Abstract
Purpose
Aurora kinase A (AURKA) is overexpressed in several cancer types, making it an attractive druggable target in clinical trials. In this study, we investigated the role of AURKA in regulating EIF4E, cap-dependent translation, and resistance to mTOR inhibitor, RAD001 (everolimus).
Experimental design
Tumor xenografts and in vitro cell models of upper gastrointestinal adenocarcinomas (UGCs) were used to determine the role of AURKA in activation of EIF4E and cap-dependent translation. Overexpression, knockdown, and pharmacologic inhibition of AURKA were used in vitro and in vivo.
Results
Using in vitro cell models, we found that high protein levels of AURKA mediate phosphorylation of EIF4E and upregulation of c-MYC. Notably, we detected overexpression of endogenous AURKA in everolimus-resistant UGC cell models. AURKA mediated phosphorylation of EIF4E, activation of cap-dependent translation, and an increase in c-MYC protein levels. Targeting AURKA using genetic knockdown or a small molecule inhibitor, alisertib, reversed these molecular events, leading to a decrease in cancer cell survival in acquired and intrinsic resistant cell models. Mechanistic studies demonstrated that AURKA binds to and inactivates protein phosphatase 2A (PP2A), a negative regulator of EIF4E, leading to activation of EIF4E and resistance to everolimus in an AKT-, ERK1/2-, and mTOR-independent manner. Data from tumor xenograft mouse models confirmed that everolimus-resistant cancer cells are sensitive to alisertib.
Conclusion
Our results indicate that AURKA plays an important role in activation of EIF4E and cap-dependent translation. Targeting AURKA-EIF4E-c-MYC axis using alisertib is a novel therapeutic strategy that can be applicable for everolimus-resistant tumors and/or subgroups of cancers that show overexpression of AURKA and activation of EIF4Eand c-MYC.
Introduction
Aurora Kinase A (AURKA) is a serine threonine kinase that is frequently amplified and/or overexpressed in several cancer types, including upper gastrointestinal adenocarcinomas (UGCs)(1–3). Interestingly, the aberrant overexpression of AURKA in cancer cells is associated with gain of novel oncogenic functions that extend beyond its normal physiological functions in forming and stabilizing mitotic spindles during cell division (4). Overexpression of AURKA in cancer cells leads to inhibition of tumor suppressors such as p53 and p73 (5–7). Recent studies have shown that overexpression of AURKA in cancer cells denotes aberrant interactions and novel oncogenic functions mediated by its kinase activity that include activation of oncogenic pathways such as NF-κB, HDM2, β-catenin, and STAT3 (1, 2, 8–12). High levels of AURKA mediate resistance to traditional first line chemotherapeutic agents such as docetaxel and 5-FU in colorectal and breast cancers (13, 14). Because of its diverse oncogenic functions, AURKA has become an attractive druggable target. Alisertib, also known as MLN8237, is an investigational small molecule inhibitor of AURKA that has shown promising efficacy in pre-clinical studies (15, 16) leading to its entry into multiple clinical trials for patients with hematologic malignancies and solid tumors (3, 17, 18).
The mammalian target of rapamycin (mTOR) signaling pathway controls several important biological functions such as translation, metabolism, cell growth, and division (19). Eukaryotic translation initiation factor (EIF4E) is a downstream target of mTOR that plays an essential role in regulating cap-dependent translation and promoting cancer cell survival (20–23). Activation of AKT and EIF4E has been shown to play a role in mediating resistance to rapamycinin non-small lung cancer cells (24). Multiple studies indicated the existence of negative and positive feedback loops between AKT and EIF4E (25), and between c-MYC and EIF4E (26), which add to the complexity of EIF4Eregulation. A classical pathway of EIF4E activation involves mitogen-activated protein kinase (MAPK)-interacting kinases (MNK) 1 (27) and MNK2 (28). However, the mechanisms by which EIF4E can be activated independent of mTOR and MAPK are poorly understood.
Chemotherapeutic resistance is a challenging problem in esophageal and gastric cancers (29, 30). Although patients receiving first line therapy may initially respond to treatment, many of them relapse and require a second line of therapy where options are often limited (31). Chemotherapeutic resistance can be attributed to an inherent intrinsic ability of cancer cells to resist the effect of anti-cancer drugs or the development of acquired resistance through mechanisms that include alternations in the target pathways and activation of pro-survival molecules (32). In a large meta-analysis study that included twenty-one studies with a total of 3475 participants, triplet therapy was suggested to be superior to doublet therapy in patients with advanced gastric or esophageal cancer. However, the survival benefit is limited with increased risks for thrombocytopenia, infection, and mucositis (33). Having several factors contribute to development of UGCs and the existence of only few treatment options add extra challenge to our ability to treat them. Currently, the only approved targeted therapies for advanced or metastatic gastroesophageal adenocarcinomas are trastuzumab and ramucirumab (34), which reflect the need to test other available targeted therapies.
In this study, we demonstrated for the first time that AURKA can phosphorylate and activate EIF4E. We also found that cell models of intrinsic and acquired resistance against everolimus have activation of AURKA-EIF4E with an increase in c-MYC protein level. We demonstrate that AURKA activates EIF4E and cap-dependent translation through binding to and inhibiting protein phosphatase 2A (PP2A), a negative regulator of EIF4E. Targeting AURKA-EIF4E-c-MYC axis using alisertib reduced cancer cell survival in vitro and induced tumor regression in a xenograft model of everolimus resistance, suggesting a novel therapeutic strategy for UGCs where other therapeutic options have failed.
Materials and Methods
Cell culture and reagents
Human esophageal FLO-1 and SK-GT-4 and gastric MKN45 adenocarcinoma cell lines were maintained in culture using Dulbecco's modified Eagle's medium (DMEM, GIBCO, Carlsbad, CA) supplemented with 5% fetal bovine serum (Invitrogen Life Technologies, Carlsbad, CA) and 1% penicillin/streptomycin (GIBCO). All cell lines were authenticated using short tandem repeat (STR) profiling (Genetica DNA Laboratories, Burlington, NC). AKT inhibitor, MK2206, MEK inhibitor, trametinib, AURKA inhibitor, alisertib, and mTOR inhibitor, RAD001 (everolimus) were purchased from Selleck chemicals (Houston, TX). p-AURKA (T288), AURKA, p-EIF4E (S209), EIF4E, p-mTOR (S2448), mTOR, p-4EB-P1 (S65), 4EB-P1, p-AKT (S473), AKT, MNK, p-MNK (T197/202), and PP2A antibodies were purchased from Cell Signaling Technology (Beverly, MA). MYC, p-PP2Ac (Y307), P-ERK 1/2 (T202/Y204), ERK antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz Biotechnology Inc. Santa Cruz, CA), and β-Actin antibody from Sigma-Aldrich (GmbH). Mouse or rabbit secondary antibodies were purchased from Promega (Madison, WI).
AURKA overexpression and plasmid
The expression plasmid for AURKA was generated by PCR amplification of the full-length coding sequence of AURKA and cloned in frame into pcDNA3.1 (Invitrogen Life Technologies). A synthetic Flag-tag sequence was added at the N-terminus of AURKA. Cloning of AURKA was confirmed by sequencing and restriction enzyme digestion. Flag-tagged coding sequence of AURKA was sub-cloned into Xba I and BamH I sites of the adenoviral shuttle vector (pACCMV), and the recombinant adenovirus was generated by co-transfecting HEK-293AD (ATCC) cells with the shuttle and adenoviral backbone (pJM17) plasmids using the Calcium Phosphate Transfection Kit (Applied Biological Materials, Inc., Richmond, BC).
Development of acquired RAD001 resistance cell models
FLO-1 and SK-GT-4 cells were treated with increasing doses of RAD001 (starting at 100 nM up to 10 µmol/L) for six months. IC50 for RAD001 was evaluated in parental and resistant cells by using CellTiter-Glo and clonogenic survival assays. Single cell colonies 1 (C1) and 2 (C2) that were developed from RAD001 resistant FLO-1 cells (FLO-1 RAD-R) were used for further investigations. RAD001 resistant SK-GT-4 cells (SK-GT-4 RAD-R (Pool)) were used as a second resistance cell model in this study.
Western blotting
Cells were scraped and centrifuged at 4°C. Pellets were re-suspended in RIPA buffer containing protease inhibitor cocktail and phosphatase inhibitor, sodium orthovanadate, (Santa Cruz Biotechnology Inc.) on ice. Protein concentration was measured using a Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA). Proteins (25 µg) from each sample were subjected to SDS/PAGE and transferred onto nitrocellulose membranes (PerkinElmer, Waltham, MA). Membranes were blocked with 5% bovine serum albumin (BSA, Sigma-Aldrich). To detect target proteins, membranes were probed with specific primary antibodies. Next day, membranes were washed for 10 minutes with TBS-T for 3 times, followed by incubation with anti-rabbit or anti-mouse secondary antibodies. Protein bands were detected using chemiluminescence reagents (Millipore, Billerica, MA).
Immunoprecipitation
Cells were scraped and centrifuged. Pellets were washed with cold Phosphate Buffered Saline (PBS), pH7.4 and re-centrifuged. 500 µL of cell lysis buffer (0.5% Triton x-100, 150 mmol/L NaCl, 5 mmol/L EDTA, and 50 mmol/L Tris supplemented with protease and phosphatase inhibitors) was added to the cells. Cell lysates were sonicated and centrifuged at maximum speed (13000 rpm) for 15 minutes at 4°C. Immunoprecipitation was performed using Dynabead Protein G (Invitrogen Life Sciences) according to the manufacturer’s instructions. AURKA antibody and IgG (Santa Cruz Biotechnology Inc.) were cross-linked to Dynabead Protein G separately. The cell lysate was added to the cross-linked beads and incubated for 90 min with rocking at room temperature. The Dynabeads were then pelleted using a magnet and washed 3 times with washing buffer. Captured proteins were recovered from the beads by elution buffer, followed by adding 7 µL of 4× protein-loading buffer to each sample and boiling for 5 min. Samples were resolved by SDS/PAGE and subjected to Western blotting. The membrane was incubated with AURKA or PP2A antibodies overnight and then with mouse IgG light chain specific antibody (Cell Signaling Technology) for 1 h, and followed by anti-mouse secondary antibody.
Luciferase reporter assays
A dual-Renilla-firefly-luciferase pcDNA3-rLuc-PolioIRES-fLuc reporter (a kind gift from Dr. John Blenis at Harvard Medical School) was used to measure cap-dependent/independent translation (35). 4XEMS-Luc reporter (a kind gift from Dr. Stephen R. Hann at Vanderbilt University School of Medicine) was used to measure the c-MYC transcriptional activity (36). Cells were seeded in 24-well plates. The next day, cells were transiently transfected with 500 ng of either the PolioIRES-fLuc or 4XEMS-Luc reporters and 12.5 ng of β-galactosidase plasmid in all experiments using lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were either transfected with siAURKA or treated with alisertib (0.5 µmol/L) with or without RAD001 (1 µmol/L) treatment for 24 h. Luciferase activity was measured using the Dual-Luciferase Reporter Assay kit (Promega) according to the manufacturer’s instructions. Firefly luciferase activities were normalized to β-galactosidase levels.
Clonogenic cell survival assay
FLO-1 Parental, FLO-1 RAD-R pool, FLO-1 RAD-R (C1 & C2), SK-GT-4, and SK-GT-4 RAD-R (pool) cells were seeded at 1000 cells/well in six-well plates for 24 h and subsequently treated with 0.02, 0.05, 0.1, 0.5, 1, 5, 10, 15, 25, or 50 µmol/L of RAD001 for 24 h. Following treatments, the wells were washed with PBS and cells were incubated in drug-free DMEM cell culture medium for ten days. Subsequently, the media were removed and cells were fixed with 2% Paraformaldehyde solution for 10 min. The cells were then gently washed with PBS and stained overnight with crystal violet (0.05% Crystal Violet in 50% Methanol). Next day, excess dye was gently washed off with PBS, plates were photographed. Colony formation and cell survival were evaluated by quantifying the dye signal in each well with ImageJ image analysis software (https://imagej.nih.gov/ij/).
Cell viability ATP-Glo assay
Cells were seeded at 1000 cells per well in 96-well plates and treated with 0.075, 0.150, 0.315, 0.625, 1.25, 2.5 and 10 µmol/L of RAD001, alisertib or combination for five days. Cell viability was measured by using the CellTiter-Glo Cell Viability Assay (Promega, Madison, WI) according to the manufacturer's protocol. Changes in absorbance were recorded in a FluolarStar luminescence microplate reader (BMG Labtech).
Protein Phosphatase assay
Cells were washed twice in Tris buffer (pH 7.5) and total cellular proteins were extracted in a lysis buffer containing 20 mmol/L imidazole-HCl, 2 mmol/L EDTA, 2 mmol/L EGTA, pH 7.0 with 10 µg/mL each of aprotinin, leupeptin, pepstatin, 1 mmol/L benzamidine, and 1 mmol/L PMSF without phosphatase inhibitors. The PP2A activity was measured using a non-radioactive immunoprecipitation malachite green phosphatase assay kit (Millipore). Okadaic acid (OA, BioVision Milpitas, CA) treatment was performed at 50 nmol/L overnight (16 h). All procedures were performed according to the manufacturer's protocol, and changes in absorbance were recorded at 650 nm in a FluolarStar luminescence microplate reader (BMG Labtech).
Gene silencing by small interfering RNA
Cells were transfected with siScramble (Coralville, Iowa), siAURKA (Ambion, Grand Island, NY), siEIF4E (Ambion), or siPP2Ac (Santa Cruz Biotechnology) in 5% FBS using Lipojet transfection kit (SignaGen Laboratories, Gaithersburg, MD) according to the manufacturer's instructions. Cells were harvested 48 h after transfection.
Translational chromatin immunoprecipitation analysis (TrIP-Chip assay) - Isolation of polysome-associated mRNA transcripts
Isolation of polysome-associated mRNA using TrIP-Chip assay was performed as described previously (37). In brief, cells were harvested with 0.05% trypsin-EDTA and washed twice with ice-cold PBS containing 100 µg/ml cycloheximide. Next, 1 × 107 cells were incubated with 800 µl DMEM, supplemented with 10% FBS and 100 µg/ml cycloheximide, at 37°C for 5 min. Next, 200 µl of cross-linker reagent dithiobis (succinimidyl propionate) (DSP) (Life Technologies) were added and followed by incubation at 37°C for 5 min. Excess DSP was quenched with 1 mol/LM Tris-HCl (pH 7.4). The cells were then washed three times with ice-cold PBS containing 100 µg/ml cycloheximide. The pellet was incubated in 500 µl of low salt buffer (LSB, 20 mmol/L HEPES, pH7.4, 100 mmol/L KCl, 2 mmol/L MgCl2, 1 mmol/L dithiotheritol) for 20 min, followed by addition of 200 µl of lysis buffer (1× LSB containing 1.2% Triton-X100). Half of the lysate was incubated with the IgG-coated beads at 4°C for 2 h. The second half of the lysate was incubated with HSP70/HSP73 antibody-conjugated magnetic beads. The polysome complexes containing translationally active mRNA transcripts were isolated and eluted from beads conjugated with HSP70/HSP73 using the Array Pure Nanoscale RNA Purification Kit (Epicentre, WI, USA).
RNA extraction and real-time RT-PCR
Extraction of total RNA from cells was performed as described previously (8). Quantitative real-time reverse transcriptase PCR (qRT-PCR) was performed using a Bio-Rad CFX Connect Real-time System (Bio-Rad), with the threshold cycle number determined by Bio-Rad CFX manager software version 3.0. Reactions were performed in triplicate, and the threshold cycle numbers were averaged. The mRNA expression results were normalized to HPRT1 housekeeping gene as described previously (8). For polysomal mRNA, the expression of c-MYC was normalized using ribosomal RNA 18S subunit as internal control. The primers used in qRT-PCR analysis are shown in Supplementary Table 1.
In vivo tumor xenograft model
MKN45 cells (5 × 106) suspended in 200 µL DMEM/growth factor-reduced Matrigel mixture (50% DMEM supplemented with 10% FBS and 50% Matrigel) were injected subcutaneously into the flank regions of female SCID Hairless Congenic (SHC) Mice (Charles River Laboratories, Inc., Stone Ridge, NY). The tumors were allowed to grow until 200 mm3 in size before starting treatments with alisertib (40 mg/kg, orally, four times weekly), RAD001 (5 mg/kg, orally, twice a week), or in combination for six weeks. Tumor xenografts were measured every alternate day, and tumor size was calculated according to the following formula: tumor volume = (length × width2)/2. Each treatment group included at least 15 tumor xenografts. The Vanderbilt Institutional Animal Care and Use Committee approved all animal work.
Immunohistochemistry
Following completion of animal treatments, the xenograft tumors were isolated and the immuno-histochemical (IHC) staining of Ki-67 and cleaved caspase 3 proteins was performed as described previously (38). Images were taken by using an Olympus BX51 microscope (Olympus Co. Center Valley, PA). The protein expression levels were evaluated by using the IHC toolbox plugin in ImageJ software (https://imagej.nih.gov/ij/plugins/ihc-toolbox/index.html).
Statistical Analysis
Two samples t-test, was used to compare the statistical difference between two groups. Data were expressed as mean ± standard deviation of 3 independent experiments. Statistical significance of the in vitro studies was analyzed by a Student's t test and analysis of variance. The differences were considered statistically significant when the p value was ≤ 0.05.
Results
AURKA mediates phosphorylation EIF4E and activation of cap-dependent translation
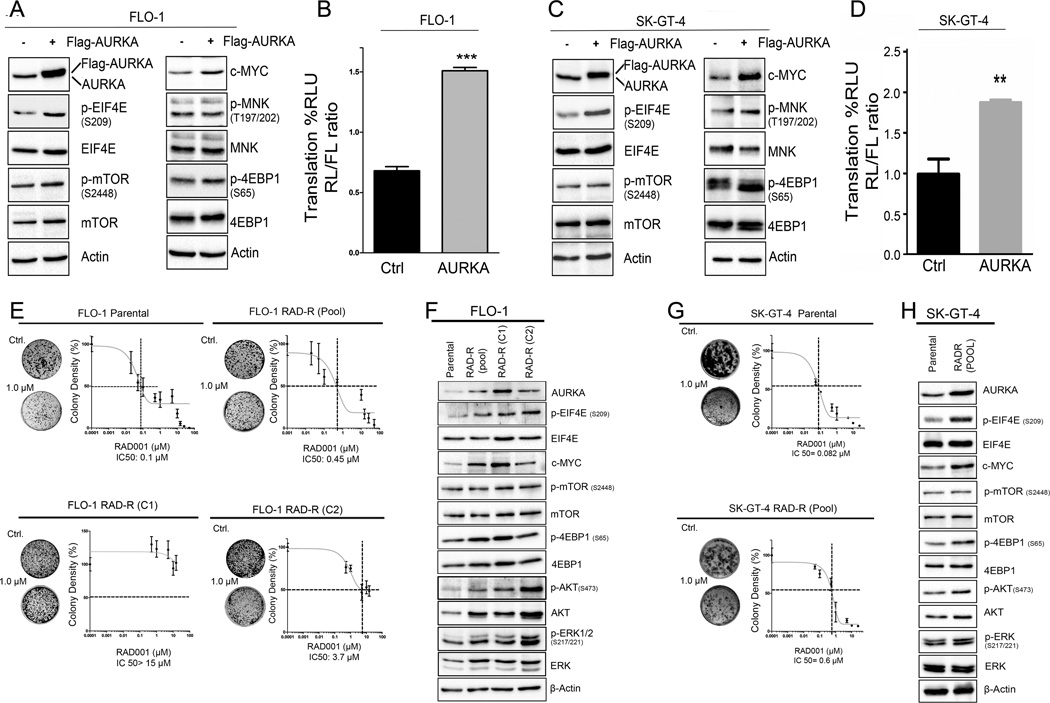
EIF4E plays an important role in cap-dependent translation. To investigate whether elevated AURKA expression can mediate phosphorylation of EIF4E, we transiently overexpressed AURKA in FLO-1 and SK-GT-4 cell lines. The results demonstrated a remarkable increase in the protein levels of phospho-EIF4E (S209) and c-MYC in both cell lines (Figure 1A &1C). Surprisingly, we did not detect obvious changes in total or phospho-protein levels of mTOR, 4EBP1, and MNK, suggesting that regulation of EIF4E by AURKA is independent of these signaling molecules (Figure 1A & 1C). To determine if the increase in phospho-EIF4E regulates cap-dependent translation, we utilized the dual-Renilla-firefly-luciferase pcDNA3-rLuc-PolioIRES-fLuc reporter to measure cap-dependent/independent translation (35). The results confirmed that AURKA-mediated phosphorylation of EIF4E increased the activity of cap-dependent translation reporter in FLO-1 (Figure 1B) and SK-GT-4 cells (Figure 1D).
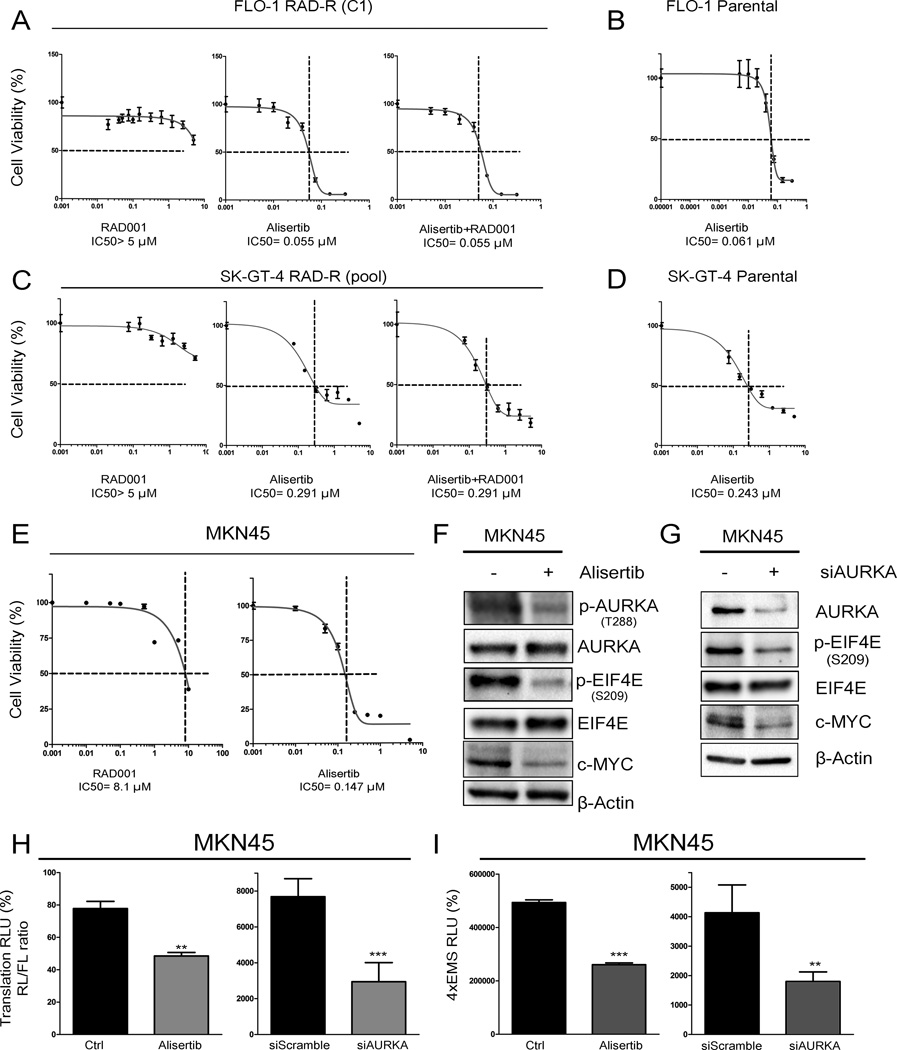
Figure 1. AURKA activates EIF4E and cap-dependent translation and becomes upregulated in cell models of acquired resistance to RAD001 (everolimus).
Overexpression of AURKA using adenoviral particles in FLO-1 (A & B) and SK-GT-4 (C & D) cells increased p-EIF4E (S209) and c-MYC protein levels, and the activity of a dual Renilla-firefly-luciferase pcDNA3-rLuc-PolioIRES-fluc reporter that measures cap-dependent/independent translation. E) FLO-1 Parental, FLO-1 RAD-R (Pool), FLO-1 RAD-R (C1), and FLO-1 RAD-R (C2) were subjected to clonogenic cell survival assay in response to everolimus. IC50 of resistant cells is significantly higher than that of parental cells. G) Clonogenic cell survival of SK-GT-4 parental and RAD-R (Pool) cells in response to RAD001indicated a significant increase of IC50 of resistant cells relative to parental cells. Western blot analysis of FLO-1 parental, FLO-1 RAD-R (Pool), FLO-1 RAD-R (C1), FLO-1 RAD-R (C2) (F), SK-GT-4 (Parental), and SK-GT-4 RAD-R (Pool) cells (H) showed an increase in p-EIF4E (S209), p-AKT (S473), p-ERK1/2 (S217/221), and c-MYC protein levels in resistant cells relative to parental cells.
Acquired resistance to everolimus requires overexpression of AURKA and phosphorylation of EIF4E independent of AKT and MAPK
Because of the role of EIF4E and cap-dependent translation in drug resistance and our findings that these are activated by AURKA independent of mTOR, we postulated that these mechanisms may be important for resistance to mTOR inhibitors. To develop cell models of acquired resistance to everolimus, FLO-1 and SK-GT-4 parental cell lines were treated with increasing concentrations of RAD001 (everolimus), starting from 0.1 up to 10 µmol/L, for six months. Using clonogenic cell survival assay, resistant pools from both cell lines showed significantly higher IC50 of RAD001, as compared with parental cells (Figure1E & 1G andSupplementary Figure S1). Single cell clones (FLO-1 RAD-R C1 & C2) were then developed from FLO-1 resistant pool (FLO-1 RAD-R pool). Clonogenic cell survival assays for these clones showed even higher IC50 for RAD001 (FLO-1 RAD-R C1, >15 µmol/L; FLO-1 RAD-R C2,4 µmol/L) (Figure 1E & S1A–F). Western blot analysis demonstrated overexpression of AURKA and an increase in phospho-EIF4E (S209) and c-MYC protein levels in resistant cells, as compared to parental cells (Figure 1F & 1H). Notably, we did not detect substantial differences in p-mTOR (S2448) or p-4EBP1 (S65) protein levels between parental and resistant cells. In addition, the RAD001-resistant FLO-1 and SK-GT-4 cells showed an increase in the protein levels of two possible regulators of p-EIF4E;p-AKT (S473) and p-ERK (S217/221)(Figure 1F & 1H). Quantitative real-time RT-PCR (qPCR) analysis did not show a significant increase in mRNA expression levels of AURKA in all resistant cells in comparison with parental cells, suggesting a post-transcription regulation of AURKA in resistant cells (Supplementary Figure S2). Of note, an earlier report indicated that resistance to mTOR inhibitor, rapamycin, involves activation of EIF4Ein non-small lung cancer cells (24). Together, our results suggest that AURKA-mediated activation of EIF4Ecould play an important role in mediating resistance to RAD001.
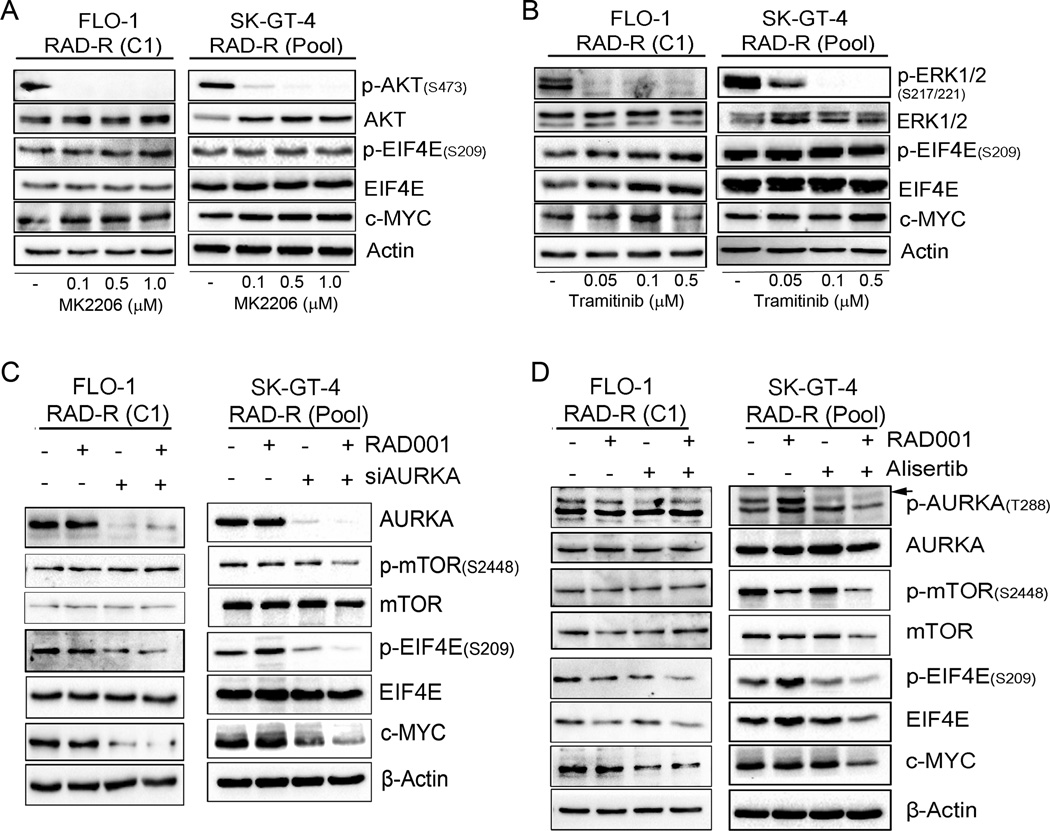
Given the known roles of AKT and ERK in regulating EIF4E phosphorylation and activity, we investigated if pharmacologic inhibition of AKT or ERK can suppress phospho-EIF4E in RAD001-resistant cells. Cells were treated with 0.1, 0.5, and 1 µmol/L of AKT inhibitor, MK2206, or with 0.05, 0.1, and 0.5 µmol/L of MEK1/2 inhibitor, trametinib, for 6 h. We achieved successful inhibition of AKT and MEK1/2 as shown by decreased phosphorylation of AKT (Figure 2A) and ERK1/2 (Figure 2B). However, the EIF4E phosphorylation and c-MYC protein levels were not decreased, suggesting that the regulation of EIF4E by AURKA is independent of AKT and MAPK signaling pathways in RAD001 resistant cells.
Figure 2. AURKA-induced phosphorylation of EIF4E and up-regulation of c-MYC is independent of AKT and MAPK/ERK pathways in RAD001 resistant cells.
A) FLO-1 RAD-R (C1) and SK-GT-4 RAD-R (Pool) resistant cells were treated with AKT inhibitor (MK2206) for 6 h, and subjected to Western blot analysis. B) The same cells were treated with MEK1/2 inhibitor (Tramitinib) for 6 h and analyzed by immunoblotting. Data showed that inhibition of AKT or MAPK/ERK signaling pathways had no effect on p-EIF4E (S209) and c-MYC protein levels. C) Knockdown of AURKA in RAD001 resistant FLO-1or SK-GT-4 cells downregulated p-EIF4E (S209) and c-MYC proteins, with or without RAD001 treatment. D) Pharmacological inhibition of AURKA using alisertib led to downregulation of p-EIF4E (S209) and c-MYC proteins in FLO-1 and SK-GT-4 resistant cells, with or without RAD001 treatment.
AURKA induces EIF4E phosphorylation and cap-dependent translation in RAD001 resistant cells
Given the reported gain of oncogenic signaling functions of AURKA in cancer cells and its recognition as a promising novel druggable target in gastrointestinal cancers (1, 8, 14), we investigated whether targeting AURKA can reverse its effects on the phosphorylation of EIF4E and c-MYC protein levels in resistant cells. AURKA knockdown markedly decreasedEIF4E phosphorylation and c-MYC protein levels in FLO-1 and SK-GT-4 resistant cells, whereas RAD001 treatment had no effect (Figure 2C and Supplementary Figure S3). Similar results were obtained using alisertib, a small molecule inhibitor of AURKA (Figure 2D). Because of the known role of MNK in phosphorylating EIF4E, we tested if AURKAmediated phosphorylation of EIF4E through MNK. Using AURKA knockdown or inhibition with alisertib, we did not detect changes in the protein levels of phospho-MNK(T197/202), albeit the remarkable decrease in phospho-EIF4E (Supplementary Figure S4). These results are consistent with our findings in Figure 1A–D where overexpression of AURKA did not change the protein levels of MNK and p-MNK (T197/202). They are also consistent with our results in Figure 2A & 2B, which demonstrated lack of effects of inhibition of AKT and MAPK pathways on phospho-EIF4E (S209) in resistant cells that express high levels of AURKA.
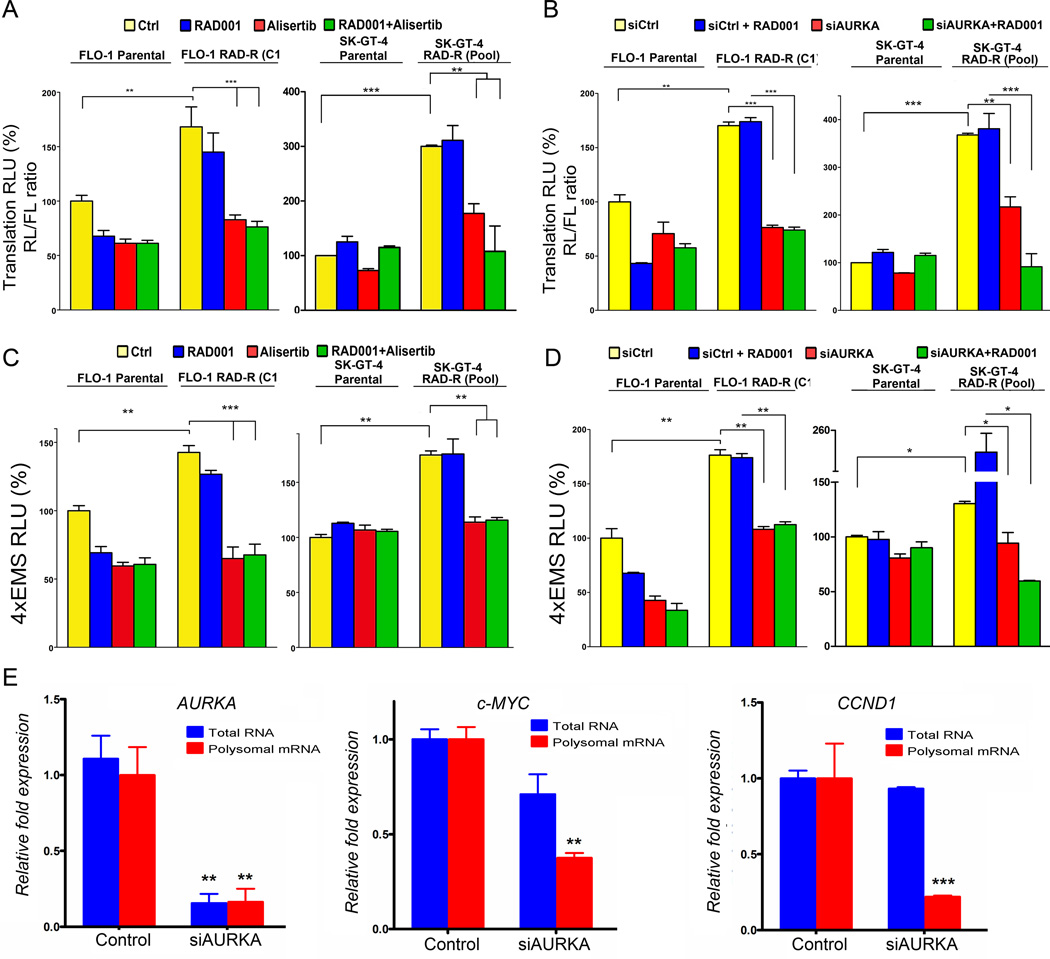
To further confirm the role of AURKA in regulating EIF4E-cap-dependent translation in resistant cells, we utilized a cap-dependent and independent protein-translation reporter assay as in Figure 1. The data showed that RAD001 resistant cells exhibited a higher cap-dependent protein translation activity than parental cells (p<0.01, Figure 3A & 3B), which are in agreement with increased p-EIF4E (S209) protein levels in resistant cells. In addition, the data indicated that inhibition or knockdown of AURKA with alisertib or siRNA, respectively, significantly decreased cap-dependent protein translation activity (p<0.01, Figure 3A & 3B). Because of the observed increase in c-MYC protein levels in RAD001 resistant cells, we utilized the4XEMS-Luc reporter, which contains transcription binding sites of c-MYC, as a measure of c-MYC transcription activity (36). The results confirmed an increase in c-MYC transcription activity in RAD001 resistant cells, as compared to parental cells (p<0.05, Figure 3C & 3D). Conversely, inhibition or knockdown of AURKA significantly decreased c-MYC transcriptional activity in RAD001 resistant cells (p<0.05, Figure 3C & 3D). Because EIF4E cap-dependent translation plays an important role in mRNA translation of c-MYC and CCND1, we used the translational chromatin immunoprecipitation (TrIP-ChIP) assay, which captures actively translated polysomal mRNAs, followed by qPCR analysis. Indeed, we detected a significant decrease in the levels of translated polysomal mRNA ofc-MYC and CCND1, as compared to their total RNA levels, in response to knockdown of AURKA in FLO-1 cells (p<0.01) (Figure 3E). Together, our data demonstrate that AURKA positively regulates EIF4E phosphorylation and cap-dependent protein translation activity in resistant cells independent of AKT and MAPK pathways.
Figure 3. AURKA mediates activation of cap-dependent protein translation and c-MYC transcriptional activity in RAD001 resistant cells.
AURKA inhibition by alisertib (A) or knockdown by siRNA (B) significantly downregulated cap-dependent translation in FLO-1 RAD-R (C1) and SK-GT-4 RAD-R (Pool) cells as compared to their respective parental cells. AURKA inhibition by alisertib (C) or knockdown by siRNA (D) significantly downregulated c-MYC transcriptional activity, as measured by 4xEMS luciferase reporter, in FLO-1 RAD-R (C1) and SK-GT-4 RAD-R (Pool) cells, relative to their respective parental cells. E) After knocking down of AURKA by siRNA, FLO-1 cells were subjected to translational chromatin immunoprecipitation (TrIP-Chip) assay and followed by qPCR analysis of total (input) and ploysomal RNA levels of AURKA, c-MYC, and CCND1. Data showed that knockdown of AURKA led to a significant decrease in the translated polysomal c-MYC and CCND1 RNA levels relative to their total RNA levels.
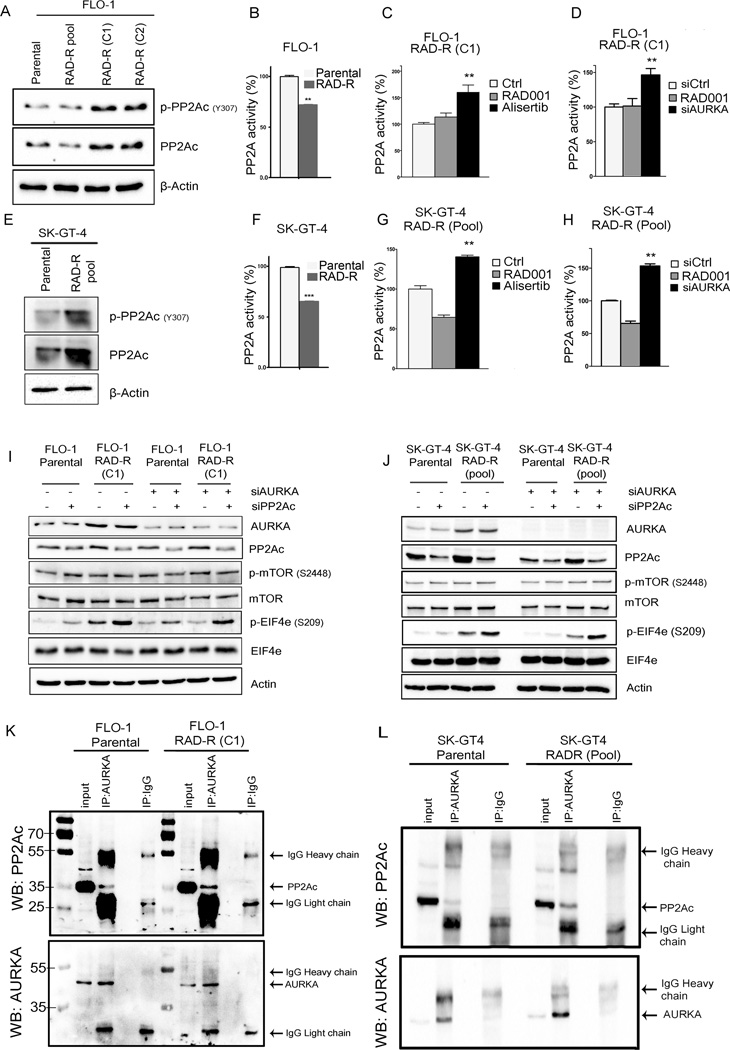
AURKA-induced activation of EIF4E is mediated by PP2A
Our results neither showed a role for AKT, MEK/ERK, or MNK in regulating EIF4E in RAD001-resistant cell models (Figure 2 and Supplementary Figure S4) nor an interaction between AURKA and EIF4E (Supplementary Figure S5); therefore we investigated whether AURKA-dependent phosphorylation of EIF4E requires the regulation of PP2A activity. It was previously shown that the catalytic subunit of PP2A, PP2Ac, is a negative regulator of EIF4E (39). We first confirmed PP2A capability to negatively regulate EIF4E in our cell model. Treatment with PP2A inhibitor, okadaic acid, increased PP2Ac phosphorylation at Y307 residue, which is indicative of inhibition of the catalytic subunit activity (Supplementary Figure S6). At the same time, PP2A inhibition increased EIF4E phosphorylation (S209) (Supplementary Figure S6A) in FLO-1 parental and RAD-R (C1) cells. These data confirm that PP2A is a negative regulator of EIF4E phosphorylation in our model. Importantly, the expression level of PP2A catalytic unit, p-PP2Ac (Y307), was markedly higher in the FLO-1 and SK-GT-4 resistant cells, as compared to parental cells (Figure 4A & 4E). Consistent with the increased phosphorylation of p-PP2Ac (Y307), we detected a significant reduction in the PP2A activity (Figure 4B & 4F). Furthermore, AURKA inhibition by alisertib decreased PP2Ac phosphorylation at Y307 in FLO-1 parental and resistant cells (Supplementary Figure S6B), suggesting restoration of PP2A activity, which was accompanied with a decrease in p-EIF4E and c-MYC protein levels (Supplementary Figure S6B). Indeed, the phosphatase activity assay data confirmed that AURKA negatively regulates PP2A activity. AURKA inhibition or knockdown significantly restored PP2A activity in the resistant cells (Figure 4C, 4D, 4G & 4H). To further establish that PP2A mediates EIF4Eactivation by AURKA, we examined EIF4E phosphorylation following knockdown of AURKA, PP2Ac, or the combination. Western blot data demonstrated that knocking down PP2Ac alone resulted in increased phosphorylation of EIF4E in resistant cells (Figure 4I & 4J), confirming PP2A as a negative regulator of EIF4E. Although knocking down AURKA downregulated p-EIF4E (S209), combined knockdown of AURKA and PP2Ac prevented this effect, suggesting that PP2A is required for EIF4E activation by AURKA (Figure 4I & 4J). In an attempt to identify the mechanism by which AURKA inhibits PP2A, we examined the potential protein binding between PP2Ac and AURKA in parental and resistant cells. Our Western blot analysis of immunoprecipitated proteins indicated that AURKA binds PP2Ac in parental and resistant cells (Figure 4K & 4L). Together, these data strongly suggest that AURKA-induced activation of EIF4E is mediated by PP2A. AURKA negatively regulates PP2A, in part, through protein binding in parental and resistant cells.
Figure 4. PP2Ac mediates EIF4E regulation by AURKA inRAD001 resistant cells.
A)Western blot analysis indicated higher p-PP2Ac (Y307) protein levels, indicative of inhibition of phosphatase activity, in the FLO-1 (A) and SK-GT-4 (E) resistant cells than their respective parental cells. The PP2A activity assay confirmed that both FLO-1 and SK-GT-4 resistant cells displayed a significant reduction in the PP2A activity (B &F). Alisertibor AURKA knockdown enhanced PP2A activity in FLO-1 RAD-R (C1) (C & D) and SK-GT-4 RAD-R (G & H) cells. Dual knockdown of AURKA and PP2Ac in FLO-1 parental and RAD-R (C1) (I) and SK-GT-4 parental and RAD-R (pool) (J) indicated that PP2A mediates AURKA regulation of EIF4E. AURKA immunoprecipitation and Western blot analysis in FLO-1 Parental and FLO-1 RAD-R (C1) cells (K) and SK-GT-4 Parental and SK-GT-4 RAD-R (Pool) cells (L) indicated a protein interaction between AURKA and PP2Ac.
Targeting AURKA with alisertib is an alternative effective therapeutic approach in RAD001 acquired and intrinsic resistant cells
Our aforementioned data strongly suggest that AURKA inhibition bypasses RAD001 resistance in UGC cell models. To confirm this finding, we performed cell viability ATP-Glo assay on FLO-1 RAD-R (C1) and SK-GT-4 RAD-R (Pool) cells treated with serial dilutions of RAD001 alone, alisertib alone, or the combination of the two drugs for five days. Our data indicated that although these cells are highly resistant to RAD001, they are sensitive to alisertib: FLO-1 RAD-R (C1) (IC50 = 0.05 µmol/L) and SK-GT-4 RAD-R (Pool) (IC50 = 0.29 µmol/L) cells. The combined treatment of alisertib and RAD001 did not show any synergistic or additive effect (Figure 5A & 5C). Notably, the sensitivity to alisertib was comparable in parental and RAD001 resistant cells, suggesting that acquired resistance to RAD001 does not alter the inherent sensitivity to alisertib in parental cells (Figure 5B & 5D).
Figure 5. Treatment with alisertib reduces cell viability of RAD001 resistant cells.
FLO-1 RAD-R (C1) cells (A) or SK-GT-4 RAD-R (Pool) cells (C) treated with alisertib or in combination with RAD001 were subjected to cell viability assay. Alisertib significantly inhibited the ability of RAD001 resistant cells to form colonies with or without RAD001 co-treatment. Parental FLO-1 cells (B) and SK-GT-4 cells (D) were treated with alisertib and subjected to cell viability assay. E) Human gastric MKN45 cell line displayed intrinsic resistance to RAD001 and sensitivity to alisertib as indicated by cell viability assay. Inhibition of AURKA with alisertib (F) or knockdown with siRNA (G) downregulated p-EIF4E (S209) and c-MYC protein levels as assayed by Western blotting. H) AURKA inhibition (left panel) or knockdown (right panel) significantly downregulated cap-dependent translation in MKN45 cells, as determined by a dual Renilla-firefly-luciferase pcDNA3-rLuc-PolioIRES-fluc reporter that measures cap-dependent/independent translation. I) AURKA inhibition (left panel) or knockdown (right panel) significantly downregulated c-MYC transcriptional activity in MKN45 cells, as measured by the 4xEMS luciferase reporter.
Next, we sought to find out if these results could be validated in intrinsic RAD001 resistance cell model. Therefore, we assayed several human UGC cell lines for their sensitivity to RAD001 and found that MKN45 cells are resistant with IC50 of 8.1 µmol/L (Figure 5E). Of note, MKN45 cells were sensitive to alisertib with IC50 of 0.14 µmol/L (Figure 5E). Western blot data showed that inhibition of AURKA with alisertib decreased p-AURKA (T288), p-EIF4E (S209), and c-MYC protein levels (Figure 5F). Similar results were obtained following knockdown of AURKA by siRNA in MKN45 cells (Figure 5G). Additionally, AURKA knockdown or inhibition decreased cap-dependent translation and c-MYC transcriptional activity in MKN45 cells as measured by reporter assays (Figure 5H & 5I). These results clearly indicate that inhibition of AURKA with alisertib is an alternative effective therapeutic approach in RAD001 acquired and intrinsic resistance cells.
Alisertib is effective in reducing tumor growth of RAD001 resistant cells
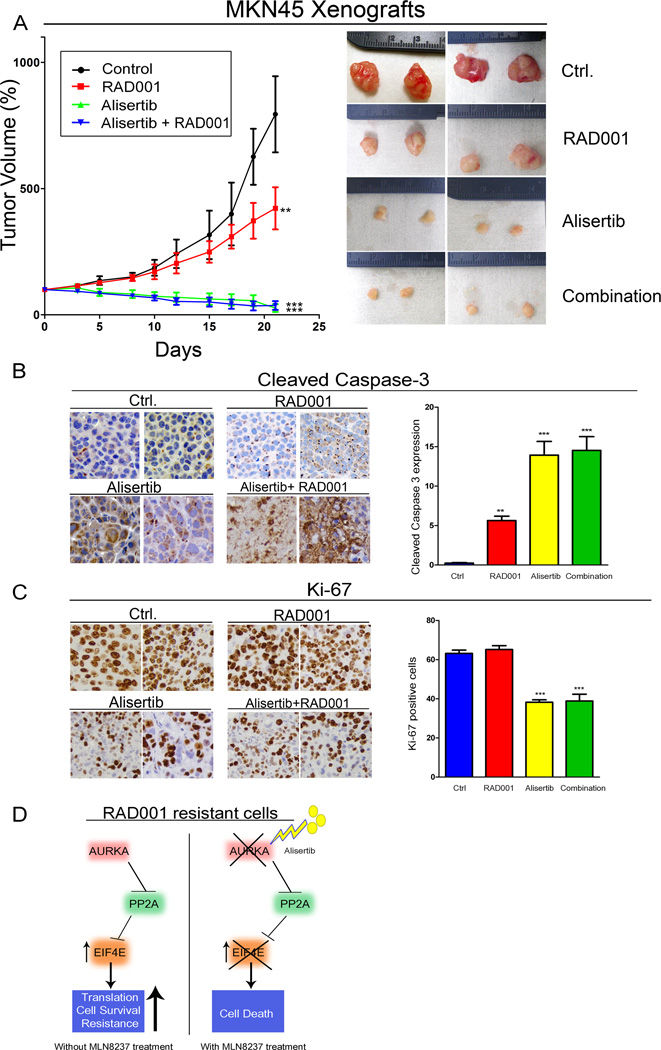
To investigate whether inhibiting AURKA with alisertib provides a potential clinical advantage in the treatment of RAD001 resistant tumors, we used a tumor xenograft mouse model of RAD001-resistant MKN45 cancer cells that were tested as shown in Figure 5E – I. After implantation, xenografted tumors were allowed to reach 200 mm3 before initiation of treatment of animals. Our data showed that alisertib abolished tumor growth and significantly reduced tumor size (p<0.001, Figure 6A). On the other hand, tumors treated with RAD001 continued to grow, though at a slower rate, as compared to control (Figure 6A). The combination treatment of RAD001 with alisertib did not have an advantage overalisertib alone (Figure 6A & 6B). Immunohistochemistry analysis of tumor xenografts demonstrated the highest cleaved caspase-3 and lowest of Ki-67 protein expression levels in groups treated with alisertib (Figure 6B & 6C).
Figure 6. Alisertib treatment reduces tumor growth in intrinsic RAD001 resistant xenograft mouse model.
A) Animals injected with MKN45 cells, and the tumors were allowed to grow until 200 mm3 in size, then treated with RAD001, alisertib or their combination for 5 weeks. Data indicated that alisertib alone or in combination with RAD001 significantly reduced tumor size in comparison with untreated or RAD001 alone treated groups. Although tumors in RAD001 treatment alone grew at a significantly slower rate than those in untreated group, they continued to grow, confirming the intrinsic resistant phenotype to RAD001. B and C) Immunohistochemistry analysis for cleaved caspase-3 expression, marker of apoptosis, and Ki-67 expression, marker of proliferation, in representative tumors of treated groups. D) A schematic diagram showing a proposed mechanism of RAD001 resistance. AURKA expression promotes RAD001 resistance through inhibition of PP2A, which leads to activation of EIF4E. Inhibition of AURKA by alisertib restores PP2A activity, thereby inhibiting EIF4E and inducing death in RAD001 resistant cancer cells.
Discussion
Amplifications and overexpression of AURKA at the 20q region are frequently detected in gastric and esophageal adenocarcinomas (6, 40). Overexpression of AURKA in cancer cells leads to inhibition of tumor suppressors such as p53 and p73 (5, 9, 41). Recent studies have shown that constitutive overexpression of AURKA in cancer cells leads to activation of several oncogenic signaling pathways, suggesting it as an important signaling hub in cancer cells (reviewed in (42, 43)). Of note, the aberrant overexpression of AURKA in cancer is associated with the gain of oncogenic functions that extend beyond its normal physiological functions in forming and stabilizing mitotic spindles during cell division. It has been reported that AURKA can activate NF-κB, HDM2, β-catenin, and STAT3 signaling networks in cancer cells (1, 9, 44–46). In this study, we have shown, for the first time, a novel oncogenic function of AURKA in cancer cells. Our results indicate that AURKA can regulate EIF4E and cap-dependent translation.
Chemotherapeutic resistance is a challenging clinical problem in gastro-esophageal cancers (29, 30). Although patients receiving first line therapy may initially respond to treatment, many of them develop recurrence within weeks or months after initial response requiring a second line treatment for disease progression where limited options are available (31). Currently, the only approved targeted therapies for advanced or metastatic gastroesophageal adenocarcinomas are trastuzumab and ramucirumab (34), which reflect the need to test other available targeted therapies. Studies on RAD001 resistant cancer models have suggested activation of EIF4E as a possible mediator of RAD001 resistance (20–22). Phosphorylation of EIF4E activates the cap-dependent translation machinery, leading to upregulation of EIF4E targets such as c-MYC (26). However, these reports fall short of describing the mechanism by which EIF4E is regulated in RAD001 resistant cancer models. Herein, we show, for the first time, an increase in AURKA overexpression in RAD001 resistant cancer cells that drives phosphorylation of EIF4E and cap-dependent translation by a mechanism that requires regulation of PP2A by AURKA.
Because of its diverse oncogenic functions, AURKA has become an attractive druggable target. To explore how AURKA mediates RAD001 resistance, we investigated EIF4E regulation by AURKA. Our results indicated that resistant cells expressed higher levels of AURKA and phospho-EIF4E, as compared to parental cells. Several reports highlighted the roles of EIF4E in regulating vital biological functions such as translation and cell death, and therefore promote tumorigenesis (20–22). Additionally, increased EIF4E expression has been associated with mTOR inhibitor resistance (24). Thus, targeting AURKA-EIF4E axis in resistant cancers is a reasonable alternative rational approach to achieve a therapeutic response. Although knocking down AURKA decreased p-EIF4Eprotein levels, our data indicated lack of direct interaction between EIF4E and AURKA, suggesting that AURKA-induced activation of EIF4E could be mediated by an effector molecule downstream of AURKA.
Earlier reports indicated that AKT can regulate mTOR pathway by inhibition of tuberous sclerosis complex 2 (TSC2), which affects mTOR/4EBP1/EIF4E signaling (47, 48), while other reports showed that AKT is regulated by EIF4E (25), suggesting a feedback loop between AKT and EIF4E. Additionally, it has been reported that MAPK signaling pathway can control EIF4E phosphorylation (27). However, in our resistant models, inhibition of AKT or MEK/ERK did not decrease EIF4E phosphorylation. Phosphorylation of EIF4E at Ser209 has been shown to be essential for EIF4E oncogenic activity. Earlier studies have shown that MNK kinase, downstream of MAPK pathway, can phosphorylate EIF4E at Ser209 (49, 50). Our results suggest that overexpression or inhibition of AURKA can affect the phosphorylation of EIF4E without changing MNK phosphorylation levels. Interestingly, we found that resistant cells displayed higher protein levels of p-PP2Ac (Y307) than parental cells, which denotes an inhibition of the PP2A activity. We show that AURKA can modulate the PP2A activity and the PP2A-EIF4E axis, whereby AURKA inhibition or knockdown restores PP2A activity and suppresses EIF4E phosphorylation. Furthermore, knockdown of both AURKA and PP2Ac restored theEIF4E activity, indicating that AURKA activation of EIF4E requires inhibition of PP2A in resistant cells. As an additional evidence of AURKA regulation of PP2A activity, our results demonstrated an interaction between the two proteins in resistant cells and the requirement of PP2A for AURKA-mediated phosphorylation and activation of EIF4E.
EIF4E-mediated cap dependent translation regulates several important oncogenes that control stress and inflammatory responses, cell survival, proliferation, and apoptosis (23). Among these, c-MYC plays a central role in tumorigenesis, stem cell properties of cancer cells, and resistance to chemotherapeutics (51). Of note, it has been shown that AURKA upregulates c-MYC mRNA expression through activation of β-catenin and STAT3 transcription factors in cancer cells (8, 44). Furthermore, a recent report has shown that AURKA-MYC interactions stabilize MYC to promote tumor cell survival presenting AURKA as an actionable drug target in MYC-amplified/overexpressed tumors (52). In this context, our data establishes the role of AURKA-EIF4E axis in regulating c-MYC translation and activity. These findings altogether suggest a multifaceted complex regulation of c-MYC by AURKA in cancer cells. We also can not ignore the possible effects of targeting AURKA on cell cycle and signaling pathways where inhibition of AURKA has been shown to lead to mitotic catastrophe (2), inhibition of NF-kB (45), β-catenin (44), and STAT3 (45). As such, our findings add to the complexity of AURKA signaling in cancer cells and support placing it as a critical signaling hub that promotes cancer cell survival and resistance to therapy.
Using an in vivo tumor xenograft mouse model, we further validated the efficacy of targeting AURKA in cancer cells that have intrinsic resistance toRAD001. While RAD001 treated tumor groups grew at slower rate, as compared to controls, the alisertib treated tumors not only stopped growing but also significantly diminished in size. Notably, the combination of alisertib and RAD001 had no advantage in further reducing tumor growth, suggesting that this combination is not a desirable approach as it could cause more toxicity to patients without a notable improvement in clinical response.
In conclusion, our results demonstrate a novel AURKA-EIF4E-MYC signaling axis in cancer cells. Based on our data, we suggest targeting AURKA as a novel approach that can be applied in tumors showing high levels of AURKA with activation of EIF4E, c-MYC, or resistance to mTOR inhibitors in upper gastrointestinal cancers and possibly other cancer types.
Supplementary Material
Translational Significance.
Upper gastrointestinal cancers (UGCs) are characterized by poor patient survival and resistance to chemotherapy. We and others have previously shown that Aurora kinase A (AURKA) is frequently overexpressed in UGCs, making it an attractive druggable target in clinical trials. Herein, we report for the first time that AURKA activates EIF4E and cap-dependent translation. We also show that resistance to everolimus, an inhibitor of mTOR, is mediated by AURKA-dependent activation of EIF4E and c-MYC. We propose targeting AURKA as a novel therapeutic strategy applicable for mTOR inhibitors resistant tumors as well as those exhibiting activation of EIF4E and c-MYC.
Acknowledgments
This study was supported by a Research Career Scientist award (1IK6BX003787) from the U.S. Department of Veterans affairs (W. El-Rifai), and grants from the U.S. National Institutes of Health (R01CA131225 and R01CA93999, W. El-Rifai). The study was also supported by National Institutes of Health grants that include Vanderbilt SPORE in Gastrointestinal Cancer (P50 CA95103), Vanderbilt Ingram Cancer Center (P30 CA68485), and the Vanderbilt Digestive Disease Research Center (DK058404). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Veterans Affairs, National Institutes of Health, or Vanderbilt University.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors have declared no conflict of interest.
Author contributions:
Ahmed Katsha: Design of experiments and acquisition of data; analysis and interpretation of data; drafting of the manuscript; technical and material support.
Lihong Wang: Assisted in acquisition of data and experimental design.
Janet Arras: Assisted in in vitro data acquisition.
Omar M. Omar: Assisted in preparation of in vitro samples.
Jeffrey Ecsedy: Provided reagents and critical reading of manuscript.
Abbes Belkhiri: Analysis and interpretation of data; experimental troubleshooting; assisted in drafting of the manuscript.
Wael El-Rifai: Study concept and design; obtained funding; study supervision; experimental troubleshooting; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content.
References
- 1.Katsha A, Soutto M, Sehdev V, Peng D, Washington MK, Piazuelo MB, et al. Aurora kinase A promotes inflammation and tumorigenesis in mice and human gastric neoplasia. Gastroenterology. 2013;145:1312–1322. e1–e8. doi: 10.1053/j.gastro.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sehdev V, Katsha A, Ecsedy J, Zaika A, Belkhiri A, El-Rifai W. The combination of alisertib, an investigational Aurora kinase A inhibitor, and docetaxel promotes cell death and reduces tumor growth in preclinical cell models of upper gastrointestinal adenocarcinomas. Cancer. 2013;119:904–914. doi: 10.1002/cncr.27801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katsha A, Belkhiri A, Goff L, El-Rifai W. Aurora kinase A in gastrointestinal cancers: time to target. Molecular cancer. 2015;14:106. doi: 10.1186/s12943-015-0375-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai MY, Wiese C, Cao K, Martin O, Donovan P, Ruderman J, et al. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nature Cell Biology. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- 5.Dar AA, Belkhiri A, Ecsedy J, Zaika A, El-Rifai W. Aurora kinase A inhibition leads to p73-dependent apoptosis in p53-deficient cancer cells. Cancer Res. 2008;68:8998–9004. doi: 10.1158/0008-5472.CAN-08-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dar AA, Zaika A, Piazuelo MB, Correa P, Koyama T, Belkhiri A, et al. Frequent overexpression of Aurora Kinase A in upper gastrointestinal adenocarcinomas correlates with potent antiapoptotic functions. Cancer. 2008;112:1688–1698. doi: 10.1002/cncr.23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katayama H, Wang J, Treekitkarnmongkol W, Kawai H, Sasai K, Zhang H, et al. Aurora kinase-A inactivates DNA damage-induced apoptosis and spindle assembly checkpoint response functions of p73. Cancer Cell. 2012;21:196–211. doi: 10.1016/j.ccr.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katsha A, Arras J, Soutto M, Belkhiri A, El-Rifai W. AURKA Regulates STAT3 Activity through JAK2 in Human Gastric and Esophageal Cancer. Mol Oncol. 2014 doi: 10.1016/j.molonc.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sehdev V, Katsha A, Arras J, Peng D, Soutto M, Ecsedy J, et al. HDM2 regulation by AURKA promotes cell survival in gastric cancer. Clin Cancer Res. 2014;20:76–86. doi: 10.1158/1078-0432.CCR-13-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang G, Chang B, Yang F, Guo XQ, Cai KQ, Xiao X, et al. Aurora Kinase A Promotes Ovarian Tumorigenesis through Dysregulation of the Cell Cycle and Suppression of BRCA2. Clinical Cancer Research. 2010;16:3171–3181. doi: 10.1158/1078-0432.CCR-09-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouchi M, Fujiuchi N, Sasai K, Katayama H, Minamishima YA, Ongusaha PP, et al. BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J Biol Chem. 2004;279:19643–19648. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- 12.Otto T, Horn S, Brockmann M, Eilers U, Schuttrumpf L, Popov N, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Lee HH, Zhu YS, Govindasamy KM, Gopalan G. Downregulation of Aurora-A overrides estrogen-mediated growth and chemoresistance in breast cancer cells. Endocrine-Related Cancer. 2008;15:765–775. doi: 10.1677/ERC-07-0213. [DOI] [PubMed] [Google Scholar]
- 14.Cammareri P, Scopelliti A, Todaro M, Eterno V, Francescangeli F, Moyer MP, et al. Aurora-A Is Essential for the Tumorigenic Capacity and Chemoresistance of Colorectal Cancer Stem Cells. Cancer Res. 2010;70:4655–4665. doi: 10.1158/0008-5472.CAN-09-3953. [DOI] [PubMed] [Google Scholar]
- 15.Carol H, Boehm I, Reynolds CP, Kang MH, Maris JM, Morton CL, et al. Efficacy and pharmacokinetic/pharmacodynamic evaluation of the Aurora kinase A inhibitor MLN8237 against preclinical models of pediatric cancer. Cancer Chemother Pharmacol. 2011;68:1291–1304. doi: 10.1007/s00280-011-1618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katsha A, Soutto M, Sehdev V, Peng D, Washington MK, Piazuelo MB, et al. Aurora Kinase A Promotes Inflammation and Tumorigenesis in Mice and Human Gastric Neoplasia. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cervantes A, Elez E, Roda D, Ecsedy J, Macarulla T, Venkatakrishnan K, et al. Phase I pharmacokinetic/pharmacodynamic study of MLN8237, an investigational, oral, selective aurora a kinase inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2012;18:4764–4774. doi: 10.1158/1078-0432.CCR-12-0571. [DOI] [PubMed] [Google Scholar]
- 18.Matulonis UA, Sharma S, Ghamande S, Gordon MS, Del Prete SA, Ray-Coquard I, et al. Phase II study of MLN8237 (alisertib), an investigational Aurora A kinase inhibitor, in patients with platinum-resistant or - refractory epithelial ovarian, fallopian tube, or primary peritoneal carcinoma. Gynecol Oncol. 2012;127:63–69. doi: 10.1016/j.ygyno.2012.06.040. [DOI] [PubMed] [Google Scholar]
- 19.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature reviews Molecular cell biology. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furic L, Rong LW, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PloS one. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, Takasu T, Perlman DM, Peterson MS, Burrichter D, Avdulov S, et al. Translation factor eIF4E rescues cells from Myc-dependent apoptosis by inhibiting cytochrome c release. The Journal of biological chemistry. 2003;278:3015–3022. doi: 10.1074/jbc.M208821200. [DOI] [PubMed] [Google Scholar]
- 23.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nature reviews Drug discovery. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 24.Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer research. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 25.Culjkovic B, Tan K, Orolicki S, Amri A, Meloche S, Borden KL. The eIF4E RNA regulon promotes the Akt signaling pathway. J Cell Biol. 2008;181:51–63. doi: 10.1083/jcb.200707018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin CJ, Malina A, Pelletier J. c-Myc and eIF4F Constitute a Feedforward Loop That Regulates Cell Growth: Implications for Anticancer Therapy. Cancer research. 2009;69:7491–7494. doi: 10.1158/0008-5472.CAN-09-0813. [DOI] [PubMed] [Google Scholar]
- 27.Shveygert M, Kaiser C, Bradrick SS, Gromeier M. Regulation of Eukaryotic Initiation Factor 4E (eIF4E) Phosphorylation by Mitogen-Activated Protein Kinase Occurs through Modulation of Mnk1-eIF4G Interaction. Molecular and cellular biology. 2010;30:5160–5167. doi: 10.1128/MCB.00448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckerdt F, Beauchamp E, Bell J, Iqbal A, Su B, Fukunaga R, et al. Regulatory effects of a Mnk2-eIF4E feedback loop during mTORC1 targeting of human medulloblastoma cells. Oncotarget. 2014;5:8442–8451. doi: 10.18632/oncotarget.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belkhiri A, El-Rifai W. Advances in targeted therapies and new promising targets in esophageal cancer. Oncotarget. 2015;6:1348–1358. doi: 10.18632/oncotarget.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang DX, Fan DM. New insights into the mechanisms of gastric cancer multidrug resistance and future perspectives. Future Oncol. 2010;6:527–537. doi: 10.2217/fon.10.21. [DOI] [PubMed] [Google Scholar]
- 31.Kanagavel D, Fedyanin M, Tryakin A, Tjulandin S. Second-line treatment of metastatic gastric cancer: Current options and future directions. World J Gastroenterol. 2015;21:11621–11635. doi: 10.3748/wjg.v21.i41.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 33.Mohammad NH, ter Veer E, Ngai L, Mali R, van Oijen MG, van Laarhoven HW. Optimal first-line chemotherapeutic treatment in patients with locally advanced or metastatic esophagogastric carcinoma: triplet versus doublet chemotherapy: a systematic literature review and meta-analysis. Cancer Metastasis Rev. 2015;34:429–441. doi: 10.1007/s10555-015-9576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah MA. Update on metastatic gastric and esophageal cancers. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:1760–1769. doi: 10.1200/JCO.2014.60.1799. [DOI] [PubMed] [Google Scholar]
- 35.Liu JY, Stevens PD, Eshleman NE, Gao TY. Protein Phosphatase PPM1G Regulates Protein Translation and Cell Growth by Dephosphorylating 4E Binding Protein 1 (4E-BP1) Journal of Biological Chemistry. 2013;288:23225–23233. doi: 10.1074/jbc.M113.492371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boone DN, Qi Y, Li ZL, Hann SR. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:632–637. doi: 10.1073/pnas.1008848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudo K, Xi Y, Wang Y, Song B, Chu E, Ju J, et al. Translational control analysis by translationally active RNA capture/microarray analysis (TrIP-Chip) Nucleic acids research. 2010;38:e104. doi: 10.1093/nar/gkq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu SM, Belkhiri A, El-Rifai W. DARPP-32 Increases Interactions Between Epidermal Growth Factor Receptor and ERBB3 to Promote Tumor Resistance to Gefitinib. Gastroenterology. 2011;141:1738–U297. doi: 10.1053/j.gastro.2011.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Yue P, Deng X, Ueda T, Fukunaga R, Khuri FR, et al. Protein phosphatase 2A negatively regulates eukaryotic initiation factor 4E phosphorylation and eIF4F assembly through direct dephosphorylation of Mnk and eIF4E. Neoplasia. 2010;12:848–855. doi: 10.1593/neo.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamada K, Yamada Y, Hirao T, Fujimoto H, Takahama Y, Ueno M, et al. Amplification/overexpression of Aurora-A in human gastric carcinoma: potential role in differentiated type gastric carcinogenesis. Oncol Rep. 2004;12:593–599. [PubMed] [Google Scholar]
- 41.Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 42.Dar AA, Goff LW, Majid S, Berlin J, El-Rifai W. Aurora kinase inhibitors--rising stars in cancer therapeutics. Mol Cancer Ther. 2010;9:268–278. doi: 10.1158/1535-7163.MCT-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katsha A, Belkhiri A, Goff L, El-Rifai W. Aurora kinase A in gastrointestinal cancers: time to target. Mol Cancer. 2015;14:106. doi: 10.1186/s12943-015-0375-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dar AA, Belkhiri A, El-Rifai W. The aurora kinase A regulates GSK-3beta in gastric cancer cells. Oncogene. 2009;28:866–875. doi: 10.1038/onc.2008.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katsha A, Arras J, Soutto M, Belkhiri A, El-Rifai W. AURKA regulates JAK2-STAT3 activity in human gastric and esophageal cancers. Mol Oncol. 2014;8:1419–1428. doi: 10.1016/j.molonc.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bornschein J, Nielitz J, Drozdov I, Selgrad M, Wex T, Jechorek D, et al. Expression of aurora kinase A correlates with the Wnt-modulator RACGAP1 in gastric cancer. Cancer Med. 2016;5:516–526. doi: 10.1002/cam4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-Kinase/Akt pathway. Molecular cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 48.Hahn-Windgassen A, Nogueira V, Chen CC, Skeen JE, Sonenberg N, Hay N. Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. Journal of Biological Chemistry. 2005;280:32081–32089. doi: 10.1074/jbc.M502876200. [DOI] [PubMed] [Google Scholar]
- 49.Hu K, Zhang J, Yu M, Xiong C. Inhibition of Mnk-eIF4E pathway sensitizes the efficacy to chemotherapy in anaplastic thyroid cancer. Future Oncol. 2016 doi: 10.2217/fon-2016-0320. [DOI] [PubMed] [Google Scholar]
- 50.Silva RL, Wendel HG. MNK, EIF4E and targeting translation for therapy. Cell Cycle. 2008;7:553–555. doi: 10.4161/cc.7.5.5486. [DOI] [PubMed] [Google Scholar]
- 51.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dauch D, Rudalska R, Cossa G, Nault JC, Kang TW, Wuestefeld T, et al. A MYC-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nature medicine. 2016 doi: 10.1038/nm.4107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.