Abstract
Bast fibres are long extraxylary cells which mechanically support the phloem and they are divided into xylan- and gelatinous-type, depending on the composition of their secondary cell walls. The former, typical of jute/kenaf bast fibres, are characterized by the presence of xylan and a high degree of lignification, while the latter, found in tension wood, as well as flax, ramie and hemp bast fibres, have a high abundance of crystalline cellulose. During their differentiation, bast fibres undergo specific developmental stages: the cells initially elongate rapidly by intrusive growth, subsequently they cease elongation and start to thicken. The goal of the present study is to provide a transcriptomic close-up of the key events accompanying bast fibre development in textile hemp (Cannabis sativa L.), a fibre crop of great importance. Bast fibres have been sampled from different stem regions. The developmental stages corresponding to active elongation and cell wall thickening have been studied using RNA-Seq. The results show that the fibres sampled at each stem region are characterized by a specific transcriptomic signature and that the major changes in cell wall-related processes take place at the internode containing the snap point. The data generated also identify several interesting candidates for future functional analysis.
Introduction
Fibre crops are important bioresources as they provide strong and long fibres (up to 100 mm in some cases1), also known as bast fibres. These extraxylary cells belong to the sclerenchyma, they support mechanically the phloem and are differentiated into xylan- and gelatinous-type2. The cell walls of xylan-type fibres are lignified, contain predominantly xylan as hemicellulose and show a typical layered structure (S1–S3) because of the different orientation of the cellulose microfibrils2. The gelatinous fibres, typically found in hemp bast fibres, are characterized by a thick cellulosic cell wall1, 3 (referred to as G-layer). Bast fibre G-layer is reminiscent of the cell walls occurring in tension wood. However, the former does not exert the same contractile function as the latter4. Fibre crops like textile hemp or flax (Cannabis sativa L. and Linum usitatissimum L.) are very attractive models to carry out investigations on cell wall processes, because their stems are characterized by tissues displaying remarkable differences in cell wall composition. The cortical tissues, which can be easily peeled off and separated, harbour the cellulosic bast fibres and are characterized by the occurrence of low amounts of lignin (ca. 2–7%)1. The core, also referred to as shivs or hurds, is instead woody. Along the stem axis it is possible to identify an empirically-determined region, called the “snap point”5, which marks the transition from elongation to fibre thickening (and resulting in changes in fibre mechanical properties). The fibres in the younger regions of the stem (at the top) first grow symplastically with the surrounding tissues6, 7, then they start to elongate actively by a mechanism known as intrusive growth, where the tip of the fibres invades the middle lamella of neighbouring cells7–11. This growth mechanism ensures that the number of fibres in a given transverse section of the stem increases, without changing the total number of cells. This gradient of fibre developmental stages is accompanied by a basipetal lignification gradient in the stem tissues, where genes involved in the production of phenylpropanoids, and more generally in the provision of metabolic precursors needed for lignin synthesis, are expressed at higher levels. In this respect, in hemp it was recently shown that genes involved in the non-oxidative phase of the pentose phosphate pathway and in the first reaction of the shikimate pathway were expressed at higher levels in the core tissues at the bottom of the stem12. The stem of fibre crops is therefore ideal to carry out high throughput molecular analyses focusing on the cell wall, because its tissue polarity and spatial lignification gradient enable the study of sequential developmental stages. Several studies have indeed been published on fibre crops, namely flax13, 14, jute15–17, ramie18, kenaf19, hemp20, 21 where a molecular approach was adopted to shed light on the mechanisms underlying bast fibre differentiation and development. These studies have identified important genes involved in bast fibre development, notably chitinases and cellulose synthases13, as well as transcription factors15, 16 and genes involved in secondary metabolism and monolignol-associated pathway15, 21. The advent of high-throughput techniques like transcriptomics has enabled huge steps forward in the study of fibre crops. For example, a very recent molecular study on flax, has shed light on the molecular mechanisms underlying advanced phases of bast fibre development, by identifying several transcription factors, as well as glycosyltransferases and unknown/not fully annotated genes14. Another recent study using transcriptomics/genomics has compared two varieties of jute differing in the cellulose/lignin fibre content and has demonstrated the expansion of lignin-biosynthetic genes with respect to flax17.
In the light of the industrial importance that gelatinous bast fibres are receiving, we here sought to investigate, via RNA-Seq, the molecular events accompanying their development in an economically important fibre crop, textile hemp. By sampling bast fibres from the top (TOP), middle (MID) and bottom (BOT) internodes of hemp stems (Fig. 1), we show that the transcriptional signature at each stem region is unique. These results are useful to identify and characterize candidate genes involved in bast fibre elongation/thickening which can be further studied functionally and used for future biotechnological applications.
Figure 1.
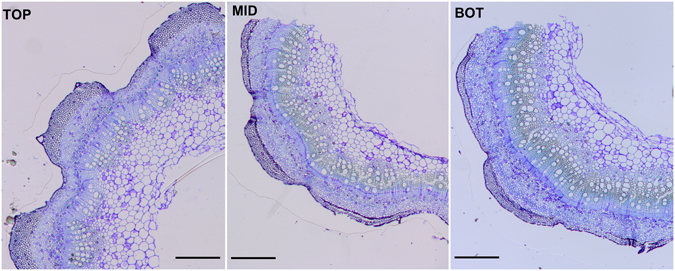
Optical microscope pictures of the different hemp stem regions (TOP, MID, BOT) analysed in this study. Staining was performed with toluidine blue. Scale bars are 500 µm.
Results and Discussion
RNA-Seq of hemp bast fibres
To analyse the transcriptional signature of hemp bast fibres at different developmental stages, RNA-Seq was performed on fibres sampled at three stem heights, i.e. top (TOP), middle (MID, containing the snap point) and bottom (BOT). Although fibres located at the top are difficult to separate because of the lack of a well-developed tertiary cell wall (G-layer), we carefully peeled the cortical tissues from the core and got rid of the majority of epidermal/parenchymatic and xylem cells by using a mortar with pestle and ethanol 80%. We reasoned that this procedure would enable us to minimize the “contamination” from the other tissues: inspection at the microscope showed that some non-glandular trichomes, parenchyma and xylem cells were still present, however those elements were sporadic (Suppl. Fig. 1).
A de novo assembly of the Santhica transcriptome was here performed by merging the reads obtained from the hypocotyl (previously published transcriptome22) with those obtained here for the bast fibres of adult plants: we reasoned that this would enable us to better capture the cell wall-related dynamism in the isolated phloem fibres of the variety under investigation. We nevertheless validated the data by comparing the results obtained after mapping using our de novo assembly with those generated after mapping against the Finola transcriptome23 (Suppl. Dataset File). As discussed in the next paragraphs, the two mapping strategies gave comparable results.
A total of 3268 differentially expressed contigs ranging from 283 to 7095 bps was obtained after data processing (see Material and Methods); of these, 2317 are annotated (Suppl. Dataset File). The Independent Component Analysis (computed with the FastICA algorithm in MetaGeneAlyse) revealed a good separation of the different stem regions when two components were used (percentage of variance explained: 98.79%) (Suppl. Fig. 2). This indicates that the bast fibres sampled for the analysis were in different developmental stages and therefore characterized by distinct transcriptomic signatures.
The RNA-Seq data were validated using targeted RT-qPCR on a subset of 12 genes (Suppl. Dataset File): the calculated coefficient is 0.9284, which indicates a very good correlation between the RNA-Seq and RT-qPCR data (Suppl. Fig. 3).
To get insights into the gene expression patterns, data clustering was performed using an arbitrary Pearson correlation coefficient threshold of 0.75. The analysis resulted in eleven clusters (Fig. 2); among these, cluster 1 and 2 account for the highest number of contigs (843 and 703 annotated contigs, respectively; Suppl. Dataset File). The hemp contigs can be assigned to three major expression patterns: progressive decrease in expression from the top to the bottom of the stem (clusters 1, 3, 4), progressive increase in expression along the stem axis (clusters 2, 5, 7) and maximum expression at the internode containing the snap point (clusters 6, 9, 10). Two additional trends were revealed with the clustering analysis: cluster 11 groups contigs showing a tendency towards decreased expression at the snap point, cluster 8 comprises genes showing no major changes along the stem axis.
Figure 2.
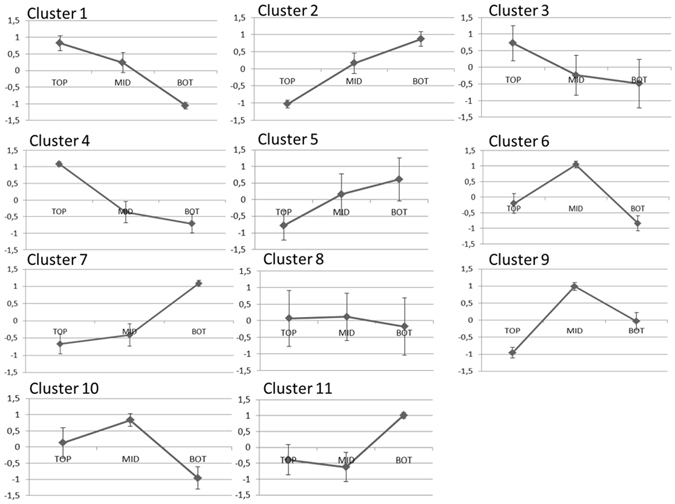
Profiles of the eleven clusters of genes obtained using a Pearson coefficient threshold of 0.75. The data represent the log2RPKM rescaled values± standard deviation (the rescaled values were calculated by subtracting to each contig expression value the average among the three stem regions and dividing by the standard deviation).
Since the bast fibres undergo progressive cell wall thickening from the top to the bottom of the hemp stem axis, we reasoned that the bulk of cell wall-related information would be obtained by focusing on the above-mentioned three major expression patterns (clusters 1-3-4, clusters 2-5-7, clusters 6-9-10). Hereafter is a description of the key gene ontologies characterizing each stem region.
Gene ontology enrichment analysis of the TOP region
The transcriptomic landscape of the TOP region is dominated by genes belonging to the DNA replication and cell cycle ontologies (Fig. 3), a result confirmed also by the analysis performed after mapping using the Finola transcriptome (Suppl. Dataset File). These findings are indicative of active nuclear division and are in agreement with what was previously reported in flax bast fibres24. Bast fibres are indeed multinucleate and during intrusive growth the number of nuclei increases, as previously documented in flax8. Compared to the MID and BOT respectively, CDC6 (cell division control 6) was expressed ca. 2 and 10 times more, DMC1 (encoding a meiotic recombination protein) was upregulated 2.4 and 7 times, MCM2, 4 and 5 (minichromosome maintenance protein 2, 4 and 5) were ca. 3 and 5 times more abundantly expressed, the ORC1A and ORC6 genes (origin recognition complex subunit 1 and 6) were between 2.5 and 5–7 times more expressed. Additionally, the key gene PCNA2 (proliferating cell nuclear antigen 2) involved in DNA replication25 was highly expressed at the TOP (ca. 99 RPKM) with respect to the BOT (18 RPKM); finally, DRT100 (coding for a DNA-damage-repair/toleration protein) was expressed ca. 12 times more at the TOP as compared to the BOT (Suppl. Dataset File).
Figure 3.
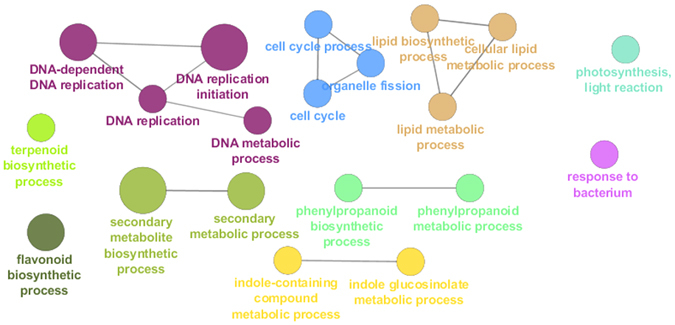
GOE analysis of the TOP region. Different colours indicate different ontologies. The bigger the circle, the higher the statistical significance.
The enrichment of genes involved in cell division was accompanied by the over-representation of candidates partaking in the lipid biosynthetic process: several 3-ketoacyl-CoA synthase isoforms (KCS10, 11, 19) were highly expressed at the TOP, together with the β-ketoacyl reductase 1 (KCR1) and the fatty acid desaturases FAD5 and 8. The enrichment of these genes can be explained by the diffuse (intrusive) growth mechanism of bast fibres8: the elongation of fibres is indeed ensured by the growth of the entire surface of the cell. Notably, among the genes belonging to the lipid biosynthetic process, there are also homogentisate phytyltransferases (HPT1), which are involved in the biosynthesis of tocopherols. These are lipid antioxidants protecting against oxidative stress (e.g. light stress26).
Genes involved in photosynthetic light reaction were likewise enriched at the TOP, notably CAB1 (chlorophyll A/B binding protein 1), LHCB4.3 and LHCA5 (light harvesting complex), expressed between 5 and 8 times more as compared to the BOT (Suppl. Dataset File). The TOP region was also characterized by genes involved in the response to pathogens: among them it is worth mentioning here UVI4 (UV-B-Insensitive 4, encoding the protein POLYCHOME), whose overexpression was shown to induce increased resistance to a bacterial pathogen via the activation of disease resistance genes27. In our dataset, several pathogenesis-related (PR) genes were also upregulated at the TOP, namely PRB1, PR-1-like, PR4 (Suppl. Dataset File).
Interestingly, the Gene Ontology term Enrichment (GOE) analysis highlighted the enrichment of genes involved in secondary metabolism biosynthetic processes too, namely terpenoids, flavonoids, indole-containing compounds and phenylpropanoids (Fig. 3). In the terpenoid biosynthetic process there are genes coding for three cytochrome P450s, i.e. CYP76C1 and C2 and CYP82G1: CYP76C1 and C2 are involved in floral linalool metabolism28, while CYP82G1 is responsible for the synthesis of homoterpene volatiles in Arabidopsis 29. Hemp trichomes are factories producing several terpenes contributing to the plant peculiar aroma30, however these metabolites can also be found in resin ducts. Since we observed the presence of non-glandular trichomes in our fibre samples, we speculate that these genes may be expressed in “contaminating” resin ducts present in our bast fibre samples.
Three genes encoding UDP glucosyltransferases were present in the “flavonoid biosynthetic process” ontology: UGT73C1 and C7 and UGT78D2. The first two belong to group D, whose members are related to stress-inducible response31; notably, UGT73C1 was shown to glucosylate cytokinins (trans-zeatin and dihydrozeatin32) and may therefore play a role in hormone homeostasis. UGT78D2 glucosylates instead flavonols in the C3 position and is strongly co-regulated with flavanone 3-hydroxylase (F3H, participating in the conversion of p-coumaroyl CoA to kaempferol/quercetin33), whose gene was also upregulated at the TOP in our dataset (Suppl. Dataset File).
Two genes encoding cytochrome P450s, i.e. CYP81D1 and D8 and two MYB transcription factors (TFs), MYB34 and MYB122, were present in the indole-containing compound metabolic process: MYB34 and 122 are two of the three TFs reported to control indole glucosinolate biosynthesis34. These TFs respond to phytohormones in different manners: MYB34 responds to both abscisic acid (ABA) and jasmonate (JA), while MYB122 plays a minor role in indole glucosinolate biosynthesis upon ethylene (ET) and JA signalling34. Given the role of indole glucosinolates in defence responses upon mechanical damage (e.g. herbivore attack35) and since the phytohormone JA is considered “the wound hormone”36, it is tempting to speculate that a mechanism involving the synthesis of JA and the subsequent activation of indole glucosinolates may be involved in the intrusive growth phase of bast fibres. The role of phytohormones in bast fibre intrusive growth remains to be confirmed.
We cannot exclude that the presence of genes belonging to the phenylpropanoid biosynthetic ontology (i.e. the laccases LAC11 and 17, the cinnamyl alcohol dehydrogenase CAD9) may be (partly) due to the traces of contaminating xylem tissues during the separation of fibre-rich peels from the TOP stem region (Suppl. Fig. 2, inset). The xylem tissue present is however in lower amount compared to the fibres; hence these transcripts may reflect an actual gene network linked to the phenylpropanoid metabolism in the TOP bast fibres. In this respect it should be noted that hemp bast fibres contain ca. 4% lignin21 and that a previous study highlighted the presence of transcripts associated with the secondary metabolism (namely peroxidases, methyltransferases) in the bast fibres sampled at the TOP20.
Among the genes grouped in the “phenylpropanoid biosynthetic process” ontology, there are five contigs annotated as PRR1 (pinoresinol reductase 1), whose expression was upregulated at the TOP (Suppl. Dataset File). In flax, lignomics unveiled a complex monolignol metabolism associated with bast fibre hypolignification, with the accumulation of both aglycone forms and glycosides of (neo)lignans37; hence PRR1 may be involved in a similar rich monolignol metabolism in hemp bast fibres. Additionally, some lignans, as dehydrodiconiferyl alcohol, are known to regulate cell division38; therefore PRR1 may also partake in the synthesis of specific lignans contributing to the regulation of cell division. Recently a transcriptome analysis performed on a wild-type and mutant jute (referred to as deficient lignified phloem fibre) identified the presence of many isoforms (more than other bast fibre-producing plants) belonging to the monolignol and shikimate-aromatic amino acid metabolism15. In particular, the identification of several shikimate O-hydroxycinnamoyl transferases demonstrated that in jute fibres there is a shunting of the phenylpropanoid metabolism from p-coumaroyl-CoA towards the production of monolignols, instead of flavonoids. Hemp bast fibres are of gelatinous-type and therefore the presence in the TOP bast fibres of transcripts belonging to both the flavonoid and phenylpropanoid pathways (Fig. 3) may reflect a difference with respect to the xylan-type fibres present in jute.
Among the transcripts showing the highest FC at the TOP with respect to the MID, we would like to draw the reader’s attention on 2 genes in particular, i.e. a jacalin-like lectin domain-containing protein orthologous to AT3G16460 and a protodermal factor 1 (PDF1). In maize, two jacalin-like lectin domain-containing proteins were shown to be involved in cell wall-related processes39; in cotton, a PDF1 gene was found to be associated with fibre initiation and early growth phases40. The exact role of these genes remains to be elucidated via functional studies; however their expression pattern strongly points towards an involvement during hemp bast fibre early growth stages.
Gene ontology enrichment analysis of the MID region
The transcriptome of the MID region is mainly characterized by processes related to secondary cell wall biogenesis (Fig. 4); this result was confirmed with the mapping against the Finola transcriptome (Suppl. Dataset File). These data are of particular relevance because they provide an overview of the key cell wall-related events responsible for the change in mechanical properties of the bast fibres observed at the snap point. In this study the MID region corresponds to the internode containing the snap point (Fig. 1); hence the fibres separated are progressively shifting from a stage of elongation to a phase of tertiary cell wall formation. According to our data, the transcriptome of the bast fibres at the MID region is characterized by genes involved in cell expansion and cell wall loosening, as well as by an active transcriptional dynamics of candidates involved in hemicellulose biosynthesis and transcriptional regulators orchestrating secondary cell wall biogenesis (Suppl. Dataset File). Alpha expansin genes (EXPA8, 10, 11 and 12) and a xyloglucan endotransglycosylase/hydrolase (XTH33) peaked at the MID region (EXPA10 and 11 showed an increase in FC > 5 with respect to the TOP; Suppl. Dataset File); their higher expression is probably due to the phase of elongation characterizing the heterogeneous fibre stages at the MID. The master regulator MYB46 and its downstream target MYB63 were expressed 4.8 and 1.4 times more at the MID as compared to the TOP (and 3.1 and 4.3 times more at the MID as compared to the BOT): these TFs activate genes involved in xylan and lignin biosynthesis and therefore the secondary cell wall biosynthetic program41. Three laccases, i.e. LAC4 (IRX1; contig_10035), LAC12 (contig_15462) and 2 contigs annotated as LAC17 (contig_19910 and contig_17371) were also highly upregulated at the MID with respect to the TOP (Suppl. Dataset File): these same genes were shown to be upregulated at older stages of development in the hemp hypocotyl22 and in the stem of adult hemp plants they may be associated with the peculiar lignification of the bast fibres. Bast fibres are hypolignified, however condensed guaiacyl (G) lignins were immunologically detected in the middle lamellas, cell wall junctions and S1 layer of phloem fibres in another fibre crop, flax42. In hemp bast fibres the presence of a similar condensed lignin was reported43; hence the laccases here detected may participate in the formation of condensed lignins in the cell wall of bast fibres.
Figure 4.
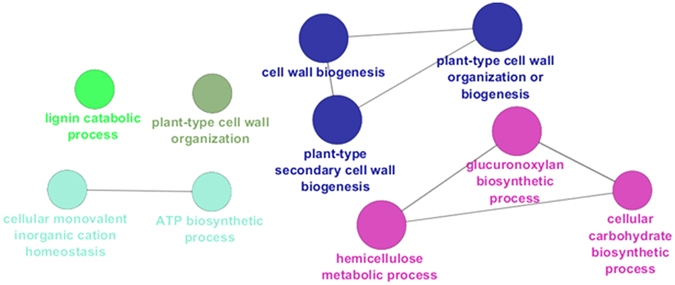
GOE analysis of the MID region. Different colours indicate different ontologies. The bigger the circle, the higher the statistical significance.
In the “hemicellulose metabolic process” ontogeny there are genes involved in glucuronoxylan biosynthesis (the principal hemicellulose in dicots’ secondary cell walls44). IRX8/GAUT12 (irregular xylem 8/galacturonosyl transferase 12), IRX9, IRX10/GUT2 (glucuronosyl transferase 2) and PGSIP3/GUX2 (plant glycogenin-like starch initiation protein 3/glucuronic acid substitution of xylan 2) were upregulated 3.2, 4.5, 4.8 and 5.4 times respectively at the MID as compared to the TOP (Suppl. Dataset File). IRX8 belongs to the glycosyltransferase (GT) family 8 and is involved in the synthesis of the reducing end tetrasaccharide of glucuronoxylans, while IRX9 is a GT43 involved in xylan backbone elongation45; IRX10 is a GT47 also involved in the elongation of the xylan backbone46. GUX2 is a Golgi-localized GT8 with xylan glucuronosyltransferase activity. At the MID, the upregulation of two contigs, annotated as ASD1/ARAF1 (which codes for a bifunctional α-L-arabinofuranosidase/β-D-xylosidase), was also observed (FC ca. 10 between MID and TOP). ARAF1 was shown to act in vivo on arabinan-containing pectins47 and its upregulation at the MID suggests cell wall remodelling associated with the deposition of secondary cell walls. Taking into account the upregulation of genes involved in xylan biosynthesis together with ARAF1 and considering the proof for the existence of a covalent link between hemicelluloses and pectins via an arabinogalactan protein (APAP148), it is reasonable to hypothesize the presence of a similar association in the cell walls of bast fibres. ARAF1 may therefore participate in the remodelling of pectins at the onset of secondary cell wall deposition.
A contig annotated as IRX1/CesA8 showed upregulation at the MID (FC MID vs TOP ca. 11): this result indicates that at the internode containing the snap point major molecular events related to cell wall biosynthesis take place with, notably, the upregulation of transcripts related to both cellulosic and non-cellulosic polysaccharide deposition.
Three genes coding for H+-ATPases (HA3, 5, 6) were upregulated at the MID: these genes may be associated with fibre elongation via the accumulation of osmotically-active compounds maintaining turgor pressure, or the acidification of the apoplast, in a manner analogous to the mechanism invoked for cotton fibre elongation49.
Among the contigs encoding TFs upregulated at the MID there is IBH1 (ILI1 binding bHLH 1 protein) with the highest FC increase in expression as compared to the TOP (ca. 34; Suppl. Dataset File). IBH1 is a negative regulator of cell elongation50 and its higher expression at the MID may indicate a function in the shift from elongation to thickening in bast fibres. A contig coding for a PLATZ TF also peaked at the MID (MID vs TOP FC > 13): PLATZ TFs were proposed to act as negative regulators of cell proliferation51 and may contribute to the transition of bast fibres from a phase of active division to secondary growth.
Among the contigs showing the highest FC increase between MID and TOP is a putative acid phosphatase (AT1G04040): notably, the corresponding protein in thale cress was found associated to the cell wall fraction52. This phosphatase may be involved in cell wall-related processes in bast fibres during the transition from elongation to thickening and is therefore an interesting candidate for further analysis.
Gene ontology enrichment analysis of the BOT region
The BOT region is characterized by ontologies related to phytohormone and non cellulosic polysaccharide biosynthesis, as well as secondary metabolic processes (Fig. 5). Mapping against the Finola transcriptome confirmed these data (Suppl. Dataset File) and additionally showed the enrichment of auxin metabolism-related genes, among which, WAT1 (WALLS ARE THIN1), an auxin efflux transporter involved in secondary cell wall deposition53, 54.
Figure 5.
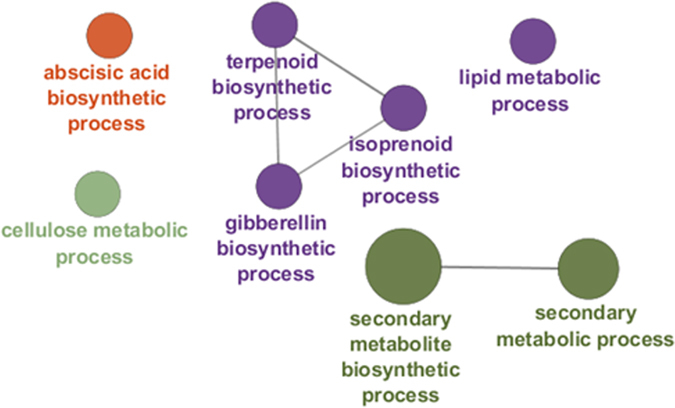
GOE analysis of the BOT region. Different colours indicate different ontologies. The bigger the circle, the higher the statistical significance.
In the “gibberellin biosynthetic process” ontogeny there are genes involved in the three steps required for gibberellin (GA) synthesis, i.e. formation of ent-kaurene from geranyl geranyl diphosphate, conversion of ent-kaurene to GA12 and synthesis of C20- and C19-GAs55. GA1/CPS (ent-copalyl diphosphate synthase) involved in the formation of ent-kaurene was expressed at the BOT > 7 times with respect to the TOP; KAO2 (ent-kaurenoic acid oxidase 2), which codes for a CYP88A member catalysing the conversion of ent-kaurenoic acid to GA12 56, showed an increase in FC > 4 as compared to the TOP; the genes encoding 2-oxoglutarate-dependent dioxygenases GA20OX2 and GA3OX1 showed a FC > 3 and >4 as compared to the TOP, respectively (Suppl. Dataset File). The expression GA2OX2 also peaked at the BOT (expressed 12-times more as compared to the TOP): this gene encodes a gibberellin 2-β-dioxygenase responsible for the conversion of GA to an inactive form and participates in GA homeostasis. In addition to that, a contig annotated as a CYP714A1 (contig_19239), which participates in GA deactivation57, was also expressed at higher levels at the BOT with respect to the TOP (FC > 7). In the BOT region of the hemp stem, characterized by bast fibres with a thick tertiary cell wall, elongation has ceased and the homeostasis of GA levels by GA2OX2 may represent an important regulatory mechanism. Transgenic tobacco plants in which the gene GA2OX2 had been silenced were taller than GA20OX1 overexpressors (the GA 20-oxidase catalyses the rate limiting step in GA biosynthesis); these results therefore show that deactivation is the key factor in the maintenance of GA homeostasis58. Concomitantly with the activation of GA2OX2, genes involved in abscisic acid (ABA) biosynthesis increased in expression at the BOT. ABA inhibits stem elongation and in our previous study it was found in higher abundance in the hemp hypocotyl at older developmental stages22. In our dataset, the ABA biosynthetic genes NCED3 and 5 were both upregulated at the BOT (FC > 5 and >11 respectively, as compared to the TOP); the gene XERICO encoding a RING-H2 zinc finger protein and involved in ABA homeostasis59 was also upregulated at the BOT (FC > 4 with respect to the TOP; Suppl. Dataset File). Additionally, three contigs annotated as CYP704A2, which was proposed to be a candidate for ABA 8′-hydroxylation (i.e. catabolism60), were expressed at higher levels at the BOT (FC BOT vs TOP between 28 and 135).
A contig annotated as a CYP82C2 was upregulated at the BOT (FC difference between BOT and TOP > 16): the A. thaliana ortholog is involved in the modulation of tryptophan-derived secondary metabolites under conditions of high JA levels61.
Genes encoding putative cytochrome P450 94 members (CYP94D2 and B2) were also upregulated at the BOT (FC BOT vs TOP between 4.6 and 38.9): the turnover of jasmonoyl-L-isoleucine (the major bioactive form of JA) is regulated by members of this class62. Our results show that a molecular control over the levels GA, ABA, JA is present at the BOT of adult hemp stems.
In the “cellulose metabolic process” ontogeny it is here worth mentioning the cellulose synthase-like genes CSLG1 and G3 (FC > 12 and >5, respectively, as compared to the TOP); recently, in fibres of flax with tertiary cell walls, the upregulation of a CSLG member was observed and it was proposed that the corresponding enzyme may catalyse the synthesis of β-1,4-galactans14. Although the LM5 antibody recognizing β-1,4-galactans does not label hemp bast fibres22, 63, it may be possible that pectic galactans in hemp fibres are masked and therefore do not react with the LM5 antibody.
Genes involved in lignin biosynthesis, i.e. PAL1 (phenylalanine ammonia lyase 1) and one contig annotated as OMT1 (caffeic acid/5-hydroxyferulic acid O-methyltransferase; contig_2172) showed, respectively, a FC > 11 and >9 as compared to the TOP (Suppl. Dataset File). Likewise, one contig encoding IRX12/LAC4 (contig_13683) and a cytochrome P450 involved in lignification (CYP71A20)64 showed a FC increase in expression > 28 and >35 at the BOT with respect to the TOP (Suppl. Dataset File). These results are in agreement with the presence of secondary fibres in the BOT region; these fibres, notably, show a positive signal at the level of the middle lamella after phloroglucinol staining43. The secondary bast fibres are present, together with the primary fibres, in the cortical peels sampled at the BOT.
Several contigs coding for MYB, NAC, bHLH, bZIP and WRKY TFs were found enriched in the bast fibres at the BOT. It is important to highlight here that, in the other fibre crop Corchorus capsularis, a WRKY TF was upregulated during the early phases of fibre development and was suggested to be a candidate gene to improve jute fibre16. Additionally, in jute, bHLH, MYB-related, WRKY and NAC TFs were among the eight most abundant families15. These TFs may therefore have overlapping functions in the development of both xylan- and gelatinous-type bast fibres.
Among the hemp TFs, notably, there is MYB4, a negative regulator of lignification which may play a role in bast fibre hypolignification37. Among the genes encoding TFs upregulated at the BOT, it is worth mentioning a LOB-domain containing protein 4 (LBD4), which showed a BOT vs TOP increase in expression of ca. 218 (Suppl. Dataset File). LOBs are key regulators of plant organ polarity which play also a role in plant secondary growth65; their role in bast fibre development is to our knowledge not yet studied, however we believe that LBDs represent interesting targets in phloem fibre development. A contig annotated as NAC047/SPEEDY HYPONASTIC GROWTH (SHYG) peaked at the BOT (BOT vs TOP FC > 78): the ortholog in thale cress was shown to be associated with hyponastic leaf movements upon waterlogging via modulation of an enzyme involved in ET biosynthesis, i.e. ACC OXIDASE566. ET may play a role during fibre thickening: several genes encoding ethylene-responsive TFs were indeed upregulated at the BOT, notably ERF1, WRI1, ERF106 (Suppl. Dataset File). Interestingly SHYG acts on expansin and xyloglucan endotransglycosylase/hydrolase genes to induce expansion of the petiole abaxial side. Its role in bast fibre thickening is yet to be unveiled, but it may entail a suite of cell wall-related events.
A contig encoding a RADIALIS-LIKE 6 TF was upregulated at the BOT ca. 18 fold with respect to the TOP: very recently a RADIALIS-LIKE TF was found upregulated in flax bast fibres and a role in phloem fibre development was therefore suggested14. RADIALIS-LIKE TFs play a role in organ (floral) symmetry; hence, together with LBDs, they may regulate aspects related with positional cues.
Conclusions
We have here provided a transcriptional fingerprinting of bast fibres from textile hemp sampled at different stem heights, which correspond to different developmental stages. We have discussed the results using, predominantly, a cell wall angle, since the development of phloem fibres requires major modifications of the cell wall. Our study has shown that each region of the stem is characterized by distinct gene expression profiles. Young stem regions are dominated by cell cycle- and photosynthesis-related genes, together with candidates involved in the biosynthesis of specific secondary metabolites, notably indole-containing compounds and oligolignols; older internodes show enrichment of phytohormone-related genes, together with genes involved in non-cellulosic polysaccharide deposition and lignification. According to our results, the bulk of cell wall-related gene dynamics in hemp bast fibres is localized at the internode containing the snap point, where the fibres shift from a phase of elongation to thickening. The data here shown contribute to the understanding of the molecular events accompanying hemp bast fibre development and identify several genes deserving further functional study.
Methods
Plant material, growth conditions and optical microscopy
A hemp monoecious fibre variety (C. sativa cv. Santhica 27) was studied in this work. Plants were grown and sampled as previously described12. Briefly, after six weeks of growth in controlled chambers, samples were taken along three stem regions localized at different heights with respect to the snap point (determined empirically by gently tilting the stem apex until a kink could be observed). The top corresponds to the internode right below the apex, the middle is the internode containing the snap point and the bottom is located two internodes below the middle sample. At the time of sampling, the plants had ca. 6–7 internodes. A segment of 2.5 cm was collected in the middle of each internode to avoid too much variation in gene expression, because of the varying developmental stages of the cell types.
Fibres were separated from the shivs by peeling the cortical tissues and by quickly processing them as described previously67. The number of biological replicates is four with 13 plants in each replicate. Sample preparation for optical microscopy was performed as previously described22.
RNA extraction and preparation of the libraries
Total RNA was extracted using a modified CTAB extraction protocol combined with an RNeasy Plant Mini Kit (Qiagen)67. The RNA concentration and quality were measured by using a Nanodrop ND-1000 (Thermo Scientific) and a 2100 Bioanalyzer (Agilent), respectively. All the RNAs had a RIN value between 8 and 9.
Libraries were prepared, quantified and their average size analysed as previously described22. The libraries were pooled at the concentration of 20 pM and sequenced on an Illumina MiSeq in five consecutive runs (MiSeq reagent kit V3, 150 cycles). Raw sequences have been deposited at the NCBI Gene Expression Omnibus (GEO) with the accession number GSE94156 (available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=itwfckosdjwdpiv&acc=GSE94156).
Processing of the reads, mapping and RNA-Seq analysis
The raw sequences obtained were uploaded in CLC Genomics Workbench 9.0.1. Sequences were filtered as follows: sequences > 35 bps, the sequence quality score was left as default value (0.05), the maximum number of ambiguities was set to 0. Adaptor trimming was performed using the Illumina adaptor sequences, then a hard trim of 14 bps at the 5′ end and 2 bps at the 3′ end was additionally carried out, resulting in a final sequence average length of 59 bps. We had previously published a de novo assembly for the variety Santhica 2722 and proven its validity by comparing the results generated with our de novo assembly and with the Finola transcriptome23. We decided to merge the reads generated in this study with those previously obtained on the hemp hypocotyl22 to get a better assembly of the transcriptome of the variety under study. We therefore uploaded in CLC Genomics Workbench 9.0.1 the reads obtained previously for the hypocotyls and those obtained in the present study for the fibres from adult plants. The parameters used are: wording size was set to 20, the bubble size to 50 and minimum contig length of 300. The reads were mapped back to the assembly with a mismatch, insertion and deletion cost of 3 (stringent criteria), and a length and similarity fraction of 0.95. The assembly was then annotated using Blast2GO PRO version 3.0 against the Viridiplantae and A. thaliana non-redundant database. However, in Suppl. Dataset only the annotation against the Arabidopsis database is shown, as it was used for the subsequent Gene Ontology term Enrichment analysis (GOE) in Cytoscape (vide infra). For each library, the mapping was performed with a maximum hits per read of 3, a similarity and length fraction of 0.95, a mismatch, insertion and deletion cost of 3. Mapping was also performed using the transcriptome of the variety Finola23, as previously described22. The expression values were then calculated using the RPKM method68.
The expression values were subjected to an ANOVA statistical test with three groups (TOP, MID, BOT), each composed of four biological replicates and, subsequently, to a false discovery rate (FDR) correction. Only the genes showing a corrected p-value < 0.05 were retained for downstream analysis. The obtained data were further filtered by removing those genes showing a maximum value of the means < 1 RPKM (this was done with the purpose of removing those contigs showing negligible changes in gene expression) and a maximum FC > 4 in absolute value. A total of 3268 contigs was obtained (Suppl. Dataset File).
Primer design
Primers were designed using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/) and verified with the OligoAnalyzer 3.1 tool from Integrated DNA technologies (http://eu.idtdna.com/calc/analyzer). Primer efficiencies were checked via qPCR using a serial dilution of cDNA (from 10 ng to 0.0032 ng/µl). The primer sequences, amplicon length and Tm, amplification efficiencies and R2 are indicated in Suppl. Dataset File.
RNA-Seq validation with RT-qPCR
The RNA extracted using the above-mentioned protocol was retrotranscribed into cDNA using the ProtoScript II RTase (NEB) and random primers, according to the manufacturer’s instructions. The cDNA was diluted to 2 ng/µl and 2 µl were used for the RT-qPCR analysis in 384-wells microplates (10 µl final volume). An automated liquid handling robot (epMotion 5073) was used to prepare the microplates. The expression of each target gene was normalized using 2 reference genes (clathrin and F-Box), after screening the 12 reference genes described previously12. To check the specificity of the amplicons, a melt curve analysis was performed. The expression of the genes was calculated using qBasePLUS 69 by using the above-mentioned 2 reference genes.
Bioinformatic analysis
The annotation of the putative transcription factors (TFs) in the de novo assembly was carried out with PlantTFcat70 (http://plantgrn.noble.org/PlantTFcat/), which gave a total of 2484 TFs (Suppl. Dataset File). The ICA was performed with the on-line program MetaGeneAlyse v1.7.171 (http://metagenealyse.mpimp-golm.mpg.de/). The Gene Ontology term Enrichment analysis (GOE) was performed as previously described72 using Cytoscape (v3.4.0) with the ClueGO v2.3.2 plugin73 (p-value < 0.05, Benjamini-Hochberg enrichment, gene ontology from level 3 to 8, kappa score set at 0.6). RNA-Seq RPKMs were log2 transformed and loaded for clustering and expression profile analysis in a data analytics software developed in-house. The software includes a Web-based user interface providing interactive data visualisation in the form of a parallel coordinates plot synchronised with 2D scatter plots of PCA projections; the user interface is backed by an R server providing the necessary statistical analyses, in particular correlation clustering and PCA projection of multidimensional data. The software allowed us to configure, execute and visually analyse the RNA-Seq RPKMs; notably, with it we were able to identify the clusters of genes shown in Fig. 1.
Electronic supplementary material
Acknowledgements
The authors acknowledge the Fonds National de la Recherche, Luxembourg, (Project CANCAN C13/SR/5774202) for financial support. This work was (partially) funded by the FNR grant BLIZAAR INTER/ANR/14/9909176. The authors thank Aude Corvisy and Laurent Solinhac for their technical support. The authors gratefully acknowledge Dr Fintan Mc Gee for fruitful discussions on Visual Analytics.
Author Contributions
G.G. conceived the idea of the study and designed the experiments. M.B. performed the RT-qPCR validation. L.M.-P. grew the plants, collected the fibres, extracted the RNA. G.G. and L.M.-P. performed library preparation, quantification and designed the primers for the RT-qPCR analysis. G.G., L.M.-P., S.L. and S.Z. carried out the bioinformatics analyses. G.G., M.B., L.M.-P., S.L., S.Z., M.G. and J.F.-H. interpreted the data and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05200-8
Accession codes: Raw reads were deposited at the NCBI Gene Expression Omnibus (GEO), under accession GSE94156.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guerriero G, Sergeant K, Hausman J-F. Integrated -Omics: A Powerful Approach to Understanding the Heterogeneous Lignification of Fibre Crops. Int. J. Mol. Sci. 2013;14:10958–10978. doi: 10.3390/ijms140610958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikshina, P. et al. Cellulosic Fibers: Role of Matrix Polysaccharides in Structure and Function. doi:10.5772/51941 (2013).
- 3.Neutelings G. Lignin variability in plant cell walls: Contribution of new models. Plant Sci. 2011;181:379–386. doi: 10.1016/j.plantsci.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Mellerowicz EJ, Immerzeel P, Hayashi T. Xyloglucan: The Molecular Muscle of Trees. Ann. Bot. 2008;102:659–665. doi: 10.1093/aob/mcn170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorshkova TA, et al. The snap point: a transition point in Linum usitatissimum bast fiber development. Ind. Crops Prod. 2003;18:213–221. doi: 10.1016/S0926-6690(03)00043-8. [DOI] [Google Scholar]
- 6.Guerriero G, Hausman JF, Cai G. No Stress! Relax! Mechanisms Governing Growth and Shape in Plant Cells. Int. J. Mol. Sci. 2014;15:5094–5114. doi: 10.3390/ijms15035094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snegireva A, Chernova T, Ageeva M, Lev-Yadun S, Gorshkova T. Intrusive growth of primary and secondary phloem fibres in hemp stem determines fibre-bundle formation and structure. AoB Plants. 2015;7:plv061. doi: 10.1093/aobpla/plv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ageeva MV, et al. Intrusive growth of flax phloem fibers is of intercalary type. Planta. 2005;222:565–574. doi: 10.1007/s00425-005-1536-2. [DOI] [PubMed] [Google Scholar]
- 9.Lev-Yadun S. Plant fibers: Initiation, growth, model plants, and open questions. Russ. J. Plant Physiol. 2010;57:305–315. doi: 10.1134/S1021443710030015. [DOI] [Google Scholar]
- 10.Snegireva AV, et al. Intrusive growth of sclerenchyma fibers. Russ. J. Plant Physiol. 2010;57:342–355. doi: 10.1134/S1021443710030052. [DOI] [Google Scholar]
- 11.Gorshkova T, et al. Plant Fiber Formation: State of the Art, Recent and Expected Progress, and Open Questions. Crit. Rev. Plant Sci. 2012;31:201–228. doi: 10.1080/07352689.2011.616096. [DOI] [Google Scholar]
- 12.Mangeot-Peter L, Legay S, Hausman J-F, Esposito S, Guerriero G. Identification of Reference Genes for RT-qPCR Data Normalization in Cannabis sativa Stem Tissues. Int. J. Mol. Sci. 2016;17:1556. doi: 10.3390/ijms17091556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mokshina N, Gorshkova T, Deyholos MK. Chitinase-Like (CTL) and Cellulose Synthase (CESA) Gene Expression in Gelatinous-Type Cellulosic Walls of Flax (Linum usitatissimum L.) Bast Fibers. PLOS ONE. 2014;9:e97949. doi: 10.1371/journal.pone.0097949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorshkov, O. et al. Transcriptome portrait of cellulose-enriched flax fibres at advanced stage of specialization. Plant Mol. Biol. 1–19, doi:10.1007/s11103-016-0571-7 (2016). [DOI] [PubMed]
- 15.Chakraborty A, Sarkar D, Satya P, Karmakar PG, Singh NK. Pathways associated with lignin biosynthesis in lignomaniac jute fibres. Mol. Genet. Genomics MGG. 2015;290:1523–1542. doi: 10.1007/s00438-015-1013-y. [DOI] [PubMed] [Google Scholar]
- 16.Samanta P, Sadhukhan S, Basu A. Identification of differentially expressed transcripts associated with bast fibre development in Corchorus capsularis by suppression subtractive hybridization. Planta. 2015;241:371–385. doi: 10.1007/s00425-014-2187-y. [DOI] [PubMed] [Google Scholar]
- 17.Islam MS, et al. Comparative genomics of two jute species and insight into fibre biogenesis. Nat. Plants. 2017;3:16223. doi: 10.1038/nplants.2016.223. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, et al. Transcriptome profiling using pyrosequencing shows genes associated with bast fiber development in ramie (Boehmeria nivea L.) BMC Genomics. 2014;15:919. doi: 10.1186/1471-2164-15-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, et al. Characterization of the Kenaf (Hibiscus cannabinus) Global Transcriptome Using Illumina Paired-End Sequencing and Development of EST-SSR Markers. PloS One. 2016;11:e0150548. doi: 10.1371/journal.pone.0150548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Pauw MAD, Vidmar JJ, Collins J, Bennett RA, Deyholos MK. Microarray analysis of bast fibre producing tissues of Cannabis sativa identifies transcripts associated with conserved and specialised processes of secondary wall development. Funct. Plant Biol. 2007;34:737–749. doi: 10.1071/FP07014. [DOI] [PubMed] [Google Scholar]
- 21.van den Broeck HC, Maliepaard C, Ebskamp MJM, Toonen MAJ, Koops AJ. Differential expression of genes involved in C1 metabolism and lignin biosynthesis in wooden core and bast tissues of fibre hemp (Cannabis sativa L.) Plant Sci. 2008;174:205–220. doi: 10.1016/j.plantsci.2007.11.008. [DOI] [Google Scholar]
- 22.Behr M, et al. Studying Secondary Growth and Bast Fiber Development: The Hemp Hypocotyl Peeks behind the Wall. Front. Plant Sci. 2016;7:1733. doi: 10.3389/fpls.2016.01733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Bakel H, et al. The draft genome and transcriptome of Cannabis sativa. Genome Biol. 2011;12:R102. doi: 10.1186/gb-2011-12-10-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roach MJ, Deyholos MK. Microarray analysis of flax (Linum usitatissimum L.) stems identifies transcripts enriched in fibre-bearing phloem tissues. Mol. Genet. Genomics. 2007;278:149–165. doi: 10.1007/s00438-007-0241-1. [DOI] [PubMed] [Google Scholar]
- 25.Strzalka W, Ziemienowicz A. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann. Bot. 2011;107:1127–1140. doi: 10.1093/aob/mcq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collakova E, DellaPenna D. The Role of Homogentisate Phytyltransferase and Other Tocopherol Pathway Enzymes in the Regulation of Tocopherol Synthesis during Abiotic Stress. Plant Physiol. 2003;133:930–940. doi: 10.1104/pp.103.026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao Z, Yang H, Hua J. Perturbation of cell cycle regulation triggers plant immune response via activation of disease resistance genes. Proc. Natl. Acad. Sci. USA. 2013;110:2407–2412. doi: 10.1073/pnas.1217024110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boachon B, et al. CYP76C1 (Cytochrome P450)-Mediated Linalool Metabolism and the Formation of Volatile and Soluble Linalool Oxides in Arabidopsis Flowers: A Strategy for Defense against Floral Antagonists. Plant Cell. 2015;27:2972–2990. doi: 10.1105/tpc.15.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, et al. Herbivore-induced and floral homoterpene volatiles are biosynthesized by a single P450 enzyme (CYP82G1) in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2010;107:21205–21210. doi: 10.1073/pnas.1009975107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andre CM, Hausman J-F, Guerriero G. Cannabis sativa: The Plant of the Thousand and One Molecules. Front. Plant Sci. 2016;7:19. doi: 10.3389/fpls.2016.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langlois-Meurinne M, Gachon CMM, Saindrenan P. Pathogen-Responsive Expression of Glycosyltransferase Genes UGT73B3 and UGT73B5 Is Necessary for Resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 2005;139:1890–1901. doi: 10.1104/pp.105.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou B, Lim E-K, Higgins GS, Bowles DJ. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 2004;279:47822–47832. doi: 10.1074/jbc.M409569200. [DOI] [PubMed] [Google Scholar]
- 33.Le Roy J, Huss B, Creach A, Hawkins S, Neutelings G. Glycosylation Is a Major Regulator of Phenylpropanoid Availability and Biological Activity in Plants. Front. Plant Sci. 2016;7:735. doi: 10.3389/fpls.2016.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frerigmann H, Gigolashvili T. MYB34, MYB51, and MYB122 Distinctly Regulate Indolic Glucosinolate Biosynthesis in Arabidopsis thaliana. Mol. Plant. 2014;7:814–828. doi: 10.1093/mp/ssu004. [DOI] [PubMed] [Google Scholar]
- 35.War AR, et al. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012;7:1306–1320. doi: 10.4161/psb.21663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koo AJK, Howe GA. The wound hormone jasmonate. Phytochemistry. 2009;70:1571–1580. doi: 10.1016/j.phytochem.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huis R, et al. Natural hypolignification is associated with extensive oligolignol accumulation in flax stems. Plant Physiol. 2012;158:1893–1915. doi: 10.1104/pp.111.192328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binns AN, Chen RH, Wood HN, Lynn DG. Cell division promoting activity of naturally occurring dehydrodiconiferyl glucosides: do cell wall components control cell division? Proc. Natl. Acad. Sci. 1987;84:980–984. doi: 10.1073/pnas.84.4.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosch M, Mayer C-D, Cookson A, Donnison IS. Identification of genes involved in cell wall biogenesis in grasses by differential gene expression profiling of elongating and non-elongating maize internodes. J. Exp. Bot. 2011;62:3545–3561. doi: 10.1093/jxb/err045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng F, et al. GbPDF1 Is Involved in Cotton Fiber Initiation via the Core cis-Element HDZIP2ATATHB2. Plant Physiol. 2012;158:890–904. doi: 10.1104/pp.111.186742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong R, Ye Z-H. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 2012;53:368–380. doi: 10.1093/pcp/pcr185. [DOI] [PubMed] [Google Scholar]
- 42.Day A, et al. Lignification in the flax stem: evidence for an unusual lignin in bast fibers. Planta. 2005;222:234–245. doi: 10.1007/s00425-005-1537-1. [DOI] [PubMed] [Google Scholar]
- 43.Crônier D, Monties B, Chabbert B. Structure and Chemical Composition of Bast Fibers Isolated from Developing Hemp Stem. J. Agric. Food Chem. 2005;53:8279–8289. doi: 10.1021/jf051253k. [DOI] [PubMed] [Google Scholar]
- 44.Zhong R, Ye Z-H. Secondary Cell Walls: Biosynthesis, Patterned Deposition and Transcriptional Regulation. Plant Cell Physiol. 2015;56:195–214. doi: 10.1093/pcp/pcu140. [DOI] [PubMed] [Google Scholar]
- 45.Peña MJ, et al. Arabidopsis irregular xylem8 and irregular xylem9: Implications for the Complexity of Glucuronoxylan Biosynthesis. Plant Cell. 2007;19:549–563. doi: 10.1105/tpc.106.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR. Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 2009;57:732–746. doi: 10.1111/j.1365-313X.2008.03729.x. [DOI] [PubMed] [Google Scholar]
- 47.Chávez Montes RA, et al. Cell Wall Modifications in Arabidopsis Plants with Altered α-l-Arabinofuranosidase Activity. Plant Physiol. 2008;147:63–77. doi: 10.1104/pp.107.110023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan L, et al. An Arabidopsis Cell Wall Proteoglycan Consists of Pectin and Arabinoxylan Covalently Linked to an Arabinogalactan Protein. Plant Cell. 2013;25:270–287. doi: 10.1105/tpc.112.107334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruan Y-L, Llewellyn DJ, Furbank RT. The Control of Single-Celled Cotton Fiber Elongation by Developmentally Reversible Gating of Plasmodesmata and Coordinated Expression of Sucrose and K + Transporters and Expansin. Plant Cell. 2001;13:47–60. doi: 10.1105/tpc.13.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. A Triantagonistic Basic Helix-Loop-Helix System Regulates Cell Elongation in Arabidopsis. Plant Cell. 2012;24:4483–4497. doi: 10.1105/tpc.112.105023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagano Y, Furuhashi H, Inaba T, Sasaki Y. A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences. Nucleic Acids Res. 2001;29:4097–4105. doi: 10.1093/nar/29.20.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bayer EM, et al. Arabidopsis cell wall proteome defined using multidimensional protein identification technology. PROTEOMICS. 2006;6:301–311. doi: 10.1002/pmic.200500046. [DOI] [PubMed] [Google Scholar]
- 53.Ranocha P, et al. Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 2010;63:469–483. doi: 10.1111/j.1365-313X.2010.04256.x. [DOI] [PubMed] [Google Scholar]
- 54.Ranocha P, et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 2013;4:2625. doi: 10.1038/ncomms3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun T. Gibberellin Metabolism, Perception and Signaling Pathways in Arabidopsis. Gibberellin Metabolism, Perception and Signaling Pathways in Arabidopsis. The Arabidopsis Book/American Society of Plant Biologists. 2008;6:e0103. doi: 10.1199/tab.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA. 2001;98:2065–2070. doi: 10.1073/pnas.98.4.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, et al. Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J. 2011;67:342–353. doi: 10.1111/j.1365-313X.2011.04596.x. [DOI] [PubMed] [Google Scholar]
- 58.Dayan J, Schwarzkopf M, Avni A, Aloni R. Enhancing plant growth and fiber production by silencing GA 2-oxidase. Plant Biotechnol. J. 2010;8:425–435. doi: 10.1111/j.1467-7652.2009.00480.x. [DOI] [PubMed] [Google Scholar]
- 59.Ko J-H, Yang SH, Han K-H. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 2006;47:343–355. doi: 10.1111/j.1365-313X.2006.02782.x. [DOI] [PubMed] [Google Scholar]
- 60.Kushiro T, et al. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′‐hydroxylases: key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu F, et al. The Arabidopsis P450 protein CYP82C2 modulates jasmonate-induced root growth inhibition, defense gene expression and indole glucosinolate biosynthesis. Cell Res. 2010;20:539–552. doi: 10.1038/cr.2010.36. [DOI] [PubMed] [Google Scholar]
- 62.Koo, A. J. K. & Howe, G. A. Catabolism and Deactivation of the Lipid-Derived Hormone Jasmonoyl-Isoleucine. Front. Plant Sci. 3, (2012). [DOI] [PMC free article] [PubMed]
- 63.Blake AW, Marcus SE, Copeland JE, Blackburn RS, Knox JP. In situ analysis of cell wall polymers associated with phloem fibre cells in stems of hemp, Cannabis sativa L. Planta. 2008;228:1–13. doi: 10.1007/s00425-008-0713-5. [DOI] [PubMed] [Google Scholar]
- 64.V D Mortel JE, et al. Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 2008;31:301–324. doi: 10.1111/j.1365-3040.2007.01764.x. [DOI] [PubMed] [Google Scholar]
- 65.Xu C, Luo F, Hochholdinger F. LOB Domain Proteins: Beyond Lateral Organ Boundaries. Trends Plant Sci. 2016;21:159–167. doi: 10.1016/j.tplants.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 66.Rauf M, et al. NAC Transcription Factor SPEEDY HYPONASTIC GROWTH Regulates Flooding-Induced Leaf Movement in Arabidopsis. Plant Cell. 2013;25:4941–4955. doi: 10.1105/tpc.113.117861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guerriero G, Mangeot-Peter L, Hausman J-F, Legay S. Extraction of High Quality RNA from Cannabis sativa Bast Fibres: A Vademecum for Molecular Biologists. Fibers. 2016;4:23. doi: 10.3390/fib4030023. [DOI] [Google Scholar]
- 68.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 69.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dai X, Sinharoy S, Udvardi M, Zhao PX. PlantTFcat: an online plant transcription factor and transcriptional regulator categorization and analysis tool. BMC Bioinformatics. 2013;14:321. doi: 10.1186/1471-2105-14-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daub CO, Kloska S, Selbig J. MetaGeneAlyse: analysis of integrated transcriptional and metabolite data. Bioinformatics. 2003;19:2332–2333. doi: 10.1093/bioinformatics/btg321. [DOI] [PubMed] [Google Scholar]
- 72.Legay S, et al. Apple russeting as seen through the RNA-seq lens: strong alterations in the exocarp cell wall. Plant Mol. Biol. 2015;88:21–40. doi: 10.1007/s11103-015-0303-4. [DOI] [PubMed] [Google Scholar]
- 73.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


