Abstract
Aldehyde dehydrogenase 7A1 (ALDH7A1) is involved in lysine catabolism, catalyzing the oxidation of α-aminoadipate semialdehyde to α-aminoadipate. Certain mutations in the ALDH7A1 gene, which are presumed to reduce catalytic activity, cause an autosomal recessive seizure disorder known as pyridoxine-dependent epilepsy (PDE). Although the genetic association between ALDH7A1 and PDE is well established, little is known about the impact of PDE-mutations on the structure and catalytic function of the enzyme. Herein we report the first study of the molecular consequences of PDE mutations using purified ALDH7A1 variants. Eight variants, with mutations in the oligomer interfaces, were expressed in Escherichia coli: P78L, G83E, A129P, G137V, G138V, A149E, G255D, and G263E. All but P78L and G83E were soluble and could be purified. All six soluble mutants were catalytically inactive. The impact of the mutations on oligomerization was assessed by analytical ultracentrifugation. Wild-type ALDH7A1 is shown to exist in a dimer-tetramer equilibrium with a dissociation constant of 16 micromolar. In contrast to the wild-type enzyme, the variants reside in monomer-dimer equilibria and are apparently incapable of forming a tetrameric species, even at high enzyme concentration. The available evidence suggests that they are misfolded assemblies lacking the three-dimensional structure required for catalysis.
Keywords: aldehyde dehydrogenase, ALDH7A1, lysine catabolism, pyridoxine-dependent epilepsy, protein oligomerization, analytical ultracentrifugation
Graphical abstract
1. Introduction
The lysine catabolic enzyme aldehyde dehydrogenase 7A1 (ALDH7A1) catalyzes the NAD+- dependent oxidation of α-aminoadipate semialdehyde (AASAL) to α-aminoadipate (Fig. 1A). Misregulation of ALDH7A1 and other members of the ALDH superfamily has been observed in cancer stem cells and linked to cancer stem cell metastasis [1, 2]. Defects in ALDH function are also associated with numerous inherited metabolic disorders. Notably, certain mutations in the ALDH7A1 gene cause pyridoxine-dependent epilepsy (PDE), a recessive, inherited disease that results in seizures [3, 4].
Figure 1.
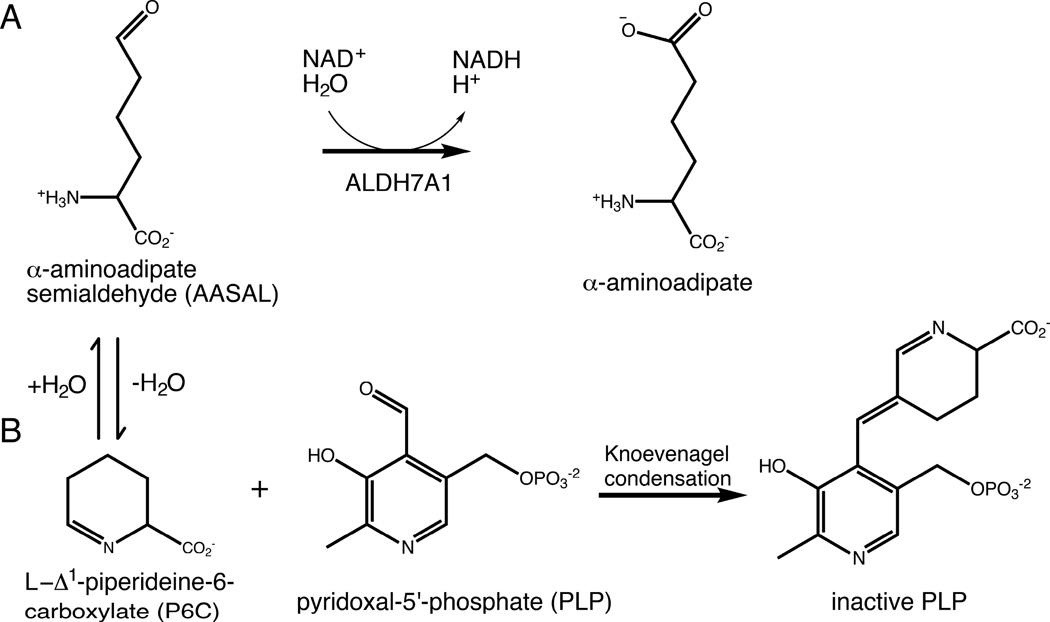
Chemical reactions relevant to ALDH7A1 and PDE. (A) The reaction catalyzed by ALDH7A1. (B) The reaction between P6C and PLP, which results in covalent inactivation of PLP. P6C forms a nonenzymatic equilibrium with AASAL.
According to the Human Gene Mutation Database [5], 47 disease-related mutations, both missense and nonsense, have been mapped to the ALDH7A1 gene. 39 of the 47 are linked to PDE. A decade ago, Mills et al. proposed that the pathology of PDE involves inactivation of the enzyme cofactor pyridoxal 5´-phosphate (PLP) [3]. Specifically, diminished ALDH7A1 activity leads to increased levels of AASAL and its cyclized form, Δ1-piperideine-6-carboxylic acid (P6C, Fig. 1). The elevated P6C concentration promotes covalent inactivation of PLP via the Knoevenagel condensation (Fig. 1B) [3]. Given that roughly 4% of all enzymes employ PLP as a cofactor [6], reduced PLP levels could have a variety of adverse physiological consequences. Increased incidence of seizures is believed to be one of these [3].
Elucidating the functional impact of ALDH7A1 mutations is key to understanding the molecular basis of PDE. High-resolution structures of ALDH7A1 complexed with NAD+ and α-aminoadipate have revealed the protein fold, the apparent quaternary structure, and the residues involved in substrate- and cofactor binding [7–9]. Despite the availability of structural information for the wild-type enzyme, it is difficult to predict the impact of ALDH7A1 point-mutations on catalytic activity and three-dimensional structure, particularly those remote from the active site. In this context, it is notable that approximately 12% of all disease-related mutations are located in protein-protein interfaces of oligomeric proteins [10]. Indeed, nearly 20% of the known PDE-associated missense mutations reside in, or near, the interfaces of the ALDH7A1 tetramer: P78L [11], G83E [12], A129P [11], G137V [12], G138V [13], A149E [11], G255D [14], and G263E [15].
To better understand the impact of these mutations, we expressed recombinant versions of the eight aforementioned PDE-related mutant enzymes in Escherichia coli and compared their self-association behavior to that of wild-type ALDH7A1. The wild-type protein forms a catalytically active, high-affinity dimer that can further associate, at higher concentrations, to form a tetramer. Two of the variant proteins did not express appreciable amounts of soluble protein (P78L, G83E), precluding detailed analysis. The other six, although soluble, are catalytically inactive and display aberrant self-association behavior. Specifically, both monomeric and dimeric states are populated at low concentration, and the dimers are apparently incapable of forming the tetrameric dimer-of-dimers observed for wild-type ALDH7A1. These results suggest that the disease-related mutations result in the production of misfolded monomers incapable of associating into active oligomers.
2. Materials and methods
2.1. Construct generation
The construct used for expression of wild-type human ALDH7A1 in the pKA8H protein expression vector was previously described [7]. Mutations in the ALDH7A1 gene were introduced with the QuikChange Lightning kit (Agilent). Mutations were validated by DNA Sanger sequencing.
2.2. Protein expression and purification
All protein expression vectors were transformed into BL21(DE3) cells (Thermo Fisher Scientific) for protein expression. An overnight starter culture was used to inoculate 1L of terrific broth. Cultures were shaken at 37°C and 250 rpm until OD600 ≈ 0.8. The temperature was then shifted to 18°C, and protein expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside. Cultures were shaken overnight. The following day, the cells were pelleted and frozen at −80°C for storage.
For purification, frozen cell pellets were resuspended in lysis buffer (50 mM Tris pH 8.0, 500 mM NaCl, 20 mM imidazole, 10% v/v glycerol, and 1% Tween-20). Cells were then lysed by sonication, and the cell-debris and unbroken cells were pelleted at 16,000 rpm for 1 hour. The cell-lysate was then loaded by gravity onto a Ni-NTA column. After washing the column with 40 bed volumes of 50 mM Tris pH 8.0, 500 mM NaCl, 20 mM imidazole, and 10% v/v glycerol, the protein was eluted with 50 mM Tris pH 8.0, 500 mM NaCl, 250 mM imidazole, and 10% v/v glycerol. Protein concentrations were estimated by A280, using the extinction coefficient of 81,360 M−1 cm−1 predicted from amino acid sequence with ExPASy ProtParam [16].
The His-tag was cleaved from the purified protein using Tobacco Etch Virus protease in the presence of 0.5 mM EDTA, employing 1 mg of protease for every 10 mg of target. After 2 hours at 28°C, the preparation was dialyzed overnight at 4°C against 50 mM Tris, pH 7.8, 50 mM NaCl, 5% v/v glycerol, 1 mM dithiothreitol. The cleaved His-tag was removed by passing the protein solution over Ni-NTA. The eluate was concentrated and further purified by size-exclusion chromatography on Superdex 200 10–30, in the presence of 50 mM Tris pH 7.8, 50 mM NaCl, 5% v/v glycerol, and 0.5 mM Tris(hydroxypropyl)phosphine.
Fractions containing the protein were pooled and concentrated. Sedimentation analysis was immediately performed on an aliquot of the resulting solution. The remainder was supplemented with NAD+ to a final concentration of 2 mM, flash frozen in liquid nitrogen, and stored at −80°C. For enzyme-activity assays, samples were thawed on ice and diluted to the desired concentration with 100 mM sodium pyrophosphate pH 8.
2.3. Kinetics measurements
ALDH7A1 activity was estimated by monitoring NADH production at 340 nm as described elsewhere [9, 17]. The assays were conducted at 25°C, in 100 mM sodium pyrophosphate pH 8 and 2.5 mM NAD+, using a Biotek EPOCH2 plate reader. The standard assay employed an ALDH7A1 concentration of 0.5 µM (based on the molecular weight of a monomer) and 2 mM AASAL. Because the variant proteins were inactive under these conditions, assays were also conducted with elevated levels of enzyme (4 µM, monomer molecular weight) and aldehyde (30 mM).
2.4. Analytical ultracentrifugation
Sedimentation velocity and equilibrium experiments were performed at 20°C, in a Beckman XL-I analytical ultracentrifuge using an An50Ti rotor. For velocity studies, aliquots of the protein solution (400 µL) and reference buffer (430 µL) were loaded into the sample- and solvent compartments, respectively, of a sedimentation-velocity cell, equipped with a dual-sector charcoal-Epon centerpiece. After temperature equilibration, the sample was centrifuged at 35,000 rpm, monitoring the radial-distribution with Rayleigh interference optics. Data were acquired at two-minute intervals until 300 radial scans had been collected. The composite data set was analyzed globally using Sedfit [17], to obtain the sedimentation-coefficient, c(s), and molecular-weight, c(M), distributions.
Sedimentation-equilibrium data were collected at three different protein concentrations and three rotor speeds – 6000, 9000, and 12000 rpm. Aliquots of the protein solutions and reference buffer were loaded into a sedimentation-equilibrium cell equipped with a six-sector charcoal-Epon centerpiece. Following a 16 h equilibration period at 6000 rpm, the absorbance, at 280 nm, was measured as a function of radial position. Data were collected at hourly intervals, until the radial distribution was unchanged. For subsequent measurements at 9000 rpm and 12,000 rpm, the samples were equilibrated for eight hours prior to acquisition of the first scan.
Sedimentation equilibrium data were initially fit to a single-species model to estimate the average molecular weight of all of the species:
| (1) |
In Eq 1, a is the total sample absorbance; ao is the absorbance at an arbitrary reference position, ro; M is the average molecular weight; ω is the radial velocity (rad/s); is the partial specific volume of the protein; ρ is the sample density; R is the gas constant; and T is the absolute temperature.
The value of M thus obtained dictated the form of the model employed for subsequent analysis. For wild-type ALDH7A1, the value of M was between that of the dimer and the tetramer. Accordingly, the following equation was employed:
| (2) |
where M2 is the dimer molecular weight and K2–4 is the association constant governing the dimer-tetramer equilibrium. For the variant enzymes, the single-species model yielded values for M below the dimer molecular weight. Thus, the data were subsequently analyzed with a model that included both the monomer-dimer and dimer-tetramer equilibria:
| (3) |
where M1 is the monomer molecular weight and K1–2 is the association constant for dimer formation.
The initial treatment of the data for the variant proteins considered just the monomer-dimer association – i.e., K2–4 was set to zero. Upon completion of the least-squares minimization, K2–4 was then allowed to vary, in order to determine whether any further reduction in χ2 could be achieved.
The least-squares analyses returned values for K1–2 and K2–4 in inverse absorbance units. The following equation was used to convert K1–2,abs to inverse concentration units:
| (4) |
where ε is the molar absorptivity of the monomer, and l is the path length of the charcoal-Epon centerpiece (1.2 cm). A similar formula was used to convert K2–4,abs to inverse concentration units:
| (5) |
where ε2 represents the molar absorptivity of the dimer, assumed to be twice that of the monomer.
3. Results
3.1. PDE-related mutations eliminate catalytic activity
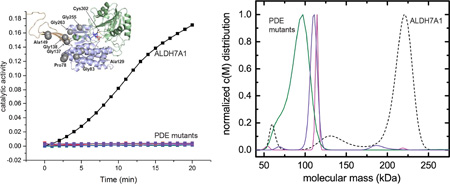
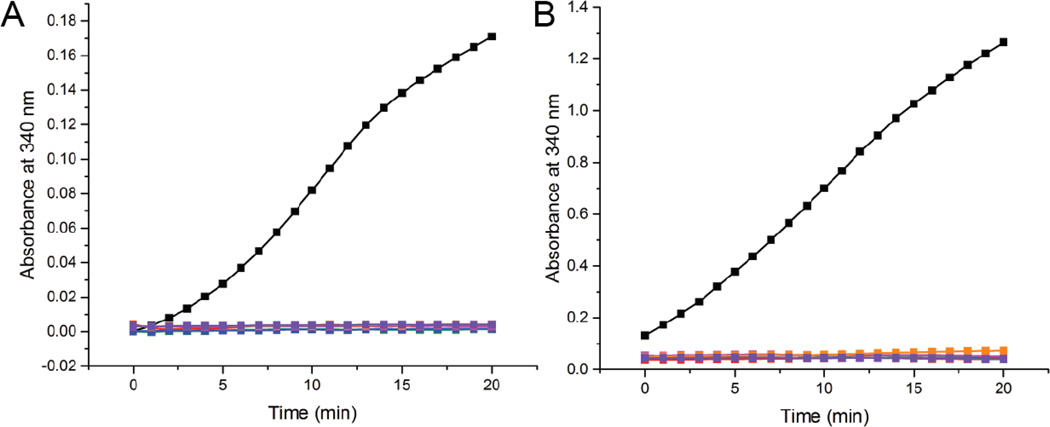
Wild-type ALDH7A1 exhibited robust activity in our standard assay (Fig. 2A), employing 0.5 µM enzyme and near-saturating concentrations of AASAL (2 mM) and NAD+ (2.5 mM). By contrast, none of the variants displayed detectable activity under the same conditions (Fig 2A). Assays were also performed with elevated levels of enzyme (4.0 µM) and AASAL (30 mM). Whereas the activity of wild-type ALDH7A1 scaled with enzyme concentration (Fig. 2B), the activity of the variant proteins remained unchanged from background (Fig. 2B). These results indicate that the mutations render ALDH7A1 catalytically inactive, consistent with clinical phenotypes observed in PDE patients.
Figure 2. Enzymatic activity of interface mutants compare to wild-type ALDH7A1.
NADH formation was monitored at 340 nm using either 0.5 µM protein and 2 mM AASAL (A) or 4 µM protein and 30 mM AASAL (B). The color scheme in both panels is identical: wild type ALDH7A1 (black), A129P (green), G137V (red), G138V (blue), A149E (magenta), G255D (orange), and G263E (purple). The NAD+ concentration was 2.5 mM. Note that the vertical scales are different in the two panels. The enzyme concentrations were calculated using the molecular weight of an ALDH7A1 monomer.
3.2. Structural context of the mutated residues
In common with all ALDH superfamily enzymes, the ALDH7A1 protomer-structure includes an N-terminal Rossmann NAD+-binding domain, C-terminal catalytic domain, and an oligomerization domain (Fig. 3). Two protomers associate to produce a domain-swapped dimer, in which the oligomerization domain from one protomer engages the β-sheet in the catalytic domain of the other (Fig. 4A). The highly conserved ALDH protomer and dimer structures are considered defining features of the superfamily.
Figure 3.
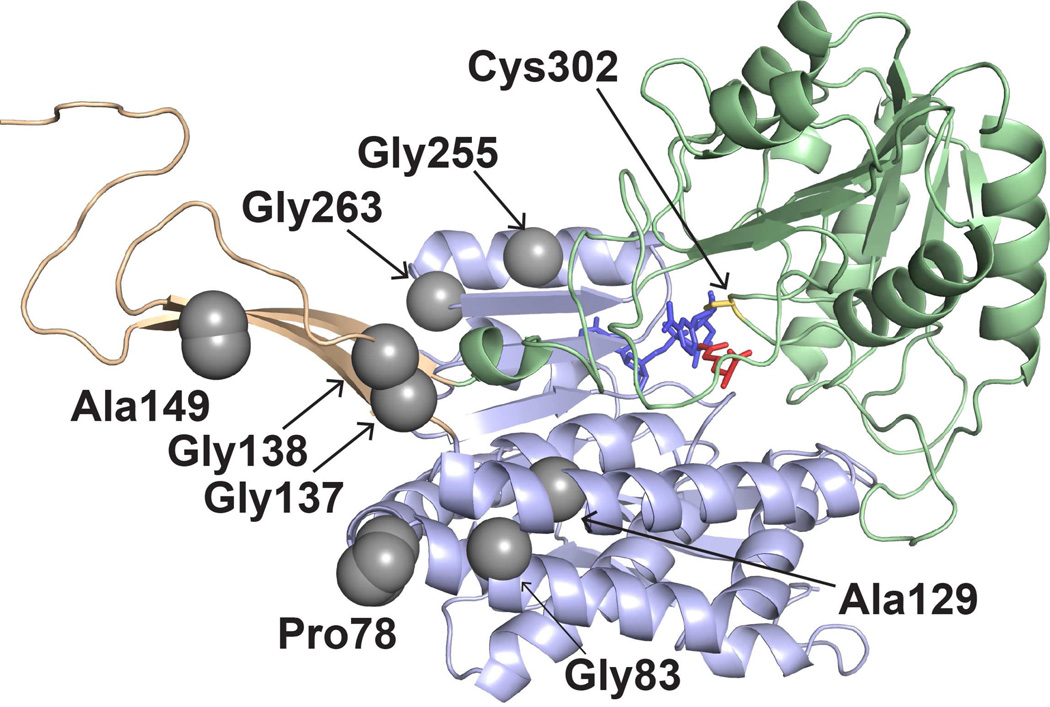
The locations of PDE-associated mutations in the ALDH7A1 protomer. The three structural domains are represented as light green (catalytic domain), light blue (NAD+-binding domain), and light orange (oligomerization domain). The active site is indicated by catalytic Cys302, NAD+ (blue), and the product α-aminoadipate (red). The gray spheres indicate the residues mutated in this study.
Figure 4.
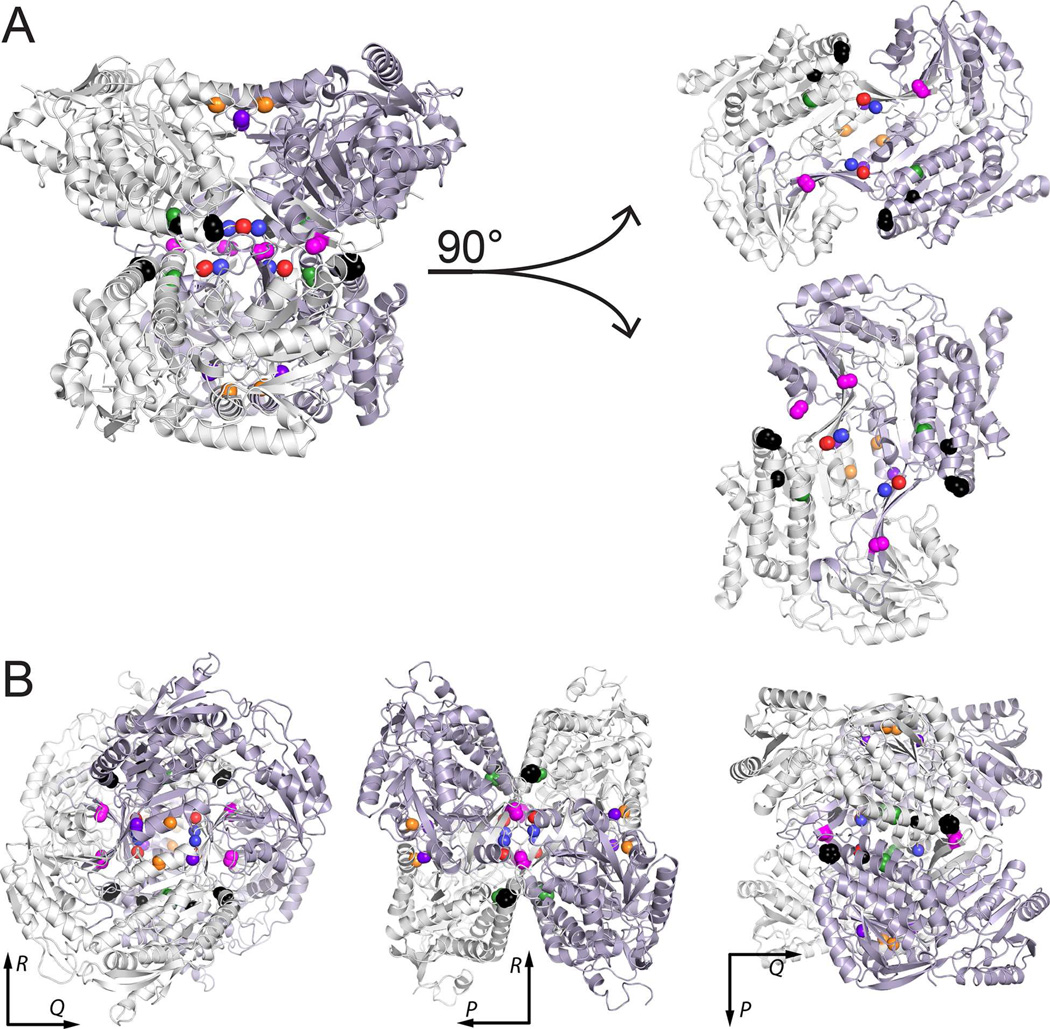
Locations of PDE-associated mutations in the protein-protein interfaces of ALDH7A1. (A) Depiction of the tetramer as a dimer of dimers, with the two dimers separated to show the interfacial locations of the mutated residues. (B) Three views of the tetramer aligned along the P, Q, and R 2-fold axes. In both panels, the locations of the residues mutated in this report are indicated by color-coded spheres: P78L (black), G83E (black), A129P (green), G137V (red), G138V (blue), A149E (magenta), G255D (orange), and G263E (purple).
In certain ALDH family members (notably ALDH7A1, ALDH1, and ALDH2), two domain-swapped dimers assemble into a tetrameric dimer-of-dimers (Fig. 4A). ALDH7A1 crystallizes as a tetramer, with point group 222 symmetry, which can be described with three mutually perpendicular 2-fold axes intersecting at the centroid of the particle. The P-axis runs parallel to the D helix of the Rossmann fold and relates the two protomers of the domain-swapped dimer (the P dimer, Fig. 4B). Rotations around the Q- and R-axes, on the other hand, relate the two dimers comprising the tetramer (Fig. 4B). Small-angle X-ray scattering [8] data suggest that the tetrameric form is also present at high concentration in solution.
The eight PDE-associated mutations selected for study are all proximal to oligomer interfaces (Figs. 3 and 4). Gly255 and Gly263 reside in a helix adjacent to the P axis within the P-dimer interface. Gly137, Gly138, and Ala149 are located in the oligomerization domain, adjacent to the Q axis, at the center of the tetramer. Their position at the centroid places these residues at the dimer-dimer interface. Pro78, Gly83, and Ala129 flank the Gly137-Gly138-Ala149 triplet and also contribute to the dimer-dimer interface.
Importantly, all of the targeted residues are remote from the active site (Fig. 3), between 18 and 28 Å (Cα – Cα distance) from the catalytic cysteine, Cys302. None of the residues directly contact NAD+ or the aldehyde substrate. Gly255, 10 Å from the adenine ring, makes the closest approach to the cofactor. Ala129, Ala149, and Gly255 make the closest approach to the aldehyde (15–16 Å). The distance from the active site suggests that the impact of the mutations on catalytic activity is indirect.
3.3. ALDH7A1 displays a dimer-tetramer equilibrium in solution
Although crystallographic and small-angle X-ray scattering data indicate that ALDH7A1 is capable of forming a tetramer both in crystallo and in solution (Fig. 4) [7–9], both techniques require relatively high protein concentrations (>1 mg/ml), which would promote formation of higher order oligomers. To obtain a more complete description of the self-association behavior, samples of wild-type ALDH7A1 were analyzed by sedimentation-equilibrium and sedimentation-velocity. In the equilibrium experiment, a protein sample is subjected to relatively low centrifugal forces so that, at equilibrium, the rates of sedimentation and diffusion balance. The resulting concentration gradient is solely dependent on the molecular weight(s) of the sedimenting species. In the velocity experiment, the sample is subjected to a relatively high centrifugal field so that there is net solute transport. The observed behavior is a function of both solute size and shape. Analysis of the velocity data returns an estimate for the distribution of sedimentation constants, c(s), which can be transformed into a corresponding molecular-weight distribution, c(M).
We collected sedimentation-equilibrium data for wild-type ALDH7A1 at three rotor-speeds and three loading concentrations (approximately 0.2, 0.4, and 0.8 mg/ml). The resulting nine radial distributions, monitored at 280 nm, were subjected to simultaneous (i.e. global) least-squares analysis.
When modeled with the assumption of a single-species (Eq 1), the optimal fit yielded an average molecular weight of 141,000 Da, intermediate between the dimer- and tetramer molecular weights (111,120 Da and 222,240 Da, respectively). This result implied that the protein resides in a dimer-tetramer equilibrium. Indeed, a substantially improved fit was obtained with a dimer-tetramer equilibrium model (Eq 2), with an association constant defined as K2–4 = [tetramer]/[dimer]2 (Figs. 5A–5C). The optimal value of the equilibrium constant derived from global fitting was 63,500 M−1, corresponding to a tetramer dissociation constant of 16 µM (Table 1). Application of a more complex model that included the monomeric form of the protein (Eq 3) did not improve the fit, suggesting that the monomer concentration is negligible under the experimental conditions.
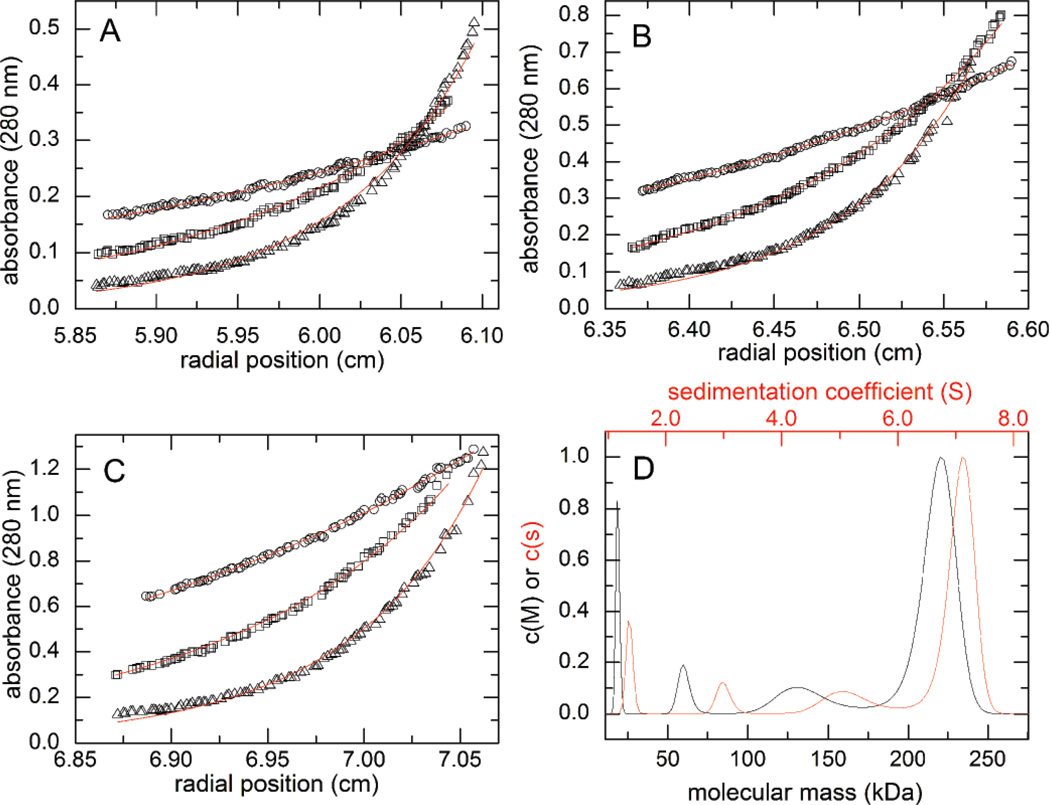
Figure 5. Analytical ultracentrifugation analysis of wild-type ALDH7A1.
(A-C) Sedimentation equilibrium data collected at three different protein concentrations: 0.2 mg/ml (A), 0.4 mg/ml (B), and 0.8 mg/ml (C). The symbols in panels A-C correspond to different centrifugation speeds: 6000 rpm (circles), 9000 rpm (squares), and 12,000 rpm (triangles). The curves represent a global fit of the data to a dimer-tetramer equilibrium model. (D) Sedimentation velocity experiment performed at 4.5 mg/ml (~80 µM, based on monomer molecular weight). The red curve corresponds to the continuous c(s) distribution, whereas the black curve corresponds to the continuous c(M) distribution. For reference, the molecular weights of the dimer and tetramer are 111 and 222 kDa.
Table 1.
Parameters obtained from sedimentation equilibrium analysis
| Average Ma(kDa) | K1–2 (M−1)b | K2–4 (M−1)c | |
|---|---|---|---|
| Wild type | 141 | N/Dd | 6.3 ± 0.3 × 104 |
| A129P | 96 | 2.3 ± 0.2 × 105 | N/D |
| G137V | 99 | 5.7 ± 0.1 × 105 | N/D |
| G138V | 103 | 14.5 ± 0.1 × 105 | N/D |
| A149E | 95 | 6.9 ± 0.7 × 105 | N/D |
| G255D | 108 | 7.5 ± 1.2 × 105 | 4.6 ± 0.5 × 104 |
| G263E | 98 | 11.5 ± 0.2 × 105 | N/D |
Average molecular weight derived from fitting with a single-species model.
Association constant for the formation of a dimer from two monomers.
Association constant for the formation of a tetramer from two dimers.
N/D indicates that the associated reaction was not included in the model used for fitting the data.
The equilibrium analysis suggested that the enzyme should be tetrameric at high concentration. Thus, a sample of ALDH7A1 was studied by sedimentation velocity at an enzyme concentration of 4.5 mg/ml (81 µM). The resulting data unequivocally indicated that ALDH7A1 is largely tetrameric in solution at this concentration (Fig. 5D), consistent with the previously determined crystal-structure data.
3.4. PDE-related mutations at protein-protein interfaces disrupt quaternary structure
Sedimentation equilibrium was performed on the ALDH7A1 variants to determine whether the mutations alter self-association behavior (Fig. 6). As described above for the wild-type protein, each of the variant enzymes was examined at three rotor speeds and three protein concentrations. Interestingly, in every case, the apparent average molecular-weight obtained with a single-species model was below that of a dimer, suggesting that the monomer concentrations were significant (Table 1). Recall that, under similar conditions, the wild-type monomer was undetectable.
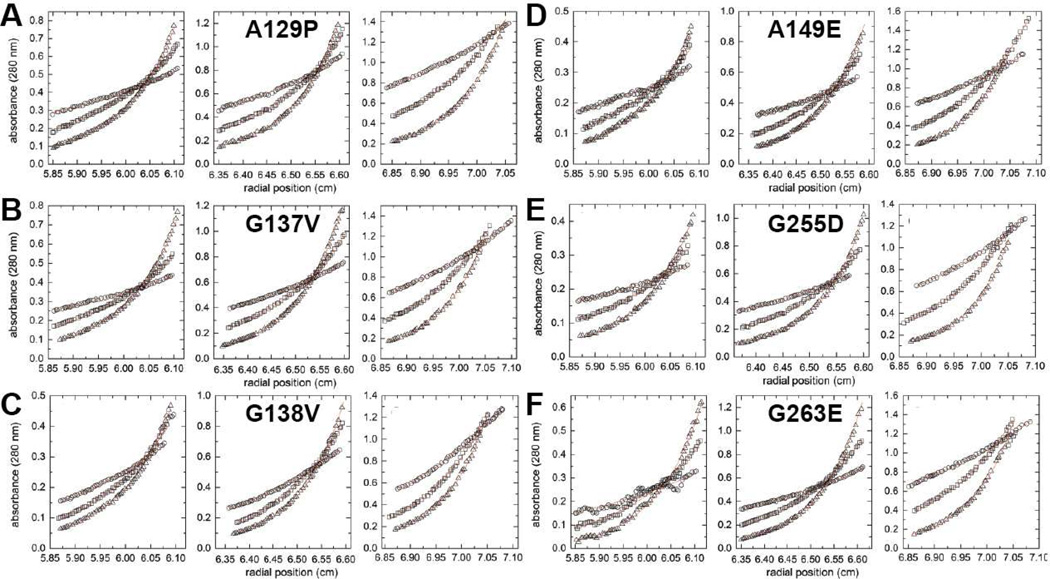
Figure 6. Sedimentation equilibrium analysis for the mutant enzymes.
For each protein, the left, middle, and right graphs show three different concentrations: 0.2 mg/ml (left), 0.4 mg/ml (middle), and 0.8 mg/ml (right). Within each graph, the three data sets correspond centrifugation speeds of 6000 rpm (circles), 9000 rpm (squares), and 12,000 rpm (triangles). The red curves represent global fits of the data to an oligomerization equilibrium model.
The equilibrium data for each the variants were also analyzed with more complex models. (Fig. 6). For all but G255D, a monomer-dimer equilibrium model yielded a satisfactory fit, implying the absence of higher order oligomers. For G255D, inclusion of a dimer-tetramer equilibrium in the model resulted in slight improvement of the fit, consistent with the possibility that higher-order oligomerization may occur in solution, (Table 1). The dimerization constants (K1–2) for the ALDH7A1 variants fall in the range between 2×105 M−1 and 15×105 M−1 (Table 1), corresponding to dissociation constants in the low micromolar range (5 µM and 0.7 µM, respectively). Altogether, the sedimentation equilibrium analyses suggest the major impact of the mutations is to perturb the interaction between monomers to make dimer formation less favorable compared to the wild-type enzyme.
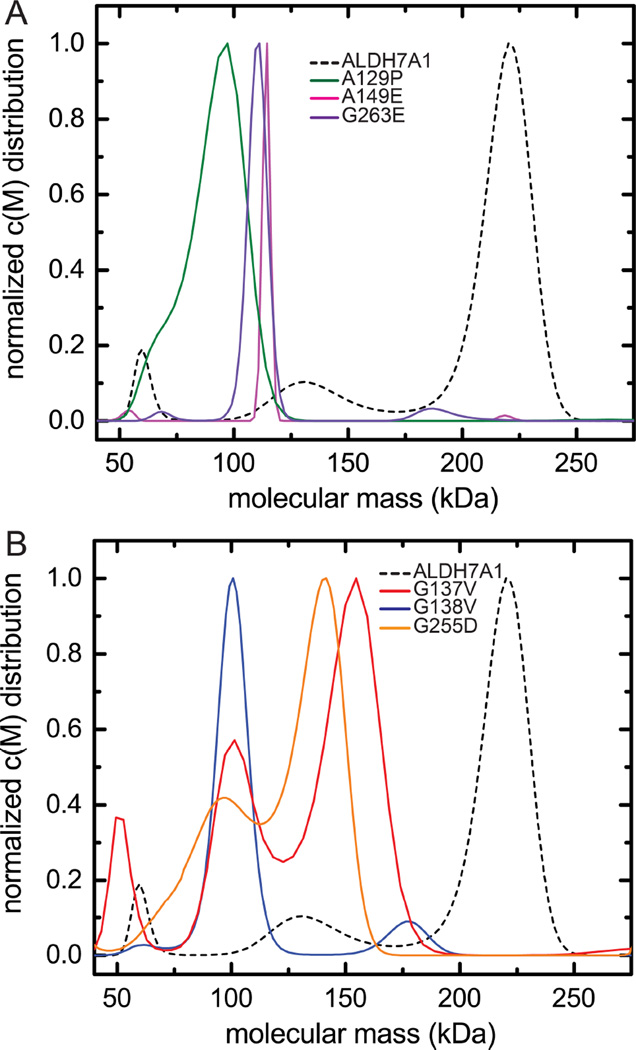
Sedimentation velocity experiments were performed to examine the oligomeric states formed at relatively high protein concentration (~80 µM). Whereas wild-type ALDH7A1 is predominantly tetrameric at this concentration (dashed line, Figure 7A-B), the velocity data for the variants suggest that the tendency for the dimers to self-associate is strongly suppressed. For A149E (Fig. 7A, magenta) and G263E (Fig. 7A purple), the c(M) maxima are close to the molecular mass expected for the dimer, implying that these variants are almost exclusively dimeric, even at the elevated protein concentration. The peak observed for A129P (Fig. 7A, green), centered just under 100 kDa, probably represents a rapidly equilibrating mixture of monomeric and dimeric species. The shoulder located near the monomer molecular mass is consistent with that idea. The major peak observed for G138V (Fig. 7B, blue) is likewise believed to reflect a rapidly equilibrating mixture of monomers and dimers.
Figure 7.
Sedimentation velocity analysis of mutant ALDH7A1 conducted at high concentration (~80 µM, monomer molecular weight). (A) Sedimentation velocity data for A129P (green), A149E (magenta), and G263E (purple). The result for wild-type ALDH7A1 (also ~80 µM) is shown for comparison (dashed black). (B) Sedimentation velocity data for G137V (red), G138V (blue), and G255D (orange). The result for wild-type ALDH7A1 is shown for comparison (dashed black).
The molecular-weight distributions obtained for G137V (Fig. 7B, red) and G255D (Fig. 7B, orange) are more complex, both variants exhibiting a prominent peak near 150 kDa. Although that value approaches the expected molecular mass for a trimeric species, it is more likely that the peaks reflect rapidly equilibrating dimer-tetramer mixtures. Recall that inclusion of dimer-tetramer association improved the least-squares fit of equilibrium data for G255D. This variant also yields a prominent peak near 100 kDa, consistent with a monomer-dimer mixture.
The molecular-weight distribution obtained for G137V resembles that of G255D. However, unlike G255D, the least-squares fit of the G137V sedimentation-equilibrium data was not improved by inclusion of dimer-tetramer association. This result suggests that tetramer formation is negligible for G137V at low µM concentrations and, in turn, that the K2–4 value for G137V is smaller than that for G255D. Evidently, none of the variant proteins forms a stable dimer-of-dimers under these experimental conditions, suggesting that association of the mutated dimers is not facile.
4. Discussion
Knowledge of the molecular basis of an inherited genetic disease is crucial for understanding the disease pathology and, ultimately, for therapeutic design. Several recent reports have stressed the need for physical characterization of disease-associated protein variants [10, 18], emphasizing the inability of structural data alone to reveal the functional impact of a particular mutation. Previous studies on the effect of PDE-related mutations on ALDH7A1 activity used either transient expression [3] or analysis of crude lysate from heterologous expression [19, 20]. In this study, we have conducted detailed biochemical and biophysical analyses on the purified variant proteins, in an effort to understand how these previously uncharacterized mutations, positioned at or near the interfaces of the ALDH7A1 quaternary structure, impact activity and structure.
Whereas previous structural studies suggested a tetrameric quaternary structure for ALDH7A1 [7–9], our results indicate more complex self-association behavior. Analysis of sedimentation-equilibrium data (Table 1) suggests that ALDH7A1 will be predominately dimeric at standard enzyme-assay concentrations (i.e., ≈ 1 µM), with only minor amounts of tetramer present. In this respect, ALDH7A1 resembles ALDH3A1 [21] and ALDH4A1 [22], previously shown to be dimers. Whether our in vitro sedimentation measurements accurately describe the self-assocation behavior of ALDH7A1 in vivo is unknown. For example, it is possible that molecular crowding effects could increase the affinity of dimers to form tetramers in vivo. Nevertheless, the ALDH7A1 activity in vitro scales with enzyme concentration between 0.5 and 4.0 µM, a range over which the percentage of tetramer increases from 3% to 17% of the total protein. This finding suggests that the specific activities of the dimeric and tetrameric forms may be comparable. Another possibility is the dimers have low or no activity, and the solution conditions of the activity assay promote formation of highly active tetramers. In this mechanism, the binding of substrates to the enzyme may facilitate tetramerization. Additional studies are needed to distinguish between these scenarios.
All examined PDE-related mutations, proximal to the protein-protein interfaces of ALDH7A1, abolish enzyme activity. Neither P78L nor G83E is expressed in appreciable amounts as soluble protein in E. coli, therefore, they were not included in these studies. Although the remaining variants (A129P, G137V, G138V, A149E, G255D, G236E) express well and are soluble, they exhibit altered quaternary structure. Whereas the wild-type protein is primarily dimeric at low µM concentrations, the variants reside in monomer-dimer equilibria, characterized by dissociation constants between 0.7 and 5 µM. The total inability of the variants to catalyze the oxidation of AASAL implies that the monomers are inactive and, moreover, that their self-association does not restore activity. The latter conclusion suggests that the association surfaces of the mutated enzymes are sufficiently altered so as to render formation of the wildtype dimer-interface unfavorable.
Parenthetically, our results are reminiscent of a previous ALDH1 study in which simultaneous mutation of two interface residues disrupted tetramer formation and severely compromised enzymatic activity [23]. This behavior is evidently not universal, given that certain mutations that disrupt the hexameric form of bacterial ALDH4A1 do not affect enzymatic activity [24].
Although the ALDH7A1 PDE variants display a range of apparent molecular masses at high concentration, ranging from approximately 50 kDa to 175 kDa, they do not approach that of the tetramer, 222 kDa. Evidently, the mutations also disrupt dimer-dimer association. The latter finding suggests that the tetramer may serve as a proxy for the capacity to form the enzymatically active domain-swapped dimer that is the hallmark of the ALDH superfamily.
5. Conclusions
In summary, our results suggest that PDE-associated mutations perturb the native protein structure, altering self-association behavior and resulting in the loss of catalytic activity. These findings provide important data for understanding the molecular basis of PDE-related genetic mutations remote from the ALDH7A1 active site.
Supplementary Material
Highlights.
ALDH7A1 catalyzes a step in lysine catabolism.
Inherited mutations in ALDH7A1 cause pyridoxine-dependent epilepsy.
Disease mutations targeting residues in the oligomer interfaces are studied.
Steady-state kinetics and analytical ultracentrifugation are used.
The mutations abolish activity and alter the self-association behavior of ALDH7A1.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM093123.
Abbreviations
- ALDH
aldehyde dehydrogenase
- AASAL
α-aminoadipate semialdehyde
- P6C
Δ1-piperideine-6-carboxylic acid
- PDE
pyridoxine-dependent epilepsy
- PLP
pyridoxal 5′-phosphate.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem. Cell. Rev. 2011;7:292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 2.Muzio G, Maggiora M, Paiuzzi E, Oraldi M, Canuto RA. Aldehyde dehydrogenases and cell proliferation. Free Radic. Biol. Med. 2012;52:735–746. doi: 10.1016/j.freeradbiomed.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 3.Mills PB, Struys E, Jakobs C, Plecko B, Baxter P, Baumgartner M, Willemsen MA, Omran H, Tacke U, Uhlenberg B, Weschke B, Clayton PT. Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat. Med. 2006;12:307–309. doi: 10.1038/nm1366. [DOI] [PubMed] [Google Scholar]
- 4.Stockler S, Plecko B, Gospe SM, Jr, Coulter-Mackie M, Connolly M, van Karnebeek C, Mercimek-Mahmutoglu S, Hartmann H, Scharer G, Struijs E, Tein I, Jakobs C, Clayton P, Van Hove JL. Pyridoxine dependent epilepsy and antiquitin deficiency: clinical and molecular characteristics and recommendations for diagnosis, treatment and follow-up. Mol. Genet. Metab. 2011;104:48–60. doi: 10.1016/j.ymgme.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Stenson PD, Ball E, Howells K, Phillips A, Mort M, Cooper DN. Human Gene Mutation Database: towards a comprehensive central mutation database. J Med Genet. 2008;45:124–126. doi: 10.1136/jmg.2007.055210. [DOI] [PubMed] [Google Scholar]
- 6.Mozzarelli A, Bettati S. Exploring the pyridoxal 5'-phosphate-dependent enzymes. Chem Rec. 2006;6:275–287. doi: 10.1002/tcr.20094. [DOI] [PubMed] [Google Scholar]
- 7.Luo M, Gates KS, Henzl MT, Tanner JJ. Diethylaminobenzaldehyde is a covalent, irreversible inactivator of ALDH7A1. ACS Chem. Biol. 2015;10:693–697. doi: 10.1021/cb500977q. [DOI] [PubMed] [Google Scholar]
- 8.Luo M, Tanner JJ. Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1. Biochemistry. 2015;54:5513–5522. doi: 10.1021/acs.biochem.5b00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brocker C, Lassen N, Estey T, Pappa A, Cantore M, Orlova VV, Chavakis T, Kavanagh KL, Oppermann U, Vasiliou V. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J Biol Chem. 2010;285:18452–18463. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao M, Zhou H, Skolnick J. Insights into Disease-Associated Mutations in the Human Proteome through Protein Structural Analysis. Structure. 2015;23:1362–1369. doi: 10.1016/j.str.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mills PB, Footitt EJ, Mills KA, Tuschl K, Aylett S, Varadkar S, Hemingway C, Marlow N, Rennie J, Baxter P, Dulac O, Nabbout R, Craigen WJ, Schmitt B, Feillet F, Christensen E, De Lonlay P, Pike MG, Hughes MI, Struys EA, Jakobs C, Zuberi SM, Clayton PT. Genotypic and phenotypic spectrum of pyridoxine-dependent epilepsy (ALDH7A1 deficiency) Brain : a journal of neurology. 2010;133:2148–2159. doi: 10.1093/brain/awq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher RC, Van Hove JL, Scharer G, Hyland K, Plecko B, Waters PJ, Mercimek-Mahmutoglu S, Stockler-Ipsiroglu S, Salomons GS, Rosenberg EH, Struys EA, Jakobs C. Folinic acid-responsive seizures are identical to pyridoxine-dependent epilepsy. Ann Neurol. 2009;65:550–556. doi: 10.1002/ana.21568. [DOI] [PubMed] [Google Scholar]
- 13.Scharer G, Brocker C, Vasiliou V, Creadon-Swindell G, Gallagher RC, Spector E, Van Hove JL. The genotypic and phenotypic spectrum of pyridoxine-dependent epilepsy due to mutations in ALDH7A1. J. Inherit. Metab. Dis. 2010;33:571–581. doi: 10.1007/s10545-010-9187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett CL, Chen Y, Hahn S, Glass IA, Gospe SM., Jr Prevalence of ALDH7A1 mutations in 18 North American pyridoxine-dependent seizure (PDS) patients. Epilepsia. 2009;50:1167–1175. doi: 10.1111/j.1528-1167.2008.01816.x. [DOI] [PubMed] [Google Scholar]
- 15.Plecko B, Paul K, Paschke E, Stoeckler-Ipsiroglu S, Struys E, Jakobs C, Hartmann H, Luecke T, di Capua M, Korenke C, Hikel C, Reutershahn E, Freilinger M, Baumeister F, Bosch F, Erwa W. Biochemical and molecular characterization of 18 patients with pyridoxine-dependent epilepsy and mutations of the antiquitin (ALDH7A1) gene. Hum Mutat. 2007;28:19–26. doi: 10.1002/humu.20433. [DOI] [PubMed] [Google Scholar]
- 16.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroncke BM, Vanoye CG, Meiler J, George AL, Jr, Sanders CR. Personalized biochemistry and biophysics. Biochemistry. 2015;54:2551–2559. doi: 10.1021/acs.biochem.5b00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coulter-Mackie MB, Tiebout S, van Karnebeek C, Stockler S. Overexpression of recombinant human antiquitin in E. coli: partial enzyme activity in selected ALDH7A1 missense mutations associated with pyridoxine-dependent epilepsy. Mol Genet Metab. 2014;111:462–466. doi: 10.1016/j.ymgme.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Coulter-Mackie MB, Li A, Lian Q, Struys E, Stockler S, Waters PJ. Overexpression of human antiquitin in E. coli: enzymatic characterization of twelve ALDH7A1 missense mutations associated with pyridoxine-dependent epilepsy. Mol Genet Metab. 2012;106:478–481. doi: 10.1016/j.ymgme.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Estey T, Chen Y, Carpenter JF, Vasiliou V. Structural and functional modifications of corneal crystallin ALDH3A1 by UVB light. PLoS One. 2010;5:e15218. doi: 10.1371/journal.pone.0015218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava D, Singh RK, Moxley MA, Henzl MT, Becker DF, Tanner JJ. The Three-Dimensional Structural Basis of Type II Hyperprolinemia. J. Mol. Biol. 2012;420:176–189. doi: 10.1016/j.jmb.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Zavala JS, Weiner H. Structural aspects of aldehyde dehydrogenase that influence dimer-tetramer formation. Biochemistry. 2002;41:8229–8237. doi: 10.1021/bi012081x. [DOI] [PubMed] [Google Scholar]
- 24.Luo M, Singh RK, Tanner JJ. Structural determinants of oligomerization of delta(1)-pyrroline-5-carboxylate dehydrogenase: identification of a hexamerization hot spot. J. Mol. Biol. 2013;425:3106–3120. doi: 10.1016/j.jmb.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.