Abstract
Background
While epithelial–mesenchymal transition (EMT) can be readily induced experimentally in cancer cells, the EMT process as manifested in human tumors needs to be better understood. Pan-cancer genomic datasets from The Cancer Genome Atlas (TCGA)—representing over 10,000 patients and 32 distinct cancer types—provide a rich resource for examining correlative patterns involving EMT mediators in the setting of human cancers.
Results
Here, we surveyed a 16-gene signature of canonical EMT markers in TCGA pan-cancer cohort. The histology or cell-of-origin of a tumor sample may align more with mesenchymal or epithelial phenotype, and non-cancer as well as cancer cells can contribute to the overall molecular patterns observed within a tumor sample; correlation models involving EMT markers can factor in both of the above variables. EMT-associated genes appear coordinately expressed across all cancers and within each cancer type surveyed. Gene signatures of immune cells correlate highly with EMT marker expression in tumors. In pan-cancer analysis, several EMT-related genes can be significantly associated with worse patient outcome.
Conclusion
Gene correlates of EMT phenotype in human tumors could include novel mediators of EMT that might be confirmed experimentally, by which TCGA datasets may serve as a platform for discovery in ongoing studies.
Introduction
Epithelial–mesenchymal transition (EMT), a reversible dynamic process by which epithelial cells acquire characteristics of mesenchymal cells, is involved in the initiation of metastasis during cancer progression (Kalluri and Weinberg, 2009). Among other things, cancer cells undergoing EMT gain migratory and invasive properties and acquire stem cell traits (Ye and Weinberg, 2015). Genes most commonly associated with EMT include those encoding markers of mesenchymal cells—including vimentin (VIM), N-cadherin (CDH2), fibronectin (FN1), integrin αvβ6 (ITGB6)—which are increased during EMT, as well as those encoding markers of epithelial cells—including E-cadherin (CDH1), desmoplakin (DSP), and occludin (OCLN)—which are decreased during EMT. Transcription factors that regulate EMT include the zinc-finger proteins Snail1 and Snail2 (SNAI1 and SNAI2 genes, respectively), the two-handed zinc-finger δEF1 family factors (δEF1/Zeb1 and SIP1/Zeb2, encoded by ZEB1 and ZEB2 genes, respectively), and the basic helix–loop–helix factors, Twist and E12/E47 (TWIST1/TWIST2 and TCF3 genes, respectively). While EMT can be readily induced in cancer cells under experimental conditions, with the effects being observed in model systems, observations of EMT in human tumor specimens can help establish the relevance of the process in the setting of cancer as it is manifested in patients (Kalluri and Weinberg, 2009; Creighton et al., 2010).
The Cancer Genome Atlas (TCGA) was a large-scale scientific effort to systematically characterize the genomic changes that occur in cancer, which involved comprehensive molecular profiling of over 10,000 cancers of various types, with the associated molecular datasets including somatic mutation, gene expression, DNA methylation, and DNA copy alteration. With the recent conclusion of the data generation phase of TCGA, there is opportunity for “second wave” analyses of the entire TCGA pan-cancer cohort, to address questions not covered by the initial TCGA-led studies that first presented the data by individual tumor type. Data from TCGA have been made available to the scientific community at large, e.g. through the Genome Data Commons (https://gdc.cancer.gov/) or through The Broad Institute’s Firehose pipeline (http://gdac.broadinstitute.org/).
This review article follows an overall format similar to that of our other recent reviews (Creighton et al., 2010; Creighton et al., 2013), whereby concepts related to EMT are discussed in light of both the current literature and results readily obtainable from publicly available genomic datasets. In particular, here we survey TCGA pan-cancer datasets for expression of genes canonically associated with EMT, in order to determine whether these genes appear coordinately expressed and in which cancer subsets. Gene “signatures” of EMT are considered here, whereby expression patterns for a set of genes associated with EMT may be summarized into a single score for each tumor profile. The data underlying the results presented here have been made available as a supplemental data file (Data File S1).
Results and Discussion
Gene signatures of epithelial–mesenchymal transition (EMT)
In many studies, individual EMT markers (e.g. vimentin or E-cadherin) are examined in human tumor specimens (e.g. using tissue microarrays or quantitative real-time polymerase chain reaction), where cancer cells that show up-regulation of mesenchymal markers or down-regulation of epithelial markers are thought to have undergone EMT. Another approach to examining EMT in tumors, where global expression profiling data are available, is to use pre-defined gene “signatures” to score each specimen for manifestation of mesenchymal- versus epithelial-associated patterns. A number of gene signatures of EMT have been defined in the literature, e.g. through the use of experimental models or through identifying gene correlates of mesenchymal or epithelial markers (Gröger et al., 2012; Byers et al., 2013; Tan et al., 2014; Zhao et al., 2015; Mak et al., 2016). In our own studies (Creighton et al., 2009; Creighton et al., 2013; Chen et al., 2016a; Chen et al., 2016b), we the authors have made use of a pre-defined EMT signature consisting of a short set of canonical EMT markers as originally put forth by Lee et al. (Lee et al., 2006), with 13 mesenchymal marker genes (VIM, CDH2, FOXC2, SNAI1, SNAI2, TWIST1, FN1, ITGB6, MMP2, MMP3, MMP9, SOX10, GCS) and three epithelial marker genes (CDH1, DSP, OCLN). The advantages of this 16-gene signature include its simplicity in focusing on some of the most well established markers, the result of the sum accumulation of a multitude of previous studies. In this article, we explore this 16-gene signature in TCGA, where the tumor profiles may be probed for degree of manifestation of the signature, allowing us to draw correlations of potential interest that may be associated with an EMT phenotype.
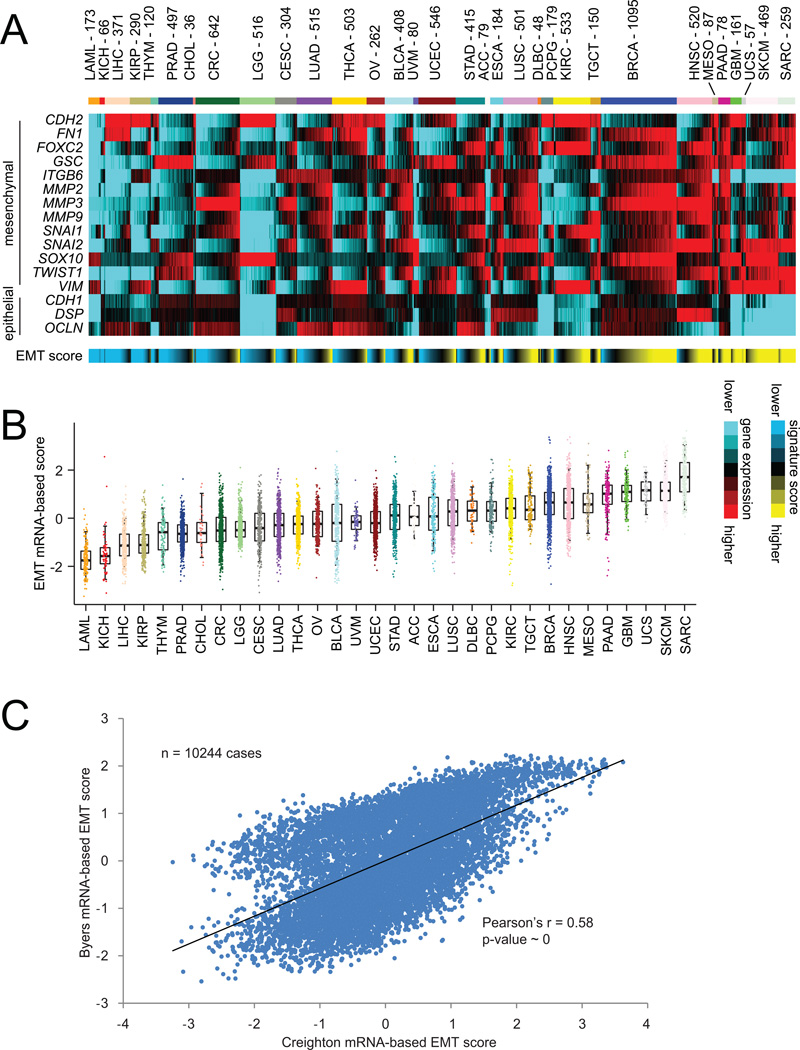
For the 10244 tumor mRNA expression profiles represented in the entire TCGA pan-cancer cohort spanning 32 different cancer types, normalized expression values (normalized to standard deviations from the median) were examined for the 16 genes in our EMT signature (Figure 1A). By carrying out a simple addition of the mesenchymal-associated gene values and a corresponding subtraction of the epithelial-associated gene values, each tumor profile was given a summary score for EMT phenotype. As would be expected, EMT scores differed considerably on average by tumor lineage, with, for example leukemia samples (“LAML”) appearing the most epithelial and with sarcomas (“SARC”) appearing the most mesenchymal; at the same time, within many tumor types a wide range of EMT manifestation levels were evident (Figure 1B). In addition to the above EMT signature, other EMT-associated signatures could be surveyed in a similar manner in TCGA data. For example, in a previous study by Byers et al. a 76-gene EMT signature was defined using gene expression profiles of lung cancer cell lines, where the genes were selected on the basis of correlation with CDH1, VIM, CDH2, and/or FN1 genes. Using this Byers signature, we scored TCGA profiles, using our previously described “t-score” metric (Cancer_Genome_Atlas_Research_Network, 2011). Across all tumors, we observe very high correlation (Pearson’s r=0.58, which can be considered quite significant, given the large number of cases involved) between the 16-gene EMT scores and the Byers EMT scores (Figure 1C), which would reflect the notion that different gene signatures reflecting the same biological process should yield largely concordant results.
Figure 1. Gene expression signatures of epithelial–mesenchymal transition (EMT) across human cancers of various types.
(A) Heat map of gene expression (mRNA) features representing canonical EMT markers (from the review article by Lee et al. (Lee et al., 2006)), across 10244 cancers represented in The Cancer Genome Atlas (TCGA) datasets. Red, higher expression (relative to median across all cancers); blue, lower expression. These features were summarized into an EMT signature score for each tumor profile (yellow, more mesenchymal-like; blue, more epithelial-like). Cancer types (denoted by TCGA project name) are ordered by low to high average EMT score. Cancer type abbreviations are as follows: LAML, Acute Myeloid Leukemia; ACC, Adrenocortical carcinoma; BLCA, Bladder Urothelial Carcinoma; LGG, Brain Lower Grade Glioma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; CRC, Colorectal adenocarcinoma (combining COAD and READ projects); ESCA, Esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and Neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; MESO, Mesothelioma; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; STAD, Stomach adenocarcinoma; TGCT, Testicular Germ Cell Tumors; THYM, Thymoma; THCA, Thyroid carcinoma; UCS, Uterine Carcinosarcoma; UCEC, Uterine Corpus Endometrial Carcinoma. (B) Box plots of EMT signature scores, by cancer type. Box plots represent 5%, 25%, 50%, 75%, and 95%. (C) For the 10244 cancer samples with mRNA data available, scatterplot comparing EMT scores based on part A (referred to here as the “Creighton” EMT signature, as previously featured in (Creighton et al., 2013)) with EMT scores based on another previously published signature by Byers et al. (Byers et al., 2013). P-value by Pearson’s correlation.
Gene correlates of EMT marker expression as observed in human tumor specimens
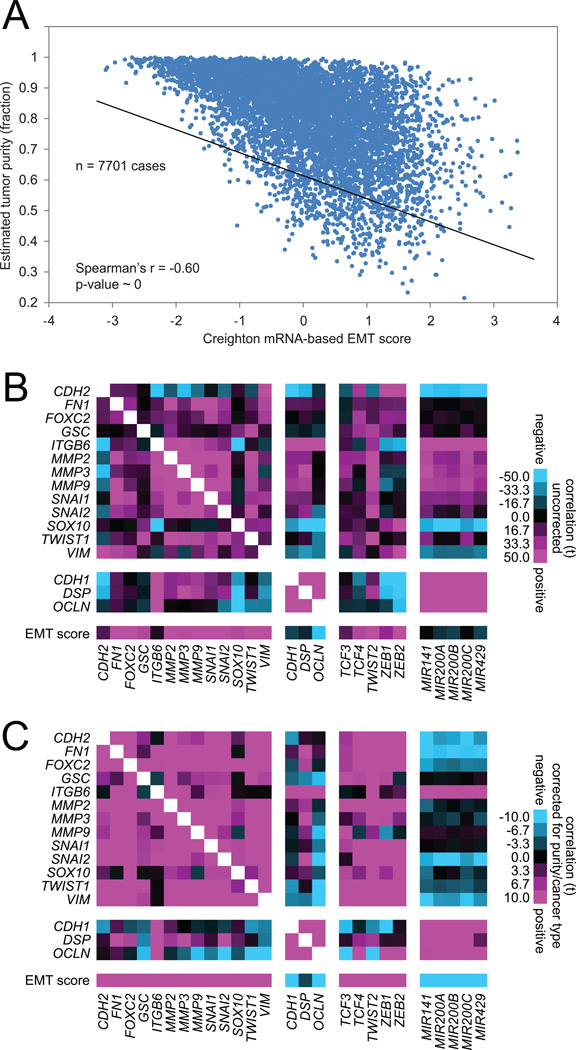
When interpreting the results of gene expression signatures of EMT as applied to human tumor specimens, a number of factors ought to be taken into consideration. As indicated above (Figure 1A), the cell of origin of a cancer can often be the main driver of manifestation of a mesenchymal versus epithelial phenotype, and so signature patterns in this case may not necessarily be reflective of cellular changes occurring within the cancer. In addition, EMT can appear manifested in only a subset of tumor cells, such as at the tumor-host interface (Paterson et al., 2013), where the tumor expression profile represents the aggregate of the various cellular populations within the sample. With regards to differing cellular populations, non-cancer as well as cancer cells contribute to the overall molecular patterns observed within a tumor sample. For example, fibroblasts or immune cells within the sample could conceivably contribute to mesenchymal-like or epithelial-like gene expression patterns, respectively, being identified. While requirements for tumor samples submitted to TCGA generally involved a minimum of 50% “purity” for cancer cells being present in the sample, tumor purities across TCGA cohort have been found to vary substantially, based on various computational- or pathology-based estimates (Aran et al., 2015). Nevertheless, EMT phenotypes can be observed in cancer cell lines where cancer cell purity would not be an issue (Gibbons et al., 2009; Byers et al., 2013; Tan et al., 2014), and analysis of human tumors can involve incorporation of such variables as tumor type and tumor purity as covariates, to identify gene correlates of EMT marker expression that remain significant and would therefore be more likely to be cancer-specific.
For this article, we obtained the sample purity scores from Aran et al. (Aran et al., 2015), for 7701 TCGA cases with corresponding mRNA expression data. As might be expected, there is a strong negative correlation (Spearman’s r=−0.60) between estimated tumor purity and scoring for our 16-gene EMT signature (Figure 2A); in other words, tumor samples that were more pure for cancer cells tended to score as more epithelial-like, and tumor samples that were less pure tended to score as more mesenchymal-like. In general, individual genes canonically associated with EMT (e.g. genes in our 16-gene signature) were strongly correlated in RNA expression with each other and with EMT signature score across cancers (Figure 2B). As the above correlations (by Pearson’s) would involve a number of factors not necessarily specific to changes within cancer cells, we also computed gene-to-gene correlations using a linear regression model incorporating both tumor purity and cancer type in addition to mRNA expression (Figure 2C). In the linear regression model, gene-to-gene correlations may remain statistically significant if they cannot be entirely accounted for by either purity or cancer type. We find that most of the EMT gene-to-gene correlations by the “uncorrected” model (Figure 2B) remain significant in the “corrected” linear regression model (i.e. corrected for both purity and tumor type, Figure 2C), though there are some notable differences; for example, miR-200 family members show negative correlations with most mesenchymal markers in the corrected model but not the uncorrected model, and N-cadherin gene (CDH2) is negatively correlated with other mesenchymal markers in the uncorrected model but positively correlated with the same markers in the corrected model.
Figure 2. Correlations between EMT-associated RNA features across human cancers.
(A) For the 7701 cancer samples with both mRNA data and estimated tumor purity data (Aran et al., 2015) available, scatterplot comparing EMT scores (from Figure 1A) with estimated tumor sample purity (fraction of cancer cells versus total cells). P-value by Spearman’s correlation. (B) Pearson’s correlations between RNA expression features across all 7701 cancers, involving canonical EMT markers (Lee et al., 2006), core transcriptional regulators of EMT (e.g. TCF3/4, TWIST, ZEB1/2), miR-200 family members (where 6491 of the 7701 samples had microRNA expression data), and the EMT signature score. (C) Correlations between EMT-associated mRNA expression features across all 7701 cancers, using a linear regression model incorporating both tumor purity and cancer type in addition to mRNA expression. For parts B and C, t-statistics greater than 2 or less than −2 would be within statistical significance (p<0.05).
Through the analysis of molecular data from human tumor specimens, many genes may be found to correlate in expression with manifestation of EMT, some of which could conceivably represent novel regulators or mediators of EMT, and others of which could represent important downstream consequences of EMT. For example, in one of our own recent studies (Ungewiss et al., 2016), we analyzed the TCGA pan-cancer datasets for genes with negative correlation to miR-200 family expression, high correlation to ZEB1, and predicted miR200b sites in the 3′ UTR by three different prediction algorithms, with hundreds of genes meeting the above criteria. When this gene list was further filtered for genes associated with poor patient prognosis in lung cancer, 29 genes were identified, including CRKL, a frequently amplified oncogene in lung cancer. Follow-up experimental studies confirmed that CRKL is a miR-200 target and furthermore found that CRKL protein regulates integrin-dependent signaling (Ungewiss et al., 2016). In similar fashion to the above example, TCGA and other public cancer genomics dataset may be interrogated for gene correlates of EMT, which may yield candidates for investigation as to their potential roles involving EMT.
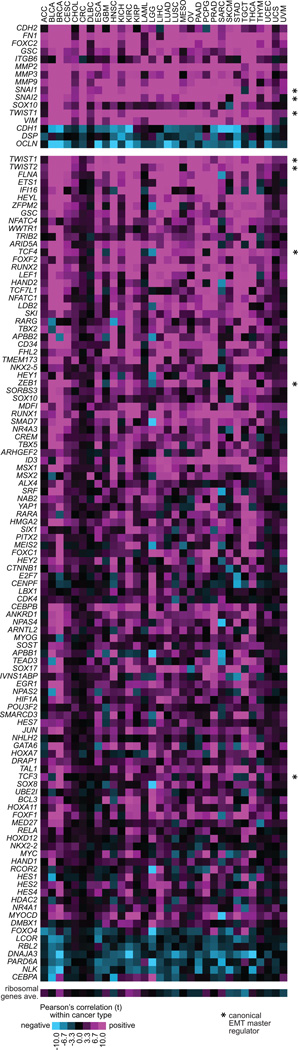
For this article, we examined a set of 377 genes with roles in regulating gene transcription (specifically, genes having a Gene Ontology annotation of “transcription factor complex” or “transcription factor binding”), for correlations with an EMT phenotype in human tumors. Gene transcriptional regulators can have important roles in cellular function, and although correlation would not in and of itself demonstrate a cause-and-effect relationship, strong correlations of a gene with EMT might suggest a hypothesis of a functional role that could be further explored in the experimental setting. In TCGA pan-cancer datasets, 107 of the above 377 gene (Figure 3) had highly significant correlations with the 16-gene EMT signature score across tumors (p<1E-20 by Pearson’s and p<1E-20 by linear regression model incorporating both tumor purity and cancer type in addition to mRNA expression). Each of the 107 genes showed significant EMT signature correlations within multiple individual tumor types (Figure 3), indicating both that these correlations are independent of cancer type and that the correlative relationships represented would span various histological or cellular backgrounds.
Figure 3. Top transcription regulators correlating with EMT phenotype in human tumors.
An initial set of 377 genes with Gene Ontology annotation of “transcription factor complex” or “transcription factor binding” were selected, and correlations for these genes with EMT phenotype across the tumors in the TCGA pan-cancer dataset were computed. Of the 377 genes, 107 (shown in the bottom panel) had highly significant correlations with the EMT signature score across tumors (p<1E-20 by Pearson’s and p<1E-20 by linear regression model incorporating both tumor purity and cancer type in addition to mRNA expression). For each cancer type (columns), the corresponding Pearson’s correlation with the EMT signature score (from Figure 1A, individual genes featured in the top panel) is shown. A t-statistic greater than 2 or less than −2 would be within statistical significance (p<0.05). Asterisk (*) denotes genes most commonly associated as master regulators of EMT. See Figure 1 legend for cancer type abbreviations. Correlations between EMT score and the averaged normalized values of a set of 25 ribosomal genes—housekeeping genes which would be presumed to not be functionally involved with EMT—are shown, to serve as a type of negative control.
A number of the correlations identified in Figure 3 are reflective of previously established functional relationships between EMT and specific gene and pathways. For example, Notch deregulation is understood to be involved in EMT and tumor aggressiveness (Wang et al., 2010; Yang et al., 2011; Hu et al., 2012), as would appear reflected in the positive correlations observed here between EMT signature and Notch pathway genes such as HEY1, HEY2, HES1, and HES2. Other genes in Figure 3 include HIF1A, YAP1, CTNNB1, and ETS1. HIF1A and hypoxia regulate EMT in cancer (Tsai and Wu, 2012; Kao et al., 2016). Recently, YAP1 has been found to transcriptionally regulate EMT in various cancer types (Shao et al., 2014; Selth et al., 2016; Yuan et al., 2016). Wnt/β-catenin pathway (represented by CTNNB1, LEF1, TCF4, etc.) has long been understood to regulate EMT (Lamouille et al., 2014; Huang et al., 2015), and ETS1 also contributes to EMT (Dittmer, 2015). We and others have also recently demonstrated a role for FOXF2 in driving EMT and metastasis in several different epithelial tumor types in vitro and in vivo (Kundu et al., 2015; Lo et al., 2016). Other genes in Figure 3 that have yet to be extensively studied in the context of EMT might be further explored in future studies.
Gene signatures of immune cells correlating with EMT marker expression in human tumor specimens
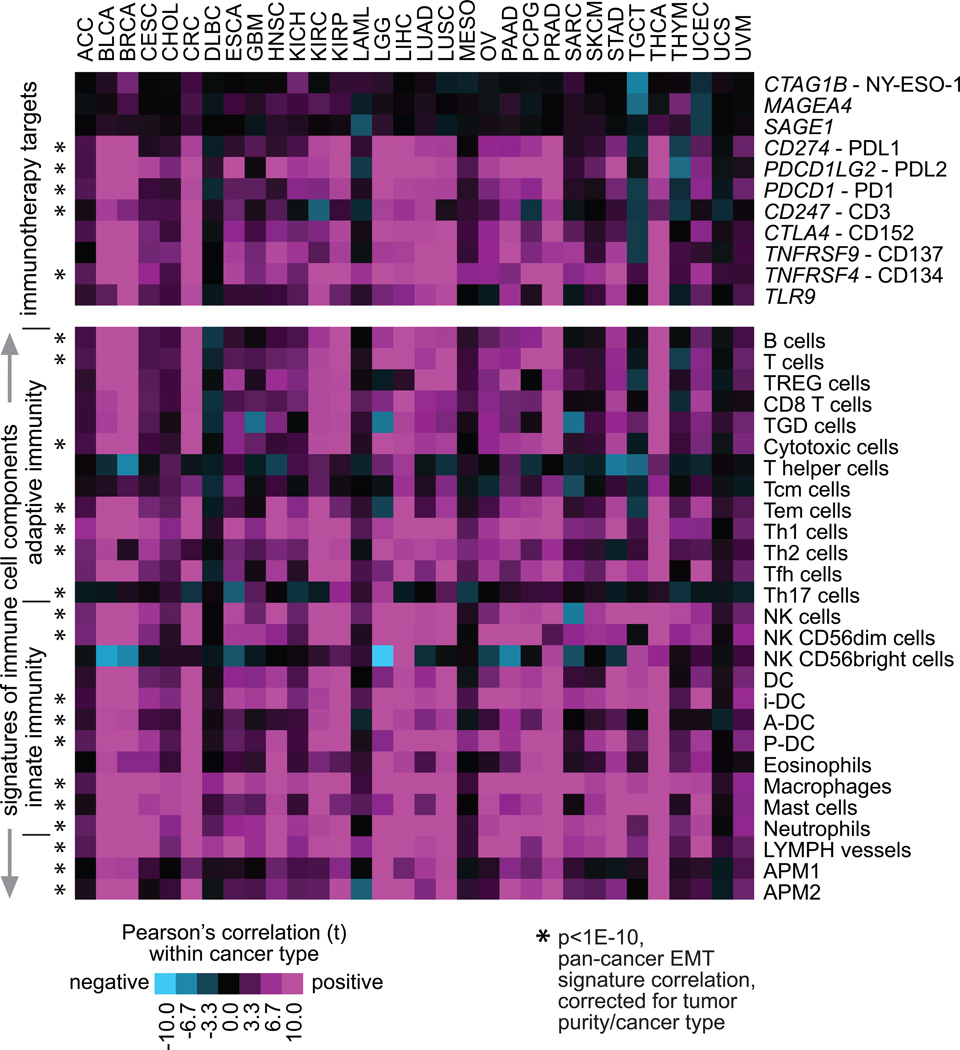
While the impact of EMT on reprogramming the tumor immune microenvironment is largely unknown, studies have demonstrated that transcription factors, such as Snail and Zeb1, that induce EMT are also associated with the activation of immunosuppressive cytokines and T-lymphocyte resistance in experimental models (Kudo-Saito et al., 2009; Chen et al., 2014; Chen et al., 2016c; Lou et al., 2016). In human lung tumors, cases displaying a “mesenchymal” phenotype are associated with distinct tumor microenvironment changes, including elevated expression of multiple immune checkpoints, such as PD-1 and PD-L1, along with evidence of preexisting immunity and increases in tumor infiltration by T cells (Kim et al., 2016; Lou et al., 2016). In a recent pan-cancer study by Mak et al. (Mak et al., 2016), an EMT signature was used to score 1934 different tumors (including breast, lung, colon, ovarian, and bladder cancers), where tumors showing high mesenchymal phenotypes tended to show high expression of immune checkpoints and other druggable immune targets. A study by Bindea et al. (Bindea et al., 2013) defined gene expression signatures of different immune cell types. In two recent studies from our group (Chen et al., 2016a; Chen et al., 2016b), we applied the Bindea signatures to expression profiles of kidney cancers and of lung cancers, respectively, and found in both studies that tumor subtypes scoring highly for EMT also scored highly for immune cell signatures.
In a similar manner to our scoring of tumor profiles for EMT by gene signature, we can score the profiles for other signatures and correlate the results with those of the EMT signature. For this article, we took the Bindea signature scores for the entire TCGA pan-cancer cohort (which were computed previously (Chen et al., 2016a)) and correlated each of these with our 16-gene EMT signature scores. Correlations were computed across all tumors (with linear regression model correcting for both tumor purity and cancer type) as well as within each individual cancer type (Figure 4). Correlations between mRNAs representing immunotherapeutic targets and EMT signature scores were also computed (Figure 4). Overall, we see strong correlations between EMT signature and immune signatures or immune checkpoint-related genes, across all cancers as well as within most individual cancer types surveyed (though notably blood cancers in TCGA such as DLBC and LAML do not show strong patterns for this). We also find here that genes encoding cancer-testis antigens, including CTAG1B (NY-ESO-1), MAGEA4, and SAGE1 do not correlate with EMT. The Bindea signatures would represent cells within the non-cancer component of the tumor sample, and samples with high immune cell infiltrate would score highly for the associated immune signatures.
Figure 4. EMT phenotype correlations with immune checkpoint-related genes and signatures across human tumors.
Genes encoding immunotherapeutic targets (represented in top panel) and gene expression-based signatures of immune cell infiltrates (Bindea et al., 2013) (bottom panel) were each correlated with EMT signature score across all cancers and within each individual cancer type. For each cancer type (columns), the corresponding Pearson’s correlation with the EMT signature score (from Figure 1A, individual genes featured in the top panel) is shown. A t-statistic greater than 2 or less than −2 would be within statistical significance (p<0.05). Features highly significant (p<1E-10) across cancers, by linear regression model incorporating both tumor purity and cancer type in addition to mRNA expression, are indicated (asterisk). TREG cells, regulatory T cells; TGD cells, T gamma delta cells; Tcm cells, T central memory cells; Tem cells, T effector memory cells; Tfh cells, T follicular helper cells; NK cells, natural killer cells; DC, dendritic cells; iDC, immature DCs; aDC, activated DCs; P-DC, plasmacytoid DCs; APM1/APM2, antigen presentation on MHC class I/class II, respectively.
Overall, the pan-cancer patterns identified here associating EMT with immune cell infiltrates and activation of the immune checkpoint pathway would be consistent with results of previous experimental studies noted above, though what significance such patterns would have with regards to immunotherapy response in patients remains to be determined. It may not necessarily follow that high EMT marker expression would predict better responsiveness to immune checkpoint inhibitors. For one thing, known checkpoint pathway-related differences would exist between cancer subtypes; therefore, specific cancer types should be individually studied, in addition to carrying out pan-cancer studies. For example, estrogen receptor-positive breast cancers respond extraordinarily poorly to checkpoint inhibitors, while triple negative breast cancers respond well (Voutsadakis, 2016). In addition to expression of immune checkpoint-related genes and immune cell signatures, responsiveness to immune checkpoint inhibition can be related to mutational burden and the emergence of novel epitopes that drive immune system recognition (Rizvi et al., 2015). More study in this area is therefore needed.
Molecular correlates of patient survival involving EMT markers
The process of EMT has been consistently shown to drive EMT tumor invasion and metastasis in the experimental setting (Eger et al., 2005; Weinberg, 2006; Aigner et al., 2007; Peinado et al., 2007; Burk et al., 2008; Gibbons et al., 2009; Yang et al., 2011; Creighton et al., 2013), and the question can be therefore raised as to whether EMT may be associated with more aggressive cancers in the setting of human patients. We might hypothesize that primary human tumors having more mesenchymal-like features would be associated with worse patient outcome (e.g. a shorter time to death or to relapse). Different studies may show a number of such trends involving EMT markers in various cancer types, but there would be some challenges regarding our ability to identify robust survival correlations, including the possible manifestation of EMT within only a subset of cancer cells within the tumor, which would represent a partial contribution to the total molecular profile (Creighton et al., 2013). Another challenge regarding correlative studies of patient outcome involves the numbers of patients and amount of follow-up data available; with smaller studies, there may not be sufficient statistical power involved in order for us to confidently identify trends in the data. TCGA pan-cancer datasets would involve a large number of patients (over 10,000 in all), but the time of patient follow-up may be fairly short for many individual cancer types (owing to the fact that many of the samples sent to TCGA for analysis were from newly-diagnosed patients). Nevertheless, it should be possible for us to leverage the very large case numbers in TCGA, while correcting for cancer type, as some cancer types would be inherently more aggressive than others (Hoadley et al., 2014).
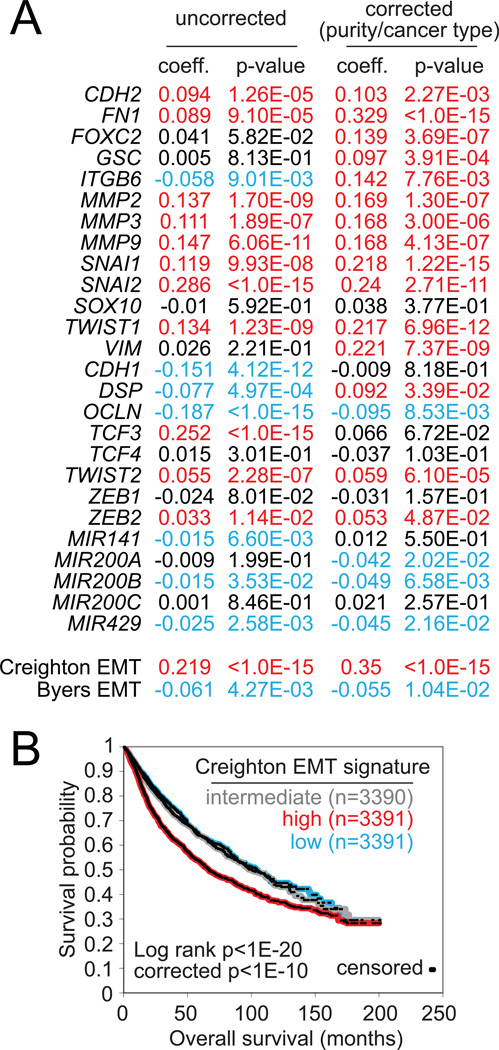
In this article, we surveyed a core set of genes involved in EMT (taken from Figure 2B) for associations with overall survival (i.e. time to death) in cancer patients, with results presented in Figure 5A. We carried out two separate tests for each gene feature examined: an “uncorrected” test across all cancers regardless of type (involving data from n=10172 patients in total) and a “corrected” test incorporating both cancer type and sample purity as covariates (n=7663 patients). Features more strongly associated with an aggressive cancer type but having a survival association that was not independent of cancer type may show significance for the uncorrected but not the corrected survival test. Most of the mesenchymal genes (i.e. genes with higher expression being associated with EMT) were associated with worse patient outcome by either corrected or uncorrected tests (i.e. higher expression was associated with a shorter time to death), while epithelial genes and miR-200 family members (including miR-200b and miR-429) tended to be associated with better patient outcome (i.e. loss of expression would be associated with a shorter time to death). Interestingly our 16-gene EMT signature score (i.e. the "Creighton" signature score) was significantly associated with worse outcome, independent of cancer type (p<1E-15, Cox model with corrections for cancer type and purity), while the EMT signature score based on the Byers signature (Byers et al., 2013) was not associated with worse outcome (which could conceivably be due to many correlates in the Byers signature being more specific to lung cancer). In a Kaplan-Meier analysis (Figure 5B), tumors in the top third of Creighton EMT signature scores (n=3391 patients) show markedly worse outcome as compared with the rest of the patients. While the absolute differences in patient survival as distinguished by this EMT signature are somewhat modest (perhaps due in part to other factors that may be at work within a given tumor), the fact differences are detectable would be in line with the notion that EMT plays a role in disease progression.
Figure 5. Pan-cancer molecular correlates of patient survival involving EMT-associated RNA features.
(A) For selected EMT-associated features, correlations with overall survival in the pan-cancer cohort from TCGA. “Uncorrected” coefficients and p-values by univariate Cox (using n=10172 cases with mRNA and survival data or n=8364 cases with microRNA and survival data); “corrected” coefficients and p-values denote significance of correlation in Cox model incorporating the molecular feature, estimated tumor purity, and cancer type (using n=7663 cases with mRNA/purity/survival data or n=6459 cases with microRNA/purity/survival data). Red, significant correlation with worse patient outcome (i.e. shorter time to patient death); blue, significant correlation with better outcome. Before computation of survival coefficients, gene features and gene signature scores were first transformed to standard deviations from the median. Patient survival data from TCGA were current as of March 31, 2016. (B) Kaplan-Meier plot of overall survival of patients stratified by Creighton EMT signature score (top third, bottom third, middle third). “Corrected” p-values by stratified log-rank test, incorporating cancer type as a confounder.
Future directions
TCGA pan-cancer datasets will continue to serve as a resource for examining correlative patterns involving EMT mediators in the setting of human cancers. As additional genes with roles in EMT are identified and functionally interrogated in the laboratory, these can also be examined in the publicly-available genomic datasets, as to whether the correlations as observed in human tumors would appear consistent with the cause-and-effect relationships as observed in experimental models. Furthermore, TCGA datasets may serve as a platform for discovery, whereby a set of gene correlates is first taken from the human tumor data, and hypotheses about the functional relationships such correlations represent are then tested in the laboratory. In addition to RNA expression, other data platforms in TCGA that might be explored in the context of EMT include DNA methylation, mutations (by whole exome or whole genome sequencing) and protein (by Reverse Phase Protein Array).
As human tumor specimens with available molecular profiling data are typically not micro-dissected to isolate cancer cells, and as the profiled samples would in fact represent a mixture of cell types, some considerations need to be taken into account when analyzing molecular correlations in the context of EMT. For example, different types of cancer notoriously have different intrinsic levels of stromal fibroblasts as part of the tumor mass, with an extreme example being pancreatic cancer (which can consist of >90% desmoplastic stroma), while some cancer subtypes have very little intrinsic fibroblastic component. Even when accounting for estimated tumor purity and for cancer type, a fibroblastic component of the molecular profile could conceivably contribute to the patterns observed. Another consideration is that some cancer types, such as ovarian, are known to behave anomalously from most other types in regards to the correlation between EMT and poor outcome, i.e., cadherin expression is elevated in more aggressive tumors, reflecting the more effective diffusion-based seeding of tumor cell clusters linked by cadherin across the peritoneal cavity (Kipps et al., 2013). Cancer type-specific phenomenon—e.g. whereby some tumors spread by dispersion of clusters, versus infiltration through stroma and subsequent intravasation, the latter of which favors EMT—may therefore be taken into consideration as necessary.
The role of the tumor microenvironment in initiating EMT and metastasis and the role of tumor cell EMT in remodeling the tumor microenvironment can also be further explored (Gibbons et al., 2009; Peng et al., 2016), using appropriate experimental model systems. The molecular profile of a tumor may be influenced by a combination of cancer cell of origin, somatic alterations, and microenvironment. Global molecular patterns representing the non-cancer component of the tumor—which component may include the involvement of immune cells, fibroblasts, or growth factors—may also be examined using TCGA datasets (Aran et al., 2015; Chen et al., 2016a; Chen et al., 2016b). The better we can understand EMT in human cancer, the more we will be able to find ways to target EMT in the clinical setting.
Experimental Procedures
Expression data underlying the results presented here were generated by TCGA Research Network (http://cancergenome.nih.gov/). All data used in this study were publicly available, e.g. from The Broad Institute’s Firehose pipeline (http://gdac.broadinstitute.org/). From TCGA, we collected molecular data on 10244 tumors of various histological subtypes (ACC project, n=79; BLCA, n=408; BRCA, n=1095; CESC, n=304; CHOL, n=36; COAD/READ, n=642; DLBC, n=48; GBM, n=161; HNSC, n=520; KICH, n=66; KIRC, n=533; KIRP, n=290; LAML, n=173; LGG, n=516; LIHC, n=371; LUAD, n=515; LUSC, n=501; MESO, n=87; OV, n=262; PAAD, n=178; PCPG, n=179; PRAD, n=497; SARC, n=259; SKCM, n=469; TGCT, n=150; THCA, n=503; THYM, n=120; UCEC, n=546; UCS, n=57; UVM, n=80) from TCGA, for which RNA-seq data (v2 platform) were available.
For computational scoring of gene expression profiles based on a given gene signature (e.g. the Creighton EMT signature or the Bindea immune signatures), log-transformed values for each gene were first normalized to standard deviations from the median across all samples. For Creighton EMT signature, as previously described (Creighton et al., 2013), the following equation was used to score/place a numerical value on tumor “EMT-ness” based on normalized values of the genes:
The Byers et al. EMT signature score (Byers et al., 2013) was computed using our previously described “t-score” metric (Creighton et al., 2012) on the normalized expression values. To computationally infer the infiltration level of specific immune cell types using RNA-seq data, we used a set of 501 genes specifically overexpressed in one of 24 immune cell types from Bindea et al. (Bindea et al., 2013). Scoring TCGA cancer samples for each of these immune cell signatures was carried out as previously described (Chen et al., 2016a), with the average of the normalized gene expression values was used to score each sample profile for each signature. In addition, samples were scored for expression of Antigen Presentation MHC class I (APM1) genes (HLA-A/B/C, B2M, TAP1/2, TAPBP) and for Antigen Presentation MHC class II (APM2) genes.
Correlation across the pan-cancer cohort between two molecular features of interest was assessed using Pearson’s correlation (with log-transformed expression values). In practice when analyzing tumor datasets, significant correlations can have r-values well below 1 and still be considered statistically and biologically significant, given all the biologically- and technically-related noise (e.g. cellular heterogeneity) that may be represented within a given tissue sample. In addition, linear regression models incorporating cancer type (one of the 30 major types listed above) as a factor in addition to gene feature, and models incorporating both cancer type and tumor purity (Aran et al., 2015) were also considered. Individual gene features were evaluated for correlation with patient survival by univariate Cox analysis; in addition, a stratified Cox model was used to evaluate survival association when correcting for both tumor type and tumor purity. For Kapan-Meier plots, a stratified Log-rank test evaluated differences between tumor groups after correction for tumor type. Patient survival data (i.e. time to death) from TCGA were current as of March 31, 2016.
The data for the specific molecular features analyzed here (representing 10244 tumors) have been made available as a supplemental data file (Data File S1). The data file is an Excel spreadsheet consisting of two tables. The first table (“EMT features”) consists of gene expression data for canonically EMT-associated genes (including the gene markers constituting the Creighton EMT signature) and microRNAs (miR-200 family members), EMT gene signature scores, estimates of tumor purity from Aran et al. (Aran et al., 2015), and associated overall survival data from the patients. The second table (“immune features”) consists of expression values for specific genes encoding immunotherapeutic targets (e.g. PD-1, PD-L1, from Figure 4) and gene expression-based signature scoring of immune cell infiltrates (based on Bindea et al. signatures).
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health (NIH) grant P30 CA125123 (C. Creighton), Cancer Prevention and Research Institute of Texas (CPRIT) grant RP120713 C2 (C. Creighton) and P2 (D. Gibbons), and CPRIT grant RP150405 (D. Gibbons). We thank Yiqun Zhang for technical assistance.
Abbreviations
- EMT
epithelial–mesenchymal transition
- TCGA
The Cancer Genome Atlas
- RNA-seq
RNA sequencing
References
- Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, Sommergruber W, Schweifer N, Wernitznig A, Beug H, Foisner R, Eger A. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26 doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Sirota M, Butte A. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf A, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman W, Becker C, Pagès F, Speicher M, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers L, Diao L, Wang J, Saintigny P, Girard L, Peyton M, Shen L, Fan Y, Giri U, Tumula P, Nilsson M, Gudikote J, Tran H, Cardnell R, Bearss D, Warner S, Foulks J, Kanner S, Gandhi V, Krett N, Rosen S, Kim E, Herbst R, Blumenschein G, Lee J, Lippman S, Ang K, Mills G, Hong W, Weinstein J, Wistuba I, Coombes K, Minna J, Heymach J. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer_Genome_Atlas_Research_Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang Y, Parra E, Rodriguez J, Behrens C, Akbani R, Lu Y, Kurie J, Gibbons D, Mills G, Wistuba I, Creighton C. Multiplatform-based Molecular Subtypes of Non-Small Cell Lung Cancer. Oncogene. 2016a doi: 10.1038/onc.2016.303. E-pub Oct 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang Y, Şenbabaoğlu Y, Ciriello G, Yang L, Reznik E, Shuch B, Micevic G, De Velasco G, Shinbrot E, Noble M, Lu Y, Covington K, Xi L, Drummond J, Muzny D, Kang H, Lee J, Tamboli P, Reuter V, Shelley C, Kaipparettu B, Bottaro D, Godwin A, Gibbs R, Getz G, Kucherlapati R, Park P, Sander C, Henske E, Zhou J, Kwiatkowski D, Ho T, Choueiri T, Hsieh J, Akbani R, Mills G, Hakimi A, Wheeler D, Creighton C. Multilevel Genomics-Based Taxonomy of Renal Cell Carcinoma. Cell Rep. 2016b;14:2476–2489. doi: 10.1016/j.celrep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gibbons D, Goswami S, Cortez M, Ahn Y, Byers L, Zhang X, Yi X, Dwyer D, Lin W, Diao L, Wang J, Roybal J, Patel M, Ungewiss C, Peng D, Antonia S, Mediavilla-Varela M, Robertson G, Jones S, Suraokar M, Welsh J, Erez B, Wistuba I, Chen L, Peng D, Wang S, Ullrich S, Heymach J, Kurie J, Qin F. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yi X, Goswami S, Ahn Y, Roybal J, Yang Y, Diao L, Peng D, Peng D, Fradette J, Wang J, Byers L, Kurie J, Ullrich S, Qin F, Gibbons D. Growth and metastasis of lung adenocarcinoma is potentiated by BMP4-mediated immunosuppression. OncoImmunology. 2016c doi: 10.1080/2162402X.2016.1234570. E-pub 26 Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton C, Chang J, Rosen J. Epithelial-mesenchymal transition (EMT) in tumor-initiating cells and its clinical implications in breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:253–260. doi: 10.1007/s10911-010-9173-1. [DOI] [PubMed] [Google Scholar]
- Creighton C, Gibbons D, Kurie J. The role of epithelial-mesenchymal transition programming in invasion and metastasis: a clinical perspective. Cancer Manag Res. 2013;5 doi: 10.2147/CMAR.S35171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton C, Hernandez-Herrera A, Jacobsen A, Levine D, Mankoo P, Schultz N, Du Y, Zhang Y, Larsson E, Sheridan R, Xiao W, Spellman P, Getz G, Wheeler D, Perou C, Gibbs R, Sander C, Hayes D, Gunaratne P Cancer_Genome_Atlas_Research_Network. Integrated analyses of microRNAs demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PLoS One. 2012;7:e34546. doi: 10.1371/journal.pone.0034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton C, Li X, Landis M, Dixon J, Neumeister V, Sjolund A, Rimm D, Wong H, Rodriguez A, Herschkowitz J, Fan C, Zhang X, He X, Pavlick A, Gutierrez M, Renshaw L, Larionov A, Faratian D, Hilsenbeck S, Perou C, Lewis M, Rosen J, Chang J. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20–38. doi: 10.1016/j.semcancer.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- Gibbons D, Lin W, Creighton C, Rizvi Z, Gregory PG, G J, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, Kurie J. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröger C, Grubinger M, Waldhör T, Vierlinger K, Mikulits W. Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS One. 2012;7:e51136. doi: 10.1371/journal.pone.0051136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley K, Yau C, Wolf D, Cherniack A, Tamborero D, Ng S, Leiserson M, Niu B, McLellan M, Uzunangelov V, Zhang J, Kandoth C, Akbani R, Shen H, Omberg L, Chu A, Margolin A, Van't Veer L, Lopez-Bigas N, Laird P, Raphael B, Ding L, Robertson A, Byers L, Mills G, Weinstein J, Van Waes C, Chen Z, Collisson E, Cancer_Genome_Atlas_Research_Network. Benz C, Perou C, Stuart J. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zheng M, Zhang R, Liang Y, Han H. Notch signaling pathway and cancer metastasis. Adv Exp Med Biol. 2012;727:186–198. doi: 10.1007/978-1-4614-0899-4_14. [DOI] [PubMed] [Google Scholar]
- Huang L, Wu R, Xu A. Epithelial-mesenchymal transition in gastric cancer. Am J Transl Res. 2015;7:2141–2158. [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg R. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;120:1786. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Wu K, Lee W. Hypoxia, Epithelial-Mesenchymal Transition, and TET-Mediated Epigenetic Changes. J Clin Med. 2016;5:E24. doi: 10.3390/jcm5020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Koh J, Kim M, Kwon D, Go H, Kim Y, Jeon Y, Chung D. PD-L1 expression is associated with epithelial-to-mesenchymal transition in adenocarcinoma of the lung. Hum Pathol. 2016 doi: 10.1016/j.humpath.2016.07.007. E-pub Jul 26. [DOI] [PubMed] [Google Scholar]
- Kipps E, Tan D, Kaye S. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat Rev Cancer. 2013;13:273–282. doi: 10.1038/nrc3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kundu S, Byers L, Peng D, Roybal J, Diao L, Wang J, Tong P, Creighton C, Gibbons D. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2015 doi: 10.1038/onc.2015.71. E-pub Mar 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Dedhar S, Kalluri R, Thompson E. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo P, Lee J, Liang X, Sukumar S. The dual role of FOXF2 in regulation of DNA replication and the epithelial-mesenchymal transition in breast cancer progression. Cell Signal. 2016;28:1502–1519. doi: 10.1016/j.cellsig.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Diao L, Parra Cuentas E, Denning W, Chen L, Fan Y, Byers L, Wang J, Papadimitrakopoulou V, Behrens C, Rodriguez J, Hwu P, Wistuba I, Heymach J, Gibbons D. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-15-1434. E-pub Feb 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak M, Tong P, Diao L, Cardnell R, Gibbons D, William W, Skoulidis F, Parra E, Rodriguez-Canales J, Wistuba I, Heymach J, Weinstein J, Coombes K, Wang J, Byers L. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson E, Kazenwadel J, Bert A, Khew-Goodall Y, Ruszkiewicz A, Goodall G. Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia. 2013;15:180–191. doi: 10.1593/neo.121828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7 doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Peng D, Ungewiss C, Tong P, Byers L, Wang J, Canales J, Villalobos P, Uraoka N, Mino B, Behrens C, Wistuba I, Han R, Wanna C, Fahrenholtz M, Grande-Allen K, Creighton C, Gibbons D. ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene. 2016 doi: 10.1038/onc.2016.358. E-pub Oct 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi N, Hellmann M, Snyder A, Kvistborg P, Makarov V, Havel J, Lee W, Yuan J, Wong P, Ho T, Miller M, Rekhtman N, Moreira A, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon E, Merghoub T, Wolchok J, Schumacher T, Chan T. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth L, Das R, Townley S, Coutinho I, Hanson A, Centenera M, Stylianou N, Sweeney K, Soekmadji C, Jovanovic L, Nelson C, Zoubeidi A, Butler L, Goodall G, Hollier B, Gregory P, Tilley W. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene. 2016 doi: 10.1038/onc.2016.185. E-pub Jun 6. [DOI] [PubMed] [Google Scholar]
- Shao D, Xue W, Krall E, Bhutkar A, Piccioni F, Wang X, Schinzel A, Sood S, Rosenbluh J, Kim J, Zwang Y, Roberts T, Root D, Jacks T, Hahn W. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T, Miow Q, Miki Y, Noda T, Mori S, Huang R, Thiery J. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med. 2014;6:1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y, Wu K. Hypoxia-regulated target genes implicated in tumor metastasis. J Biomed Sci. 2012;19:102. doi: 10.1186/1423-0127-19-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewiss C, Rizvi Z, Roybal J, Peng D, Gold K, Shin D, Creighton C, Gibbons D. The microRNA-200/Zeb1 axis regulates ECM-dependent β1-integrin/FAK signaling, cancer cell invasion and metastasis through CRKL. Sci Rep. 2016;6:18652. doi: 10.1038/srep18652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutsadakis I. Immune Blockade Inhibition in Breast Cancer. Anticancer Res. 2016;36:5607–5622. doi: 10.21873/anticanres.11145. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li Y, Kong D, Sarkar F. The role of Notch signaling pathway in epithelial-mesenchymal transition (EMT) during development and tumor aggressiveness. Curr Drug Targets. 2010;11:745–751. doi: 10.2174/138945010791170860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The Biology of Cancer. New York: Garland Science; 2006. [Google Scholar]
- Yang Y, Ahn Y, Gibbons D, Zang Y, Lin W, Thilaganathan N, Alvarez C, Moreira D, Creighton C, Gregory P, Goodall G, Kurie J. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011;121:1373–1385. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Weinberg R. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Li D, Li H, Wang L, Tian G, Dong Y. YAP overexpression promotes the epithelial-mesenchymal transition and chemoresistance in pancreatic cancer cells. Mol Med Rep. 2016;13:237–242. doi: 10.3892/mmr.2015.4550. [DOI] [PubMed] [Google Scholar]
- Zhao M, Kong L, Liu Y, Qu H. dbEMT: an epithelial-mesenchymal transition associated gene resource. Sci Rep. 2015;5:11459. doi: 10.1038/srep11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.