) polymerase. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;872(1–2):141–147. doi: 10.1016/j.jchromb.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheymol G. Effects of obesity on pharmacokinetics implications for drug therapy. Clin Pharmacokinet. 2000;39(3):215–231. doi: 10.2165/00003088-200039030-00004. [DOI] [PubMed] [Google Scholar]
- 19.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16(1):31–41. doi: 10.1159/000180580. [DOI] [PubMed] [Google Scholar]
- 20.Mosteller RD. Simplified calculation of body-surface area. N Engl J Med. 1987;317(17):1098. doi: 10.1056/NEJM198710223171717. [DOI] [PubMed] [Google Scholar]
- 21.Jadhav PR, Gobburu JVS. A new equivalence based metric for predictive check to qualify mixed-effects models. AAPS J. 2005;7(3):E523–E531. doi: 10.1208/aapsj070353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Certara USA, Inc. Phoenix WinNonlin® User’s Guide. [Google Scholar]
- 23.Mehrotra S, JF, Gobburu J. Don’t Get Boxed In: Commentary on the Visual Inspection Practices to Assess Exposure-Response Relationships From Binary Clinical Variables. J Clin Pharmacol. 2012;52(12):1912–1917. doi: 10.1177/0091270011429568. [DOI] [PubMed] [Google Scholar]
- 24.Kummar S, Ji J, Morgan R, et al. A phase I study of veliparib in combination with metronomic cyclophosphamide in adults with refractory solid tumors and lymphomas. Clin Cancer Res Off J Am Assoc Cancer Res. 2012;18(6):1726–1734. doi: 10.1158/1078-0432.CCR-11-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phase I safety and pharmacokinetic (PK) study of veliparib in combination with whole brain radiation therapy (WBRT) in patients (pts) with brain metastases. [Accessed September 28, 2015];J Clin Oncol. http://meetinglibrary.asco.org/content/96956-114.
- 26.Kummar S, Ji J, Morgan R, et al. A Phase I Study of Veliparib in Combination with Metronomic Cyclophosphamide in Adults with Refractory Solid Tumors and Lymphomas. Am Assoc Cancer Res. 2012;18(6):1726–1734. doi: 10.1158/1078-0432.CCR-11-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Kim S, Sha X, Wiegand R, Wu J, LoRusso P. Complex Disease–, Gene–, and Drug–Drug Interactions: Impacts of Renal Function, CYP2D6 Phenotype, and OCT2 Activity on Veliparib Pharmacokinetics. Clin Cancer Res. 2014;20(15):3931–3944. doi: 10.1158/1078-0432.CCR-14-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones P. Profiling PARP inhibitors. Nat Biotechnol. 2012;30(3):249–250. doi: 10.1038/nbt.2138. [DOI] [PubMed] [Google Scholar]
- 29.Morgan DJ, Bray KM. Lean body mass as a predictor of drug dosage. Implications for drug therapy. Clin Pharmacokinet. 1994;26(4):292–307. doi: 10.2165/00003088-199426040-00005. [DOI] [PubMed] [Google Scholar]
- 30.Penning TD, Zhu G-D, Gandhi VB, et al. Discovery of the Poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J Med Chem. 2009;52(2):514–523. doi: 10.1021/jm801171j. [DOI] [PubMed] [Google Scholar]
- 31.Brown JS, Kaye SB, Yap TA. PARP inhibitors: the race is on. Br J Cancer. 2016;114(7):713–715. doi: 10.1038/bjc.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.