Abstract
The existence of multipartite viruses is an intriguing mystery in evolutionary virology. Several hypotheses suggest benefits that should outweigh the costs of a reduced transmission efficiency and of segregation of coadapted genes associated with encapsidating each segment into a different particle. Advantages range from increasing genome size despite high mutation rates, faster replication, more efficient selection resulting from reassortment during mixed infections, better regulation of gene expression, or enhanced virion stability and cell-to-cell movement. However, support for these hypotheses is scarce. Here we report experiments testing whether an evolutionary stable equilibrium exists for the three genomic RNAs of Alfalfa mosaic virus (AMV). Starting infections with different segment combinations, we found that the relative abundance of each segment evolves towards a constant ratio. Population genetic analyses show that the segment ratio at this equilibrium is determined by frequency-dependent selection. Replication of RNAs 1 and 2 was coupled and collaborative, whereas the replication of RNA 3 interfered with the replication of the other two. We found that the equilibrium solution is slightly different for the total amounts of RNA produced and encapsidated, suggesting that competition exists between all RNAs during encapsidation. Finally, we found that the observed equilibrium appears to be host-species dependent.
Introduction
The highest level of physical organization of the genome is the division of the hereditary material into multiple segments. Genome segmentation is a ubiquitous feature of eukaryotes, with nuclear chromosome numbers covering an enormous range: from 2n to 630n 1, 2. In contrast, bacteria and archaea typically have a single chromosome3. Although many viruses also have a single genome segment, in some species the genome has been partitioned into multiple segments4–6. Whereas most viruses package multiple segments into a single virus particle (e.g., reovirus and orthomyxovirus), some plant and fungal viruses package each segment into a separate virus particle, a property known as multipartition. By contrast, the only multipartite animal viruses are the single-stranded DNA bidensoviruses infecting silkworms7 and the very recently discovered single-stranded positive-sense RNA virus in mosquitoes related to the Flaviviridae 8. In the extreme case, plant nanoviruses have up to eight DNA genome segments plus several satellite-like segments packaged up into different viral particles, although not all segments must enter a cell to cause infection4. For multipartite RNA viruses, the number of segments is typically lower, ranging from two to five. It is thought that all genome segments must enter the same cell to produce all the RNAs and proteins that are required to complete the process of infection and release the progeny that will infect new cells and transmit to new individual hosts5, 6.
The evolution of segmented genomes revolves around tradeoffs between potential costs and benefits inherent to different genome architectures. An obvious cost of multipartition is the necessity of coinfecting cells with at least one particle of each kind to ensure the presence of at least one copy of each segment, a cost that increases with the number of segments and particles9. All else being equal, an equimolecular composition of particles would maximize the probability of initiating the infection of a host cell successfully. Deviations from this situation would increase the cost of multipartition. Another potential cost of genome segmentation would be the breakage of co-adapted groups of genes during coinfection with several strains of the virus10. Several advantages have been proposed to compensate for these costs: (i) for the high mutation rates of most RNA viruses, smaller segments are more likely to be copied without errors than larger segments11, (ii) smaller genomic segments should be replicated faster12, (iii) segmentation favors genomic reassortment and thus increases genetic variability by rapidly bringing together beneficial mutations that have occurred in different lineages13, 14, minimizing the effect of clonal interference15 and speeding up the rate of adaptation, (iv) encapsidation of smaller genomes results in enhanced capsid stability16, (v) particularly in the case of plant viruses, smaller capsids would facilitate trafficking throughout the size-limiting plasmodesmata17, and (vi) segmentation represents an efficient yet simple way to control gene expression by regulating gene copy numbers18.
Starting on the mid-seventies, a number of publications have addressed different aspects of the replication and regulation of gene expression of plant multipartite viruses, especifically for members of the Bromoviridae family such as Brome mosaic virus (BMV) and Cowpea chlorotic mottle virus (CCMV). Many interesting conclusions were drawn in these studies, but particularly relevant for the problem of the evolution of multipartite virus are: (i) the ratio of RNA segments varies among closely related viral species (BMV and CCMV)19 and even among different BMV isolates20. (ii) The relative abundances of genomic segments for a particular bromovirus species varies among host species21, 22. (iii) Mutations in coding and noncoding sequences of different genomic segments have a profound impact on the accumulation of the other segments23–27.
Sicard et al. monitored the frequency of the eight single-gene-encoding segments, made of circular DNA, that constitute the genome of the nanovirus Faba bean necrotic stunt virus (FBNSV) during infection of single host plants18. They observed that regardless of the initial ratio of segments in the inocula, the ratio of segments always evolved towards a constant composition that the authors designated as the “setpoint genome formula” (hereafter referred as SGF), which did not represent an equimolecular mixture of genomic segments. They also found that the SGF corresponds to a state of maximal viral accumulation and of enhanced symptoms, thus suggesting that segmentation has evolved as a mechanism to regulate gene expression. Finally, in agreement with the bromoviruses results, they also found that the exact stoichiometry of the SGF depends on the host plant species. FBNSV, although being a well-suited model system for addressing questions related to genome segmentation and multipartition, is not very representative of most multipartite plant viruses: most are RNA viruses and have a lower number of genome segments.
As mentioned above, the molecular biology of multipartite RNA viruses has been extensively studied19–27, although it is not known for these viruses whether an SGF exists and whether it is evolutionarily stable. In other words: we still miss experimental evidences to support or reject some of the evolutionary genetic mechanism brought forward to explains the evolution of multipartition in RNA virus populations9, 11–14. If no stable SGF exists, or if multiple stable equilibria are possible, genome segment ratios could be in a state of perpetual flux, and some hypotheses for the advantages of segmentation would need to be discarded or revisited. Similarly, if different SGF exist in different host species, this would suggest that segmentation can facilitate adaptation by rapidly altering the proportion of RNAs –and possibly their expression– in a mutation-independent manner18.
The aim of our study is threefold. First, we sought to explore whether a SGF also exists for a prototypical multipartite RNA virus. Second, we also set out to determine the effect of host species on the stoichiometry of the SGF. Third, we also propose a novel analytical and computational framework, of universal applicability, to the evolutionary analysis of the abundance of any number of segments in segmented viral genomes. To tackle these questions, we have chosen Alfalfa mosaic virus (AMV; genus Alfamovirus, family Bromoviridae), whose genome is composed of three single-stranded positive-sense RNA molecules (RNA1, RNA2 and RNA3). Briefly, RNA1 and RNA2 encode proteins essential for replication (P1 and P2) while RNA3 encodes for the movement (MP) and coat (CP) proteins, the latter being translated from a subgenomic RNA4 (sgRNA4) produced by transcription of the negative-sense strand of RNA328. Our results show that an evolutionary stable but host-species-dependent SGF occurs for this prototypical plant RNA virus.
Results
Determination of the SGF for AMV
We first set out to determine whether an SGF exists for AMV, and if so to determine its characteristics. To this end we performed inoculation experiments with a range of RNA1, RNA2 and RNA3 ratios. Nicotiana benthamiana plants were mechanically inoculated with a constant amount of RNA (1 μg) but varying proportions of the three segments as detailed in the Methods section. The seven ratios employed were 1:1:1, 10:1:1, 1:10:1, 1:1:10, 10:10:1, 10:1:10, and 1:10:10. If a SGF exists, then we expect that these different combinations will all evolve towards it as infection progresses. We sampled different tissues (Fig. 1A) at different stages of infection and estimated the abundance of each RNA on the samples by RT-qPCR. Preliminary control experiments confirmed the specificity of each set of primers as well as their almost identical and high amplification efficiencies. Two types of RNA samples were prepared from each tissue: total RNA and encapsidated RNA from purified viral particles. The first sample represents the total amounts of RNA1, RNA2 and RNA3 synthetized during infection (the averages from n = 3 different plants are shown in Fig. 1B) whereas the second sample represents the amount of each RNA that has been encapsidated (the averages from n = 3 different plants are shown in Fig. 1D) and thus is expected to be the relevant figure in terms of horizontal virus transmission. Table 1A shows the results of the multivariate analysis of variance (MANOVA) analysis for the RNA frequencies from total RNA extractions fitted to equation (1). This equation, described in detail in the Methods section, relates all three experimental factors with the observed RNA segments frequencies: the RNA mixtures inoculated (M), the replicate plants inoculated with each RNA mixture (P) and the different tissues sampled (S). All three factors contribute in a highly significant manner to the observed variability in RNA segments frequency (in all cases P < 0.001). The resulting relative accumulations of RNA1, RNA2 and RNA3 differ among the seven input mixtures (factor M). Overall, plants inoculated with the same mixture also differ in the estimated output mixtures (factor P nested within factor M, P(M)). An overall difference also exist among tissue samples from plants inoculated with the same RNA mixture (factor S nested within factor M, S(M)). Finally, significant differences exist among the four sample types taken from each one of the three different plants inoculated with the same mixture of RNAs (factor (P × S)(M)).
Figure 1.
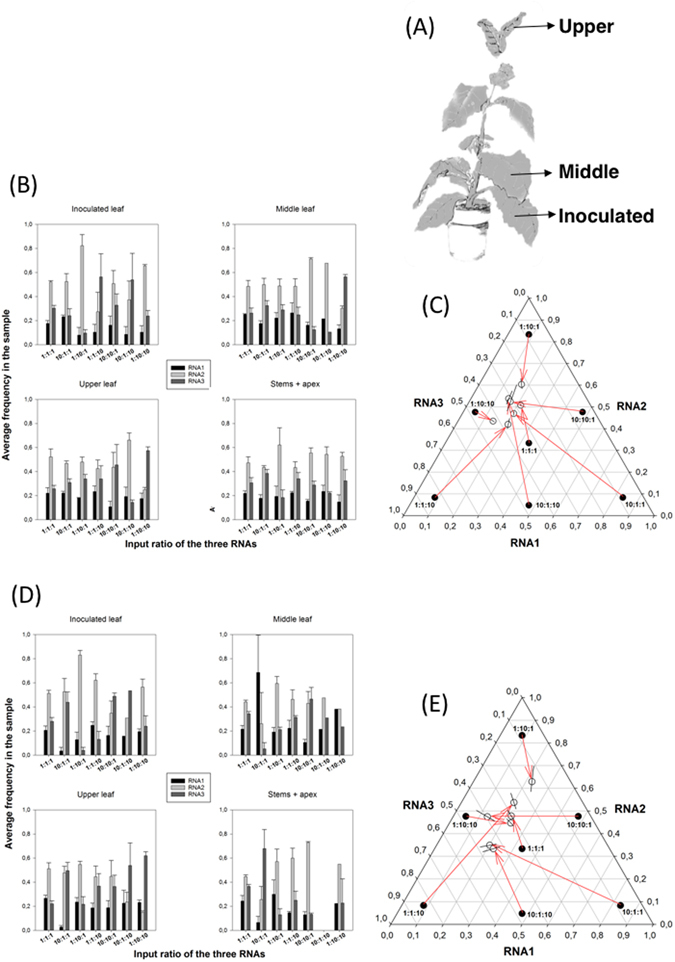
Effect of differences in the input ratio of RNA segments on the outcome of AMV infection. (A) Scheme of the sampling process, with indication of the three leafs sampled. (B) Experimental determinations (by RT-qPCR) of the frequency of each one of the three RNA segments in samples of total RNA for different input ratios. Bars represent the mean of n = 3 plants; error bars represent ±1 SEM. (C) Normalized frequency ternary plot showing the estimated SGF. Solid circles represent the indicated inoculation rates. Open circles show the marginal mean estimates of relative ratios at the end of the experiment corresponding to each input ratio. Lines crossing the open circles represent the 95% Cis. Red arrows connect initial and final ratios. (D) As in (B) but determined for encapsidated RNAs. (E) As in (C) but for encapsidated RNAs.
Table 1.
Results of the MANOVA analysis for the RNA frequencies estimated from total and from virion RNA extractions in experiments with variable input ratios done in N. benthamiana.
| Effect | Wilk’s Λ | F | Hypothesis df | Error df | P | Power | |
|---|---|---|---|---|---|---|---|
| (A) Total RNA | |||||||
| Intercept | 0.002 | 16600.650 | 3 | 76 | <0.001 | 1 | 0.999 |
| M | 0.026 | 31.742 | 18 | 215.446 | <0.001 | 1 | 0.705 |
| P(M) | 0.015 | 16.813 | 42 | 226.218 | <0.001 | 1 | 0.754 |
| S(M) | 0.003 | 21.563 | 63 | 227.701 | <0.001 | 1 | 0.855 |
| (P × S)(M) | 0.008 | 8.224 | 111 | 229 | <0.001 | 1 | 0.799 |
| (B) Virion RNA | |||||||
| Intercept | 0.027 | 754.120 | 3 | 62 | <0.001 | 1 | 0.973 |
| M | 0.325 | 4.760 | 18 | 175.848 | <0.001 | 1 | 0.312 |
| P(M) | 0.499 | 1.162 | 42 | 184.687 | 0.248 | 0.958 | 0.207 |
| S(M) | 0.126 | 2.955 | 63 | 185.911 | <0.001 | 1 | 0.499 |
| (P × S)(M) | 0.186 | 1.675 | 84 | 186.356 | 0.002 | 1 | 0.429 |
The model fitted is shown in equation (1). M: inoculated mixture of RNA segments; P(M): replicate plant inoculated with each mixture; S(M): type of sample from a plant inoculated with a given mixture; and (P × S)(M): interaction term between sample type and replicate plant inoculated with a given mixture M.
However, a significant P-value tells nothing about the magnitude of the effect that a factor has on the measured variables; a small effect may still be significant from an statistical point of view, while being too small to be biologically relevant. To assess the magnitude of effects we used the statistic. Table 1A shows that significant differences exist among plants inoculated with the same mixture (P(M) term in equation (1) in the Methods section) and among equivalent samples from different plants ((P × S)(M) term in equation (1)) and that the effect associated with these two factors is large ( = 0.754 and = 0.799, respectively). In statistics, > 0.15 is usually considered as a large effect; all values reported in Table 1 are far larger than this threshold. These differences appear as an unavoidable consequence of the stochastic events that take place during inoculation of different plants as well as during the progression of infection (e.g., bottlenecks during cell-to-cell and systemic movement of viral particles29). Despite these differences, significant effects have been detected for the other factors.
The largest effect = 0.855 is associated to differences among samples (i.e., tissues; S(M) term in equation (1)). Differences among samples may result from a complex combination of factors. To mention a few, not necessarily being exhaustive, (i) different tissues being colonized by different subpopulations of RNAs simply by chance (i.e., bottlenecks and founder effects29), (ii) different tissues being at different developmental stages may impose differences in susceptibility to AMV infection, (iii) variability in the antiviral defense status of new tissues compared to old ones (e.g., antiviral RNAi-mediated response30), and even (iv) genetic differences in the composition of viral populations at different stages in their within-host evolution as a consequence of interplay between mutation and variable selection pressures on different tissues (i.e., the quasispecies nature of AMV)31. The weakest effect, = 0.705, corresponds to differences among inoculation ratios (M term in equation (1)), though it can still be considered as a very strong effect.
The grand mean estimate for the relative frequencies corresponds to a SGF total of 1.00:2.88:1.87 (±1 SEM: 0.00:0.24:0.22). Figure 1C is a normalized ternary plot showing the marginal average of output ratios estimated for each input ratio. Regardless of the initial conditions (included in the plot to illustrate the coordinates of the different starting points), after infection all RNA populations tend to a rather limited region of the possible space of solutions, which contains the SGF total ratio.
Table 1B shows the results from the MANOVA analysis run for the RNA frequencies estimated from encapsidated RNAs fitted to equation (1). The only difference with the results just reported for the total RNAs is the lack of differences among replica plants (term P(M) in equation (1); P = 0.248). The magnitude of significant effects is 0.312, which is smaller than found for the case of total RNA but still considered as large (>0.15). In this case, the grand mean estimated for relative frequencies corresponds to a SGF encap of 1.07:2.55:1.75 (±1 SEM: 0.07:0.39:0.26), a value that does not differ from the ~1:3:2 reported above for total RNAs, and in both cases shows that RNA2 is the most abundant one. As above, Fig. 1E summarizes the evolution of segments ratio from the input mixture to the average values obtained at the time of analyzing the encapsidated RNAs extracted: values converge to a particular region of the space that contains the SGF encap value.
Overall, the results summarized in Fig. 1C and E suggest the existence of an attractor region in the frequencies phase diagram to which RNA populations converge after infection. In the following section, we will explore whether (i) this equilibrium is driven by frequency-dependent selection (FDS) and (ii) it is stable32.
AMV SGF is driven by FDS to a stable equilibrium
Figure 2 shows the graphical analysis of FDS as a driving force of the observed SGF. Marginal mean frequency data shown in Fig. 1C and E for each RNA segment were transformed into relative abundances as described in the Methods section and then log-transformed. The plots show the output log-relative abundances as a function of the input log-relative abundances (the seven inoculation mixtures). Figure 2A shows the results for the RNA segments quantified in the total RNA samples (from Fig. 1C). The dashed line represents the expectation under the null hypothesis of no-FDS32. For each RNA, we computed the output and input log-relative abundance and plotted them (different symbols). First, the data were fitted to a set of models with increasing number of parameters, but in all cases the linear regression was the best-fitting model (shown as solid lines). In all three cases, statistical significance of the FDS is tested by the deviation of the slope of the linear regression from 1 (the dashed diagonal). All three regression lines in Fig. 2A have a slope significantly less than 1 (t 5 ≥ 6.964, P ≤ 0.001). The analysis of Fig. 2A provides additional information of considerable biological interest, i.e., whether the FDS is linear or not, how strong it is, and if an equilibrium point exists. A point of equilibrium occurs if and where the regression line crosses the diagonal32. At such point the focal RNA segment frequency is at the same frequency as expected in the absence of FDS. In all three cases, the slope is less than one, meaning that the equilibrium is evolutionarily stable; the system evolves towards an equilibrium SGF total that is stable against random perturbations of any of the three RNA components. For instance, perturbations may be associated with the inoculation process or by bottlenecks inherent to systemic movement and colonization of new growing tissues in the apical meristem29.
Figure 2.
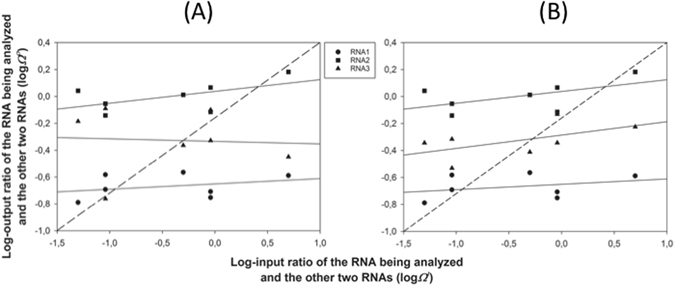
Graphical analysis of FDS as a mechanism to explain the relative abundance of the three RNA segments. (A) Abundances estimated from total RNA samples. (B) Abundances estimated from RNA extracted from viral particles. The dashed line corresponds to the null hypothesis of no-FDS. The continuous lines show the best fitting linear model to the relative abundances of each RNA.
Figure 2B presents the graphical analyses of FDS for the encapsidated RNA ratios shown in Fig. 1E. The conclusions are qualitatively the same as those described in the previous paragraph for the total RNA samples: all three relationships are linear, with slopes significantly less than 1 (t 5 ≥ 13.610, P ≤ 0.001) and thus the SGF encap also corresponds to an evolutionarily stable equilibrium. Notice that in the case of encapsidated RNAs, the three curves show similar positive slopes while in the case of the total RNAs the curves for RNA1 and RNA2 are similar in slope and both positive wherease the curve for RNA3 is negative.
To further characterize the nature of this FDS, we have analyzed the particular relationship between the marginal mean abundances of RNAs in both types of samples, from total and encapsidated RNAs. To do so, we have computed partial correlation coefficients among RNA abundances using as control variables the input ratios (M in equation (1)), plant replicate (P in equation (1)) and tissue sampled (S in equation (1)). Table 2 shows the hemi-matrix of correlations (notice that the matrix is symmetrical and thus the upper half has been removed). Focusing first in the quantifications from the total RNA extractions, we found that the synthesis of RNA3 negatively correlates with the production of both RNA1 and RNA2, while the levels of RNA1 and RNA2 production do not affect each other (lack of significant correlations). Looking now at the correlations between encapsidated RNAs, we found that all are negatively correlated with each other, thus suggesting that they compete for available capsids, which should then become a limiting factor. Finally, looking at correlations between total and encapsidated RNAs (non-gray cells in the hemi-matrix), with exception of RNA1, positive correlations exist between the amount of total and encapsidated RNAs (although the correlation for RNA3 becomes non-significant after accounting for multiple tests of the same hypothesis). RNA2 and RNA3 seem to strongly compete for encapsidation: the more RNA2 produced, the less RNA3 encapsidated and vice versa. However, RNA1 does not seem to be involved in this competition.
Table 2.
Results from the partial correlation analyses among abundances of different RNA segments.
| Total RNA extraction | Virion RNA extraction | |||||
|---|---|---|---|---|---|---|
| RNA1 | RNA2 | RNA3 | RNA1 | RNA2 | ||
| Total | RNA2 | r = 0.024, P = 0.835 | ||||
| RNA3 | r = −0.445, P < 0.001* | r = −0.895, P < 0.001* | ||||
| Virion | RNA1 | r = −0.001, P = 0.991 | r = −0.049, P = 0.661 | r = 0.052, P = 0.645 | ||
| RNA2 | r = −0.021, P = 0.850 | r = 0.319, P = 0.004* | r = −0.282, P = 0.011* | r = −0.425, P < 0.001* | ||
| RNA3 | r = −0.020, P = 0.863 | r = −0.278, P = 0.012* | r = −0.238, P = 0.032 | r = −0.418, P < 0.001* | r = −0.644, P < 0.001* | |
All tests have 79 df. Asterisks indicate cases significant after the Holm-Bonferroni correction of multiple tests of the same null hypothesis.
AMV SGF varies among host species
Next, we explored to which extent the host species determines the value of SGF. To do so, we inoculated five different susceptible hosts (N. benthamiana, Nicotiana tabacum, Cucurbita pepo, Medicago sativa, and Capsicum annuum) with a 1:1:1 mixture of the genomic RNA segments and evaluated the output frequency of each segment 7 days post inoculation days (dpi) for N. benthamiana and 12 dpi for the rest of species, following the same sampling scheme than in the experiments previously described. Frequency data are shown in Fig. 3A (averages from n = 3 different plants). These data were fitted to equation (2) in Methods using MANOVA and the results from this analysis are shown in Table 3. In case of segment frequencies in the total RNA extraction, all factors had a highly significant effect, with magnitudes being in all cases 0.483 (Table 3A). There is great variation in the segments ratio among host species (solid symbols in Fig. 3C), although the estimates for both Nicotiana spp. remain closer among them than they are relative to the other species analyzed. The grand mean value SGF total across hosts is 1.05:3.36:9.04 (±1 SEM: 0.05:0.84:3.27), a value that sharply contrasts to the above stable equilibrium value found for N. benthamiana (~1:3:2) due to the larger bias in the synthesis of RNA3 that characterizes the infection of C. pepo (1.00:6.70:16.11 (±1 SEM: 0.39:1.86:3.42)), M. sativa (1.00:2.37:12.45 (±1 SEM: 0.27:0.70:1.87)) and C. annuum (1.00:2.61:14.35 (±1 SEM: 0.35:0.96:2.78)).
Figure 3.
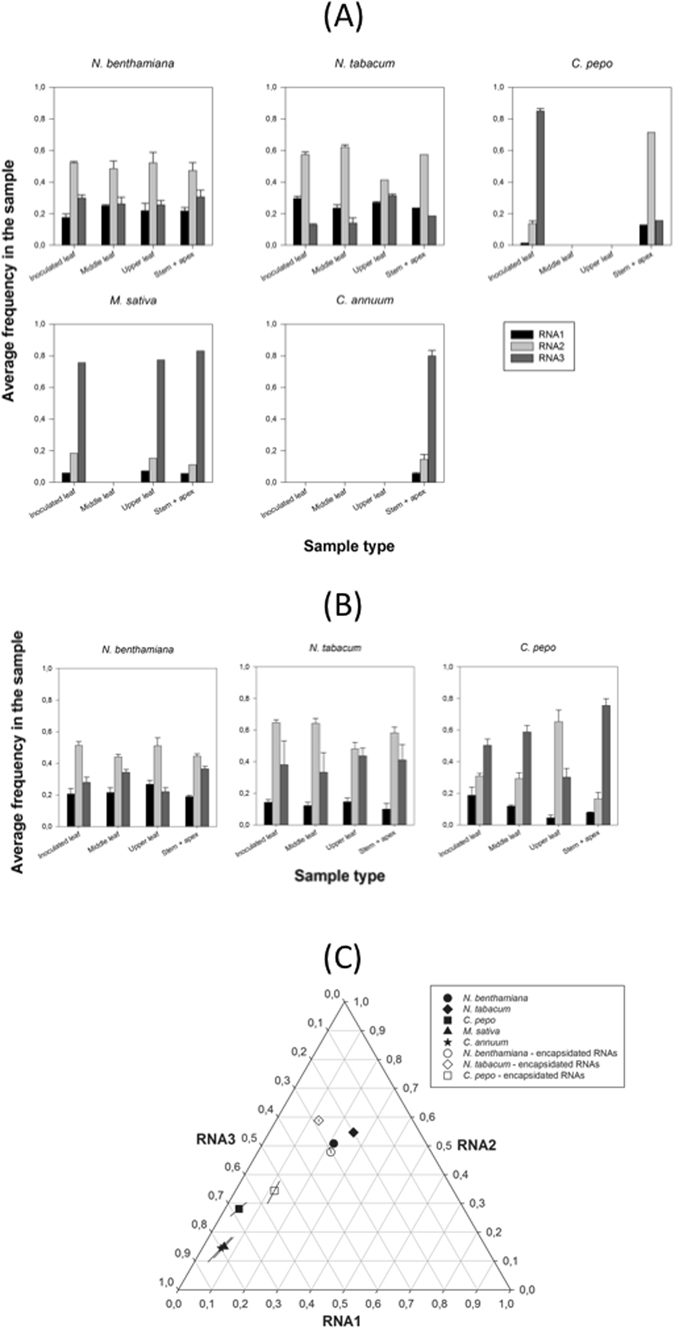
Effect of host species in the accumulation of each genomic segment of AMV. (A) Experimental determinations (by RT-qPCR) of the frequency of each AMV genomic in total RNA extractions from five different host species. Bars represent the mean of n = 3 plants per hsot species; error bars represent ±1 SEM. (B) As in (A) but determined for encapsidated RNAs. No data are available for C. annuum and M. sativa. (C) Normalized frequency ternary plot showing the effect of host species in the estimated SGF. Solid symbols represent the marginal mean frequencies estimated from total RNA samples. Open symbols represent the marginal mean frequencies estimated from virion RNA samples. Lines crossing symbols represent 95% CIs.
Table 3.
Results of the MANOVA analysis for the RNA frequencies estimated from total and from virion RNA extractions in experiments with 1:1:1 input ratio done in five different host species.
| Effect | Wilk’s Λ | F | Hypothesis df | Error df | P | Power | |
|---|---|---|---|---|---|---|---|
| (A) Total RNA | |||||||
| Intercept | 0.002 | 5932.782 | 3 | 39 | <0.001 | 1 | 0.998 |
| E | 0.004 | 61.443 | 12 | 103.456 | <0.001 | 1 | 0.842 |
| P(E) | 0.139 | 2.828 | 39 | 116.235 | <0.001 | 1 | 0.483 |
| S(E) | 0.035 | 11.953 | 21 | 112.537 | <0.001 | 1 | 0.675 |
| (P × S)(E) | 0.017 | 6.014 | 57 | 117.111 | <0.001 | 1 | 0.743 |
| (B) Virion RNA | |||||||
| Intercept | 0.007 | 1631.200 | 3 | 32 | <0.001 | 1 | 0.994 |
| E | 0.038 | 44.242 | 6 | 64 | <0.001 | 1 | 0.806 |
| P(E) | 0.373 | 2.108 | 18 | 90.995 | 0.011 | 0.957 | 0.280 |
| S(E) | 0.059 | 5.714 | 27 | 94.099 | <0.001 | 1 | 0.611 |
| (P × S)(E) | 0.215 | 1.352 | 48 | 95.970 | 0.106 | 0.976 | 0.401 |
The model fitted is shown in equation (2). E: plant species; P(E): replicate plant from species E; S(E): type of sample from a plant of a given species; and (P × S)(E): interaction term between sample type and replicate plant from species E.
Frequency data obtained from encapsidated RNAs are shown in Fig. 3B. Quantifications obtained for encapsidated RNAs from M. sativa and C. annuum were not different from negative controls and thus were not considered for the following analyses. In case of segment frequencies in the encapsidated RNAs, the only not significant factor was the interaction between plant replicate and type of sample ( term in equation (2)) (Table 3B), although highly significant differences exist among host species. In this case, the grand mean value SGF encap across hosts is 1.00:3.24:2.74 (±1 SEM: 0.00:0.71:0.97), which also differs from the stable equilibrium value found for N. benthamiana but to a lesser extent; e.g., for C. pepo 1.00:2.95:4.62 (±1 SEM: 0.12:0.54:0.44). Figure 3C (open symbols) also shows that estimates obtained from total and encapsidated RNA extractions render values that are close in the normalized ternary plot, thus showing a good correlation among them. Therefore, we can conclude that the segments ratio at 12 dpi strongly depends on the host species which is being infected, which suggests the SGF is host-species dependent.
Total RNA production is maximized at the SGFtotal
We have observed that SGF total is maintained by a FDS mechanism, and that the actual value taken by the SGF total depends on the host wherein AMV replicates. Has SGF total been optimized in each host by natural selection to maximize the total accumulation of the three genomic RNAs? To tackle this question, we have computed a partial correlation coefficient between the total RNA accumulation (summing up the accumulations of the three RNA segments) and the Euclidean distance (other multivariate distances have been tested, with identical results) from the SGF values obtained for each individual sample (i.e., a particular tissue from a given plant from each host species) and their corresponding evolutionarily stable SGF total estimates using as control variables the host species (E in equation (2)), input ratios (M in equation (1)), plant replicate (P in equation (2)) and tissue sampled (S in equation (2)). The rationale behind this test is as follows: if SGF total has been optimized to maximize the production of AMV genomic RNAs, then the farther from the evolutionarily stable SGF total equilibrium the replicating AMV population, the lower the accumulation of genomic RNAs. Conversely, the closer to the equilibrium SGF total a replicating viral population would be, the higher the accumulation of AMV RNAs. A low yet highly significant negative correlation exists between distance to the optimal SGF total and total RNA accumulation (r = −0.249, 120 d.f., 1-tailed P = 0.003), thus backing up the hypothesis that RNA accumulation is maximal at the equilibrium SGF total. Notice that this test implicitly assumes that no defective genomes are being produced and hence every RNA molecule that has been counted would eventually contribute to successfully complete the infection process. If a fraction of the accounted molecules was defective, then the test would still be valid as far as the number of defective molecules for each of the three RNAs would distribute proportionally to their rate of production. If an RNA segment produces more defective copies than others, the validity of the test would be jeopardized.
By contrast, despite being negative as expected under the above hypothesis, the correlation observed between the distance to the evolutionary stable value of SGF encap and the total amount of the three RNAs encapsidated was not significant (r = −0.133, 89 d.f., 1-tailed P = 0.105), suggesting that the strength of selection for encapsidation has been weaker than for replication or, alternatively, that selective constraints for encapsidation may differ among tissues and that by pooling together data from different tissues we have lost the statistical power to detect a significant correlation.
Transgenic expression of P1 and/or P2 proteins disrupts SGF
In the next set of experiments, we explored the effect on the segments ratio of constitutive overexpression of RNA1 and RNA2 encoding for proteins P1, P2 or both proteins by transgenes inserted in the plant. Our hypothesis is that by providing these proteins transgenically in a large excess, the SGF will be largely perturbed in a non-random manner. This last qualification is important, since we found that SGF represents an evolutionary stable equilibrium and thus small random perturbations will result in the system returning to this equilibrium. To test this hypothesis, we inoculated transgenic N. tabacum plants expressing P1, P2 or P1 and P2 (P12 plants)33 with a 1:1:1 input ratio of the three segments. As in all experiments described so far, we quantified the output segments frequencies in both total RNA extracts (data shown in Fig. 4A) and in RNA preparations from previously purified viral particles (data shown in Fig. 4C). These frequency data are then fitted to the MANOVA model shown in equation (3) in the Methods section. Table 4 shows the results of the analysis. Regardless the type of RNA sample analyzed, significant differences exist in the output frequencies among genotypes (term E in equation (3)). The magnitude of the effects associated with all factors is large (in all cases 0.821 for estimates from total RNAs extraction and 0.715 for estimates from encapsidated RNAs). The grand mean value estimated SGF total is 1.55:5.58:10.94 (±1 SEM: 0.40:1.36:9.48), which departs from the stable equilibrium described above, yet with a large excess of RNA3. Paying attention to the situation in different transgenic plants, the marginal means are 1.58:4.19:1.00 (±1 SEM: 0.01:0.04:0.03) for P1, 2.68:9.53:1.00 (±1 SEM: 0.01:0.04:0.03) for P2 and 1.00:5.10:39.36 (±1 SEM: 0.01:0.04:0.03) for P12, suggesting large departures from the equilibrium SGF total (Fig. 4B). Regarding encapsidated RNAs, the marginal SGF encap means vary even widely: 7.20:11.71:1.00 (±1 SEM: 0.01:0.03:0.02) for P1, 13.60:18.58:1.00 (±1 SEM: 0.01:0.04:0.03) for P2 and 1.00:4.08:10.40 (±1 SEM: 0.01:0.04:0.03) for P12 (Fig. 4D). The grand mean value in this case is 6.7:11.11:3.35 (±1 SEM: 2.63:2.98:2.35).
Figure 4.
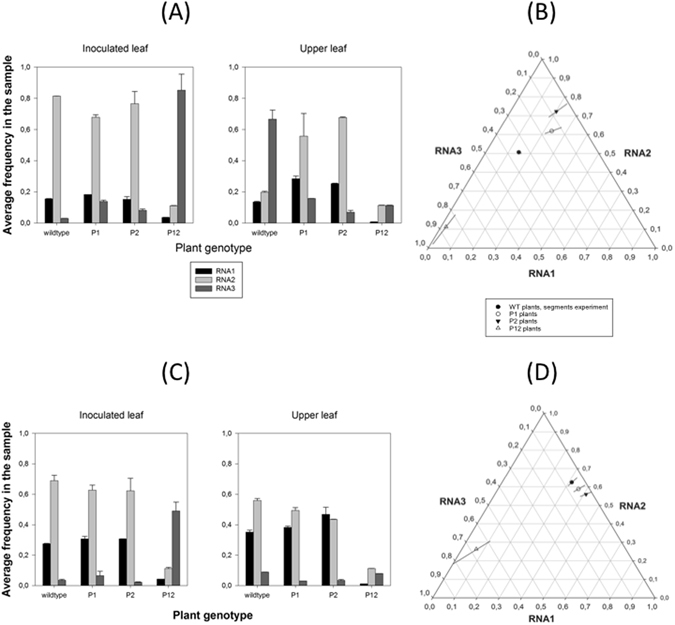
Accumulation of the three genomic segments of AMV on transgenic N. benthamiana plants that express viral proteins P1, P2 or both. (A) Experimental determinations (by RT-qPCR) of the frequency of each AMV genomic in total RNA extractions from five different host species. Bars represent the mean of n = 3 plants; error bars represent ±1 SEM. (B) Normalized frequency ternary plot showing the effect of transgenic expression of viral RNAs in the estimated SGF. (C) As in (A) but determined for encapsidated RNAs. (D) As in (B) but for encapsidated RNAs.
Table 4.
Results of the MANOVA analysis for the RNA frequencies estimated from total and from virion RNA extractions in experiments with 1:1:1 input rations done in wildtype and P1, P2 and P12 transgenic N. benthamiana.
| Effect | Wilk’s Λ | F | Hypothesis df | Error df | P | Power | |
|---|---|---|---|---|---|---|---|
| (A) Total RNA | |||||||
| Intercept | 0.002 | 1114.421 | 3 | 6 | <0.001 | 1 | 0.998 |
| E | 0.002 | 20.386 | 9 | 14.753 | <0.001 | 1 | 0.879 |
| S | 0.095 | 19.062 | 3 | 6 | 0.002 | 0.996 | 0.905 |
| E × S | 0.006 | 12.063 | 9 | 14.753 | <0.001 | 0.998 | 0.821 |
| (B) Virion RNA | |||||||
| Intercept | 0.002 | 924.673 | 3 | 6 | <0.001 | 1 | 0.998 |
| E | 0.003 | 16.705 | 9 | 14.753 | <0.001 | 1 | 0.859 |
| S | 0.081 | 22.714 | 3 | 6 | 0.001 | 1 | 0.919 |
| E × S | 0.023 | 6.064 | 9 | 14.753 | 0.001 | 0.933 | 0.715 |
The model fitted is shown in equation (3). E: N. benthamiana genotype; S: sample type; E × S: plant genotype by sample type interaction term.
We have checked whether transgenic expression of RNA1 and/or RNA2 may trigger antiviral RNAi. To do so, we have analyzed the total accumulation of RNA1 and RNA2 in infected P1, P2 and P12 plants relative to the accumulation of these two viral RNAs in wildtype plants. If RNAi is at play, then RNA accumulation in the transgenic plants would be reduced relative to the accumulation in wildtype plants. These analyses show that RNA1 accumulates, on average, 54.14% less in P1 plants, 25.34% less in P2 plants and up to 99.66% less in P12 plants than in wildtype plants. Likewise, RNA2 accumulates, on average, 76.89% in P1 plants, 50.75% less in P2 plants and 99.66% less in P12 plants than in WT plants. Notice that the accumulation of RNA2 in P2 plants is less affected than accumulation in P1 plants, which is not fully explained by the operation of RNAi itself, and other mechanisms may at work as well. Therefore, antiviral RNAi is probably at play in the transgenic plants; alas it would be hard to fully separate its effect from the effect of a non-optimal segments ratio. In any case, whether or not antiviral RNAi may be turned on in these transgenic plants prior to virus inoculation, is not invalidating our conclusion that a strong non-random perturbation of the SGF does not allow the virus to return to the equilibrium condition. At best, these results illustrate that the magnitude of the perturbation results from the contribution of two opposite forces: mRNA transcription from the transgene and RNAi-mediated degradation of these mRNAs and the viral RNAs.
In conclusion, constitutive and high expression of viral RNA components from a transgene strongly perturb the system and take it far away from its stable equilibrium, which cannot be reached again because the constant input of RNA1 and/or RNA2. It is striking that in most cases, the SGF total shows greater propensity for change than SGF encap. There are many mechanisms that could underlie this observation, although it suggests that there might be a tradeoff between within-host spread and virus particle yield per cell.
Discussion
Here, we have explored the within-host evolution of the ratio of the three genomic segments of the multipartite plant virus AMV. We found that regardless the ratio used at inoculation, an evolutionary stable equilibrium is reached in which the three RNA segments are represented in a ~1:3:2 stoichiometric ratio, in N. benthamiana. The occurrence of a stable genome-segment formula has been dubbed as the setpoint genome formula, or SGF, by Sicard et al., who described within-host evolution towards a stable composition for a multipartite DNA virus, FBNSV18. Here, we have extended this observation to a second multipartite virus, which is more representative of the vast majority of multipartite viruses by virtue of being an RNA virus. We have observed that the ratio of encapsidated segments also represents an evolutionary stable equilibrium with a very similar composition: ~1:3:2. Related to these observations, we found that the production and encapsidation of the different segments are linked in a non-trivial manner, at least in N. benthamiana plants. We speculate that RNA1 and RNA2 cooperated during replication whilst competing with RNA3 (supported by the significant negative correlations between total amounts of RNA1 and RNA2 and of RNA3), and that all three RNAs competed with each other for the CP for encapsidation (supported by the significant negative correlations between the amount of all three encapsidated RNAs). With the only exception of RNA2, no positive correlation exists between the accumulation of total and encapsidated RNAs, suggesting that the process of encapsidation is not only determined by the accumulation of the corresponding RNAs. Interestingly, negative correlations exist between the total accumulation of RNA2 and encapsidated RNA3 and vice versa but not with total and encapsidated forms of RNA1. Although these inferences on the interactions between the genome segments are based on a correlation analyses, they do reflect a biologically relevant association between viral traits. However, we have not tested the underlying mechanisms, and do not know whether there is a direct causal link between both traits or the correlation is mediated by a third yet unknown factor. Future work will explore the mechanisms of these correlations.
We found that the SGF appears to be dependent on the host species, suggesting the involvement of host factors that differ among host species play a role in its regulation. In agreement with our findings, Ni et al. also found that the relative abundances of encapsidated and total RNA segments of BMV were also dependent on the host species34. Indeed, when the ratios of the three segments were followed during the progression of infection in two monocot hosts, they converge into stable SGF total (1:2:3 for barley and 1:2:2 for wheat). However, at odds with our findings, these authors concluded that no relationship existed between the total RNAs produced and their relative encapsidation. Nonetheless, a significant correlation exists between the relative frequencies of total and encapsidated RNAs in wheat (Spearman’s ρ = 1, 2 d.f., P < 0.001) but not in barley (ρ = 0.400, 2 d.f., P = 0.600).
Multiple theories have been proposed to explain the existence of multipartite viruses, most commonly found in plants. The most recent and tantalizing proposal is that genome segmentation represents an efficient and rapidly adaptable way of regulating gene expression throughout manipulation of gene copy numbers18. As the changes in SGF observed here in alternative host species occurred within a narrow time window, multipartition might be advantageous for rapid adaptation to new hosts in a manner that is largely nucleotide-sequence independent, and therefore also mutation independent. Such an approach to adaptation could be especially advantageous in alternative hosts, where founder numbers may be small due to low infection probabilities and effective population sizes might also not be large initially due to poor replication. The hypothesis of segmentation as a mechanism to regulate gene expression has been proposed for segmented DNA viruses6, 18. Unfortunately, in the case of RNA viruses, it is not trivial to distinguish between RNA molecules that are serving as templates for replication from those that are serving as mRNAs at different stages during infection, therefore, we cannot directly test this hypothesis with our data. Additional experiments quantifying the number of RNA segments that either serve as mRNA, are encapsidated, or simply not used in any way, would be necessary for directly testing this possibility for a segmented RNA virus.
On the other hand, according to this hypothesis, a tight link must exist between the necessity of producing a given protein and the abundance of the RNA segment that encodes for it. At first glance, this hypothesis does not apply to AMV for two reasons. Firstly, as one may imagine that CP, necessary for producing infectious virions and encoded by the RNA3, would be required in larger numbers than the replicase complex, encoded by RNA1 and RNA2. However, it is important to recall at this point that CP is translated from a sgRNA4. We have not quantified the abundance of this subgenomic RNA. Interestingly, however, we observed that the ratio of RNA1 and RNA2 remains more or less constant in all experimental conditions tested (see below). Secondly, our observation of RNA2 accumulating more than RNA1 may suggest that P2 should also accumulate more than P1. Unfortunately, no quantitative data are available on the accumulation of AMV P1 and P2 in virus infected tissues. Comparing with other members of the Bromoviridae family, it has been shown that BMV and Cucumber mosaic virus (CMV) 1a protein accumulates to larger amounts than the corresponding 2a protein in purified replication complexes35–37, in spite that the SGF total for BMV was 1:2:3 for barley and 1:2:2 for wheat, suggesting that either trans elements are controlling the translation of viral RNAs or that cis-acting elements may affect the efficiency of each RNA in recruiting ribosomes. In this sense, it has been observed that AMV CP enhances the translational efficiently of viral RNAs in vivo 38 via the interaction with the 3′ termini, which adopts two alternative structures for translation (a linear array of hairpins with high affinity for CP) and replication (a pseudo-knotted structure)39. A similar mechanism has been reported as regulator for translation of the replication complex proteins of BMV40. The assumption that the amount of protein expressed is always proportional to the amount of messenger RNA, although appealing, has been proven wrong. For example, during mixed phage infections of bacterial cells, increasing the number of genomic copies results in switches between lytic and lysogenic states and the concomitant production of viral proteins41. Indeed, in such instances the regulation of is an emerging property of the structure of regulatory networks rather than directly resulting from gene copy number41. Translation efficiency and RNA stability are inexorably linked42, further challenging simple interpretations of the effects of observed RNA levels on actual protein expression levels. Indeed, it would not be surprising that RNA stability and translational productivity of the three RNA segments of AMV may be affected by the actual host species.
We found significant differences between the SGF estimated from total RNA production and from encapsidated RNA. These results may stem in part from different interactions between the different 3′ untranslated regions of the RNA segments and the CP, and their effects on encapsidation43. Furthermore, selection may operate in distinct ways here, eventually resulting in an evolutionary tradeoff. On the one hand, within-cell selection on replication will result in a SGF total that maximizes replication of the three RNA segments, likely by producing an optimal combination of RNAs and proteins, thus a segments ratio that would necessarily depart from the 1:1:1 as more proteins encoded by one segment are needed than proteins from other segments. This possibility is clearly supported by the negative correlation that we have observed between the total amount of genomic RNA produced and the distance to the SGF total. On the other hand, selection operating at the systemic movement and at the between-host levels will result in a SGF encap that maximizes the probability of successful transmission. The SGF encap values that maximize within-cell replication and transmission might be different, resulting in a tradeoff. However, our observations do not back up this possibility, as we have not observed the predicted negative correlation between total encapsidated RNAs and the distance to the SGF encap.
The mechanisms that may determine AMV’s SGF remain elusive. Probably the evolutionarily stable SGF results from complex molecular interactions between viral and host components, inextricably intertwined with viral population dynamics. Some evidences available in the literature may help to bring light into this complex question. For instance, results from transient expression experiments of proteins P1 and/or P2 revealed that replication of RNA1 and RNA2 depends on the presence of these proteins in cis and that, within infected cells, the replication of RNA1 and RNA2 is strictly coordinated through the encoded proteins rather than by RNA-RNA interactions44. This coordination may ensure the expression of proteins P1 and P2 in the correct ratio to form the replication complex. However, the replication of RNA3 is not linked to the replication of RNA1 and RNA245, 46. In this sense, these interactions explain the results reported in Table 2, namely, the negative correlation observed between replication of RNA3 and production of RNA1 and RNA2: replication of both RNA1 and RNA2 is coupled and not interfering each other, while the replication of RNA3 must use the full replicase complex in trans and, thus, competes with the replication of RNA1 and RNA2. These observations lead to the prediction that the ratio between RNA1 and RNA2 could be constant (see below) but also suggest that the different RNAs of segmented viruses could be considered as independent molecules unless they replicate coordinately, with cis elements that constraint the accumulation of the corresponding viral RNAs. Independent of whether different viral RNAs are coupled or not, we observed that providing a specific viral protein in trans (P1, P2 or P12 transgenic plants) alters the AMV SGF, and thereby probably the relationship between accumulation, virulence and transmission. It is tempting to speculate that virus resistance mediated by the expression of viral proteins –normally the CP in transgenic plants– could be at least partially due to a drastic alteration of the genome segment ratio and the negative effects thereof on viral replication.
Regarding the results obtained from hosts other than N. benthamiana, first we must acknowledge a limitation of our experimental design: as we did not consider different starting ratios or multiple time points post inoculation, one could question whether virus populations have reached a stable SGF. Conservatively speaking, we can only conclude that at advanced stages of infections in all hosts (i.e., 12 dpi), the ratio of segments significantly differs among hosts and significantly departs from the one value estimated for N. benthamiana. This being said, we observed that the ratio between RNA1 and RNA2 remains constant (ca. 1 RNA1 molecule per 3 RNA2 molecules) probably due to the coordinated replications between both RNAs, as mentioned in the previous paragraph. The coordinated replication of both RNA1 and RNA2 may determine the ratio of both viral RNAs independently of the host species. In this sense, we observed that the ratio of both viral RNAs oscillated between 1:2–1:3. Even in the highly artificial situation in which one of the replicase components was constitutively provided in trans by the plant but the other necessarily was translated from viral RNAs (P1 or P2 plants), the ratio between both RNA1 and RNA2 was maintained between 1:2 (2:4:1 for P1) and 1:3 (3:9:1 for P2). This suggests that the RNA1, that encodes for the methyltransferase-helicase P1 protein, is actually the most limiting factor in the system. Protein P1 is required to form a complex with the P2 RNA-dependent RNA polymerase protein (encoded by RNA2), acting in trans for replication and transcription of RNA 2, RNA3 and sgRNA4. However, efficient replication of RNA1 may also require translation of RNA1 into P1 protein in cis, indicating a linkage between translation and replication of this segment, as shown for BMV47, 48. Only in the case of P12 plants, the ratio was completely altered between the three RNAs, probably due to the transgenic presence of the full replicase complex in trans (P12 plants) and the incapacity of the virus to modulate the transgenes or its expression. It is interesting to note the low accumulation of RNA3 in P1 and P2 plants but not in P12 plants, which is consistent with the observation that coordinated expression of BMV proteins 1a and 2a from RNA1 and RNA2, respectively, is also required to complete the synthesis of RNA3 in transgenic yeast cells49. Apparently, the system dynamically evolves to maintain the correct ratio between RNA1 and RNA2, to the detriment of the accumulation of the RNA3. Furthermore, the accumulation of RNA3 was significantly altered depending of the host species, indicating that the virus may use this RNA to accommodate its life cycle to the presence/absence of different host factors, for instance, the transcription factor promoting salicylic-dependent defense signaling response recently reported to interact with the AMV CP50.
All three AMV RNAs contain binding sites for the CP at the 3′UTR and bind it with an equal distribution between all viral RNAs43, 51. In solution, AMV CP occurs as dimers and these dimers are the building blocks of viral capsids51. N-terminal peptides of CP bind to the 39 nucleotides of the 3′UTR RNAs in a 2:1 stoichiometric ratio52. Binding of the CP to the 3′UTR also enhances translation of viral RNAs by mimicking the function of the host poly(A)-binding protein53, 54. Altogether, these evidences point to the idea of a CP with multiple functions that are critical at different steps of the virus infectious cycle. The results obtained in the present work support the idea that the RNAs are competing for the CP, and it is therefore a limiting factor that could be used for interventions aimed at controlling virus infection. In agreement with this result, it has been recently observed that AMV CP accumulated at the nucleus and nucleolus, an observation interpreted as a mechanism to control virus expression by the cytoplasmic/nuclear balance of CP accumulation55. The sequestering of the CP at the nucleus may represent a defensive strategy to control the virus cycle, but also the accumulation of the viral RNAs containing this open reading frame.
Methods
Host species and virus inoculation
Plants from the experimental hosts C. annuum L., C. pepo L., N. benthamiana Domin, N. tabacum L. cv. Samsun, and M. sativa L. were all mechanically inoculated with 5 μL of a mixture of 5′-capped transcripts corresponding to AMV strain 425 RNA1, RNA2 and RNA3 plus a few μg of purified AMV CP as described previously56. In addition, transgenic N. tabacum plants that express AMV polymerase proteins P1 (P1 plants) and P2 (P2 plants) or both (P12 plants)33, were also assayed. For the transcription reactions, clones pUT17A, pUT27A and pAL3-NcoP3, containing full-length cDNAs of AMV RNA1, RNA2 and RNA3, respectively, were linearized with appropriate restriction enzymes and transcribed with mMESSAGE mMACHINE® T7 kit (Ambion, USA). The quantification of the AMV RNAs was performed with a ND-1000 spectrophotometer (Thermo Scientific, USA) and agarose gel eletrophoresis using an RNA ladder (RiboRuler High Range RNA Ladder 200 to 6000, Thermo Scientific) and several dilutions of the transcribed RNAs.
Before addressing the specific questions of this study, we estimated the minimal amount of AMV transcripts required to initiate an infection in the different hosts by performing serial dilutions of an initial inoculum mixture with a ratio 1:1:1. Henceforth, all ratios of AMV genomic RNA segments are given as RNA1:RNA2:RNA3 (±1 SEMs). For N. benthamiana plants we selected a final concentration of total RNAs of 40 ng/μL × 5 μL = 0.2 μg each, whereas for the rest of hosts it was necessary to increment the transcripts concentration five times (200 ng/μL × 5 μL = 1 μg total RNA).
All species were inoculated with the AMV RNAs ratio of 1:1:1 (three plants per ratio) except N. benthamiana plants that were also inoculated with ratios: 10:1:1, 1:10:1, 1:1:10, 10:10:1, 10:1:10, and 1:10:10. For each of these experiments, at least three plants were inoculated. All plants were grown in a biosafety level-2 greenhouse at 24/20 °C day/night temperature with 16 h light. After 7 dpi (N. benthamiana) or 12 dpi (rest of species), all inoculated plants were analyzed for the abundance of each RNA segment in both total RNA extraction and virus particle purification from inoculated (always the 3rd true leave), 5th and ≥ 8th leaves (Fig. 1A) and from the remaining tissues of the plants (i.e, four samples per plant).
Virus particles purification and total RNA extraction
Leaves (Fig. 1A) or entire plants were homogenized with mortar and pestle in liquid N2 to minimize the putative irregular virus distribution in the tissue. Total RNA extraction was performed using 0.1 g of tissue and the Plant RNA Isolation Mini Kit (Agilent, USA) following the manufacturer’s protocol. All samples were diluted to a final concentration of 50 ng of total RNA/µL. Virus particles purification was performed using 0.5 g of the homogenized tissue, following the protocol previously described57. The fraction of enriched virus particles was resuspended in 100 μL of PE buffer (10 mM NaH2PO4, 1 mM EDTA, pH 7.0), that was subsequently subjected to RNA extraction using the Plant RNA Isolation Mini Kit (Agilent, USA). All RNA samples were stored at −80 °C until use.
Quantification of AMV RNAs by RT-qPCR
The standard curves to quantify the AMV RNA1, RNA2 and RNA3 in the samples by RT-qPCR were prepared using known amounts of DNase-treated transcripts derived from the linearized pUT17A, pUT27A and pAL3-NcoP3 plasmids, respectively. To ensure a correct estimation of the transcripts concentration, all sample were analyzed with a ND-1000 spectrophotometer (Thermo Scientific, USA) and by agarose gel electrophoresis. To construct the standard curve for each RNA, we selected six (RNA2 and RNA3) or seven (RNA1) different viral RNAs concentrations, calculated in terms of molecules/μL (www.endmemo.com/bio/dnacopynum.php), that were generated by 5-fold serial dilutions of a starting solution containing 1010 (RNA1) or 2 × 109 (RNA2 and RNA3) molecules of the corresponding viral RNA per μL. All dilutions were made in a solution containing 50 ng/μL of total RNA extracted from healthy N. benthamiana plants.
The primers used for amplifying RNA1, RNA2 and RNA3 were designed using PrimerQuest® Design Tool version 2.2.3 (IDT Inc., USA), selecting the parameters GC% = 40–60%, T m = 57–60 °C, and size = 100–150 bp. The primers used for the RT-qPCR reactions for AMV RNA1, RNA2 and RNA3 are listed in Supplementary Table S1 online. The specificity of each primer set was confirmed with independent reactions using each one of the three RNAs as templates and with an additional reaction in which total RNA extracted from healthy plants was used as template. All these control reactions rendered negative results, except for the appropriate combination of primer and template RNA. To estimate the number of genome equivalents present and their frequencies, all data for the standard curve were first log-transformed to ascertain the range over which the response was linear. The dynamic range was limited to one dilution before the response appeared to saturate. Linear regression of the log-transformed data was then performed, rendering high values for the determination coefficient (R 2 > 0.98) and of the slope-derived amplification efficiency (90–110%). For those samples that fell within the dynamic range, the estimated linear regression parameters were used to estimate the unknown concentrations in the virus samples.
Duplicated RT-qPCR reactions were carried out in 10 μL reaction volume using the GoTaq® 1-step RT-qPCR system (SYBR® Green) (Promega, USA) and the StepOnePlus Real-Time PCR System (Applied Biosystems, USA). Each reaction contained 50 ng RNA sample, 5 μL of the 2× master mix, 10 μM of both the forward and reverse primer, 0.2 μL of GoScriptTM RT Enzyme Mix and 0.155 μL of CXR reference Dye (30 μM). The reactions were incubated at 42 °C for 15 min, followed by 95 °C during 10 min and 40 cycles of 95 °C for 10 s, 62 °C for 34 s and 72 °C for 30 s. After the RT-qPCR reaction, the melting curve stage was determined by incubating 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s. The quantification of RNAs 1, 2 and 3 copy number was calculated using the StepOne Software v.2.2.2 (Applied Biosystems, USA).
Supplementary File S2 online contains the absolute quantifications of the three RNA segments for all the experimental samples used in this study.
Statistical methods
The number of copies of RNA segment i, RNA i, on each sample were transformed into relative frequencies, f i, by dividing them by the sum of the values estimated for every RNA segment on the corresponding sample, averaged across the two technical replicates of RT-qPCR: . To analyze the effect that different inocula mixtures of the three RNA segments had on the outcome of infection, frequency data shown in Fig. 1B and C were fitted to a multivariate linear model using MANOVA techniques. The model equation fitted reads:
| 1 |
where is the vector of frequencies measured for technical replicate l ∈ {1,2} of sample S k (k ∈ {inoculated leaf (3rd true leaf), 4th leaf, 5th leaf, and the rest of the plant (stems + apical tissues)}) taken from plant replicate P j (j ∈ {1,2,3}) that was inoculated with a mixture M i (i ∈ {1:1:1, 10:1:1, 1:10:1, 1:1:10, 10:10:1, 10:1:10, 1:10:10}) of RNA segments. Factors P and S, as well as their interaction, were treated as orthogonal, and nested within factor M. measures the experimental error and was assumed to be normally distributed. is the vector of grand mean frequency values and represents a statistical estimate of the SGF. Wilk’s Λ distribution was used for the multivariate tests of each factor in the model.
To assess the magnitude of effects we used the statistic that represents the proportion of total variability attributable to a given factor while controlling for all other factors. The advantage of respect other measures of effect magnitude is that it allows for comparisons among different experimental designs. Conventionally, values of < 0.05 are considered as small, 0.05 0.15 as medium and 0.15 as large
To analyze whether SGF depends on the host species inoculated with a 1:1:1 mixture, the corresponding frequency data (Fig. 2A and B) were fitted to the following multivariate linear model:
| 2 |
In this case, factor E i represents the plant species (i ∈ {C. annuum, C. pepo, M. sativa, N. benthamiana, N. tabacum} and all other factors are as described for equation (1).
To analyze whether SGF is affected by the transgenic expression of viral proteins P1, P2 and P12 in plants inoculated with a 1:1:1 mixture, the estimated segment frequencies (shown in Fig. 4A and C) were fitted to the following multivariate linear model:
| 3 |
where E i now represents the N. benthamiana genotype and i ∈ {wildtype, P1, P2, P12} and all other factors are as described for equation (2). In this case, only one plant per genotype was assessed.
Next, we considered whether there was frequency-dependent evolution of the ratio of RNA segments infection of plants. In other words, we considered whether the frequency of one segment depends in a positive or negative manner on the abundance of the other two segments. Here, we made use of the classic population genetic approach described by Ayala & Campbell32. In short, the ratio of the j th RNA segment to its two counterparts was computed as for both the input mixture and the observed output mixture. In the absence of frequency-dependent selection (FDS), it is expected that the regression of the output on the input would be linear with slope one32. Significant deviations from the slope one relationship are taken as evidence of positive or negative FDS.
If FDS exists, then it can be evaluated whether (i) one or more equilibrium points exist and (ii) their stability. If the relationship between and is linear, a single equilibrium point exists. If the relationship is not linear, then the number of equilibria equals the number of times the best-fitting function intersects with the diagonal of the - phase diagram (i.e., the equation of slope one and intercept zero). Equilibria stability can be assessed by evaluating the value of the derivative at the corresponding equilibrium point. < 1 corresponds to a stable equilibrium in which the three segments coexist whereas > 1 corresponds to the case of a non-stable one in which the abundances of the three segments may experience changes due to very small perturbations.
In all cases, segment frequency data obtained from total RNA extractions and from virus preparations were analyzed separately. MANOVA and other statistical analyses were done using IBM SPSS version 23 (Armonk, NY, USA).
Electronic supplementary material
Acknowledgements
We thank Francisca de la Iglesia, Paula Agudo and Lorena Corachan for their dedication and expert technical assistance. This project was funded in part by grants BFU2015-65037P from the Spanish Ministry of Economy and Competitiveness-FEDER, PROMETEOII/2014/021 from Generalitat Valenciana and EvoEvo (ICT610427) from the European Commission 7th Framework Program to S.F.E. The China Scholarship Council and the Chinese Academy of Agricultural Sciences provided funding to B.W.
Author Contributions
M.P.Z., J.A.S.N. and S.F.E. designed research; B.W. and J.A.S.N. performed research; S.F.E. analyzed data; M.P.Z., J.A.S.N. and S.F.E. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05335-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Crosland WJ, Crozier RH. Myrmecia pilosula, an ant with only one pair of chromosomes. Science. 1986;231:1278. doi: 10.1126/science.231.4743.1278. [DOI] [PubMed] [Google Scholar]
- 2.Grubben, G. J. H. & Denton, O. A. Plant Resources of Tropical Africa 2. Vegetables. (PROTA Foundation/Backhuys Publishers, 2004).
- 3.Allers T, Mevarech A. Archaeal genetics – the third way. Nat. Rev. Genet. 2005;6:58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- 4.Gronenborn B. Nanoviruses: genome organization and protein function. Vet. Microbiol. 2004;98:103–109. doi: 10.1016/j.vetmic.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Jaspars EM. Plant viruses with a multipartite genome. Adv. Virus Res. 1974;19:37–149. doi: 10.1016/S0065-3527(08)60659-4. [DOI] [PubMed] [Google Scholar]
- 6.Sicard A, Michalakis Y, Gutiérrez S, Blanc S. The strange lifestyle of multipartite viruses. PLoS Pathog. 2016;12:e1005819. doi: 10.1371/journal.ppat.1005819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayakawa T, et al. Analysis of proteins encoded in the bipartite genome of a new type of parvo-like virus isolated from silkworm – structural protein with DNA polymerase motif. Virus Res. 2000;66:101–108. doi: 10.1016/S0168-1702(99)00129-X. [DOI] [PubMed] [Google Scholar]
- 8.Ladner JT, et al. A multicomponent animal virus isolated from mosquitoes. Cell Host Microb. 2016;20:357–367. doi: 10.1016/j.chom.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iranzo J, Manrubia SC. Evolutionary dynamics of genome segmentation in multipartite viruses. Proc. R. Soc. B. 2012;279:3812–3819. doi: 10.1098/rspb.2012.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escriu F, Fraile A, García-Arenal F. Constraints to genetic exchange support gene coadaptation in a tripartite RNA virus. PLoS Pathog. 2007;3:e8. doi: 10.1371/journal.ppat.0030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pressing J, Reanney DC. Divided genomes and intrinsic noise. J. Mol. Evol. 1984;20:135–146. doi: 10.1007/BF02257374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nee S. The evolution of multicompartimental genomes in viruses. J. Mol. Evol. 1987;25:277–281. doi: 10.1007/BF02603110. [DOI] [PubMed] [Google Scholar]
- 13.Chao L. Evolution of sex in RNA viruses. J. Theor. Biol. 1988;133:99–112. doi: 10.1016/S0022-5193(88)80027-4. [DOI] [PubMed] [Google Scholar]
- 14.Chao L. Levels of selection, evolution of sex in RNA viruses, and the origin of life. J. Theor. Biol. 1991;153:229–246. doi: 10.1016/S0022-5193(05)80424-2. [DOI] [PubMed] [Google Scholar]
- 15.Miralles R, Gerrish PJ, Moya A, Elena SF. Clonal interference and the evolution of RNA viruses. Science. 1999;285:1745–1747. doi: 10.1126/science.285.5434.1745. [DOI] [PubMed] [Google Scholar]
- 16.Ojosnegros S, et al. Viral genome segmentation can result from a trade-off between genetic content and particle stability. PLoS Genet. 2011;7:e1001344. doi: 10.1371/journal.pgen.1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbertson RL, Sudarshana M, Jiang H, Rojas MR, Lucas WJ. Limitations on Geminivirus genome size imposed by plasmodesmata and virus-encoded movement proteins: insights into DNA trafficking. Plant Cell. 2003;15:2578–2591. doi: 10.1105/tpc.015057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sicard A, et al. Gene copy number is differentially regulated in a multipartite virus. Nat. Commun. 2013;4:2248. doi: 10.1038/ncomms3248. [DOI] [PubMed] [Google Scholar]
- 19.Allison RF, Janda M, Ahlquist P. Infectious in vitro transcripts from Cowpea chlorotic mottle virus cDNA clones and exchange of individual RNA components with Brome mosaic virus. J. Virol. 1988;62:3581–3588. doi: 10.1128/jvi.62.10.3581-3588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahlquist P, French R, Janda M, Loesch-Fries S. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc. Natl. Acad. Sci. USA. 1984;81:7066–7070. doi: 10.1073/pnas.81.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajimorad MR, Kurath G, Randles JW, Francki RIB. Change in phenotype and encapsidated RNA segments of an isolate of Alfalfa mosaic virus: an influence of host passage. J. Gen. Virol. 1991;72:2885–2893. doi: 10.1099/0022-1317-72-12-2885. [DOI] [PubMed] [Google Scholar]
- 22.Dzianott A, Bujarski JJ. Infection and RNA recombination of Brome mosaic virus. Arabidopsis thaliana. Virology. 2004;318:482–492. doi: 10.1016/j.virol.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 23.French R, Ahlquist P. Intercistronic as well as terminal sequences are required for efficient amplification of Brome mosaic virus RNA3. J. Virol. 1987;61:1457–1465. doi: 10.1128/jvi.61.5.1457-1465.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.French R, Ahlquist P. Characterization and engineering of sequences controlling in vivo synthesis of Brome mosaic virus subgenomic RNA. J. Virol. 1988;62:2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pacha RF, Allison RF, Ahlquist P. cis-Acting sequences required for in vivo amplification of genomic RNA3 are organized differently in related bromoviruses. Virology. 1990;174:436–443. doi: 10.1016/0042-6822(90)90097-B. [DOI] [PubMed] [Google Scholar]
- 26.Traynor P, Young BM, Ahlquist P. Deletion analysis of Brome mosaic virus 2a protein: effects on RNA replication and systemic spread. J. Virol. 1991;65:2807–2815. doi: 10.1128/jvi.65.6.2807-2815.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacha RF, Ahlquist P. Use of Bromovirus RNA3 hybrids to study template specificity in viral RNA amplification. J. Virol. 1991;65:3693–3703. doi: 10.1128/jvi.65.7.3693-3703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bol JF. Alfalfa mosaic virus: coat protein-dependent initiation of infection. Mol. Plant Pathol. 2003;4:1–8. doi: 10.1046/j.1364-3703.2003.00146.x. [DOI] [PubMed] [Google Scholar]
- 29.Tromas N, Zwart MP, Lafforgue G, Elena SF. Within-host spatiotemporal dynamics of plant virus infection at the cellular level. PLoS Genet. 2014;10:e1004186. doi: 10.1371/journal.pgen.1004186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melnyk CW, Molnar A, Baulcombe DC. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011;30:3553–3563. doi: 10.1038/emboj.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuevas JM, Willemsen A, Hillung J, Zwart MP, Elena SF. Temporal dynamics of intrahost molecular evolution for a plant RNA virus. Mol. Biol. Evol. 2015;32:1132–1147. doi: 10.1093/molbev/msv028. [DOI] [PubMed] [Google Scholar]
- 32.Ayala FJ, Campbell CA. Frequency-dependent selection. Annu. Rev. Ecol. Syst. 1974;5:115–138. doi: 10.1146/annurev.es.05.110174.000555. [DOI] [Google Scholar]
- 33.Van Dun CM, Van Vloten-Doting L, Bol JF. Expression of Alfalfa mosaic virus cDNA1 and 2 in transgenic tobacco plants. Virology. 1988;163:572–578. doi: 10.1016/0042-6822(88)90298-X. [DOI] [PubMed] [Google Scholar]
- 34.Ni P, Vaughan RC, Tragesser B, Hoover H, Kao CC. The plant host can affect the encapsidation of Brome mosaic virus (BMV) RNA: BMV virions are surprisingly heterogeneous. J. Mol. Biol. 2014;426:1061–1076. doi: 10.1016/j.jmb.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes RJ, Buck KW. Complete replication of a eukaryotic virus-RNA in vitro by a purified RNA-dependent RNA-polymerase. Cell. 1990;63:363–368. doi: 10.1016/0092-8674(90)90169-F. [DOI] [PubMed] [Google Scholar]
- 36.Kao CC, Quadt R, Hershberger RP, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a form a complex in vitro. J. Virol. 1992;66:6322–6329. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quadt R, Kao CC, Browning KS, Hershberger RP, Ahlquist P. Characterization of a host protein associated with Brome mosaic virus RNA-dependent RNA-polymerase. Proc. Natl. Acad. USA. 1993;90:1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neeleman L, Olsthoorn RCL, Linthorst HJM, Bol JF. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc. Natl. Acad. Sci. USA. 2001;98:14286–14291. doi: 10.1073/pnas.251542798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsthoorn RC, Mertens S, Brederode FT, Bol JF. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 1999;18:4856–4864. doi: 10.1093/emboj/18.17.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi G, Letteney E, Kim CH, Kao CC. Brome mosaic virus capsid protein regulates accumulation of viral replication proteins by binding to the replicase assembly RNA element. RNA. 2009;15:615–626. doi: 10.1261/rna.1375509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mileyko Y, Jon RI, Weitz JS. Small-scale copy number variation and large-scale changes in gene expression. Proc. Natl. Acad. Sci. USA. 2008;105:16659–16664. doi: 10.1073/pnas.0806239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radhakrishnan A, Green R. Connections underlying translation and mRNA stability. J. Mol. Biol. 2016;428:3558–3564. doi: 10.1016/j.jmb.2016.05.025. [DOI] [PubMed] [Google Scholar]
- 43.Bol JF. Replication of alfamo- and ilarviruses: role of the coat protein. Annu. Rev. Phytopathol. 2005;43:39–62. doi: 10.1146/annurev.phyto.43.101804.120505. [DOI] [PubMed] [Google Scholar]
- 44.Van Rossum CMA, García ML, Bol JF. Accumulation of Alfalfa mosaic virus RNAs 1 and 2 requires the encoded proteins in cis. J. Virol. 1996;70:5100–5105. doi: 10.1128/jvi.70.8.5100-5105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vlot AC, Laros SM, Bol JF. Coordinate replication of Alfalfa mosaic virus RNAs 1 and 2 involves cis- and trans-acting functions of the encoded helicase-like and polymerase-like domains. J. Virol. 2003;77:10790–10798. doi: 10.1128/JVI.77.20.10790-10798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vlot AC, Bol JF. The 5′ untranslated region of Alfalfa mosaic virus RNA 1 is involved in negative-strand RNA synthesis. J. Virol. 2003;77:11284–11289. doi: 10.1128/JVI.77.20.11284-11289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz M, et al. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell. 2002;9:505–514. doi: 10.1016/S1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 48.Yi G, Kao C. cis- and trans-acting functions of Brome mosaic virus protein 1a in genomic RNA 1 replication. J. Virol. 2008;82:3045–3053. doi: 10.1128/JVI.02390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of Brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-D. [DOI] [PubMed] [Google Scholar]
- 50.Aparicio F, Pallás V. The coat protein if Alfalfa mosaic virus interacts and interferes with the transcriptional activity of the bHLH transcription factor ILR3 promoting salicylic-dependent defense signaling response. Mol. Plant Pathol. 2017;18:173–186. doi: 10.1111/mpp.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaspars, E. M. J. Interaction of Alfalfa mosaic virus nucleic acid and protein In Molecular Plant Virology (ed. Davies, J. W.) 155–225 (CRC Press, 1985).
- 52.Ansel-McKinney P, Gehrke L. RNA determinants of a specific RNA-coat protein peptide interaction in Alfalfa mosaic virus: conservation of homologous features in ilarvirus RNAs. J. Mol. Biol. 1998;278:767–785. doi: 10.1006/jmbi.1998.1656. [DOI] [PubMed] [Google Scholar]
- 53.Krab IM, Caldwell C, Gallie DR, Bol JF. Coat protein enhances translational efficiency of Alfalfa mosaic virus RNAs and interacts with the eIF4G component of initiation factor eIF4F. J. Gen. Virol. 2005;86:1841–1849. doi: 10.1099/vir.0.80796-0. [DOI] [PubMed] [Google Scholar]
- 54.Neeleman L, Linthorst HJM, Bol JF. Efficient translation of alfamovirus RNAs requires the binding of coat protein dimers to the 3′ termini of the viral RNAs. J. Gen. Virol. 2004;85:231–240. doi: 10.1099/vir.0.19581-0. [DOI] [PubMed] [Google Scholar]
- 55.Herránz MC, Pallás V, Aparicio F. Multifunctional roles for the N-terminal basic motif of Alfalfa mosaic virus coat protein: nucleolar/cytoplasmic shuttling, modulation of RNA-binding activity, and virion formation. Mol. Plant Microbe Interact. 2012;25:1093–1103. doi: 10.1094/MPMI-04-12-0079-R. [DOI] [PubMed] [Google Scholar]
- 56.Neeleman L, Bol JF. Cis-acting functions of Alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology. 1999;254:324–333. doi: 10.1006/viro.1998.9568. [DOI] [PubMed] [Google Scholar]
- 57.Van Vloten-Doting L, Jaspars EM. Uncoating of Alfalfa mosaic virus by its own RNA. Virology. 1972;48:699–708. doi: 10.1016/0042-6822(72)90154-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


