Abstract
Identifying molecular mechanisms that regulate insulin expression in bone marrow-derived mesenchymal stem cells (bmMSCs) can provide clues on how to stimulate the differentiation of bmMSCs into insulin-producing cells (IPCs), which can be used as a therapeutic approach against type 1 diabetes (T1D). As repression factors may inhibit differentiation, the efficiency of this process is insufficient for cell transplantation. In this study, we used the mouse insulin 2 (Ins2) promoter sequence and performed a DNA affinity precipitation assay combined with liquid chromatography-mass spectrometry to identify the transcription factor, chicken ovalbumin upstream promoter transcriptional factor I (COUP-TFI). Functionally, bmMSCs were reprogrammed into IPCs via COUP-TFI suppression and MafA overexpression. The differentiated cells expressed higher levels of genes specific for islet endocrine cells, and they released C-peptide and insulin in response to glucose stimulation. Transplantation of IPCs into streptozotocin-induced diabetic mice caused a reduction in hyperglycemia. Mechanistically, COUP-TFI bound to the DR1 (direct repeats with 1 spacer) element in the Ins2 promoter, thereby negatively regulating promoter activity. Taken together, the data provide a novel mechanism by which COUP-TFI acts as a negative regulator in the Ins2 promoter. The differentiation of bmMSCs into IPCs could be improved by knockdown of COUP-TFI, which may provide a novel stem cell-based therapy for T1D.
Keywords: siRNAs, differentiation, stem cell transplantation, diabetes, mesenchymal stem cells
Introduction
Type 1 diabetes (T1D) is an insulin-dependent disease, resulting from destruction of β cells of the pancreatic islets.1 Modern treatment plans for patients with T1D involve regular administration of insulin injections.2 Although other therapeutic strategies have been used in clinical practice (e.g., pancreatic islet transplantation), the lack of a cell source remains challenging.3 Recently, stem cell therapies have shown promise as treatments for neurodegenerative disorders,4 spinal cord injury,5, 6 stimulation of hematopoiesis,7, 8 and other applications. Bone marrow-derived mesenchymal stem cells (bmMSCs) can be obtained from autologous bone marrow and induced to differentiate into insulin-producing cells (IPCs) in vitro as a potential therapy for T1D.9, 10, 11 Therefore, it is critical to identify the factors that regulate the differentiation of bmMSCs into IPCs.
The transcriptional activators NeuroD/BETA2, MafA, Ngn3, and Pdx1 bind to special sites within 300–400 bp from the transcription start site of the insulin promoter and they interact synergistically.12, 13, 14, 15 Previous reports have shown that bmMSCs can be differentiated into IPCs by single or combined transfection of these transcriptional activators.10, 16, 17 However, most differentiated cells cannot secrete sufficient amounts of insulin.18 Therefore, other regulatory mechanisms may exist, which may impact the differentiation of bmMSCs into IPCs. MafA is a member of the large Maf family of transcription factors. It has been shown to be a key regulator of tissue-specific expression of insulin 2 (Ins2) in β cells, but its effect is weak unless it is coexpressed with other key transcription factors.19, 20 We hypothesize that some repressors may reduce MafA-induced Ins2 activation. Recently, many studies have shown that the transcriptional repressors also play a key role in the process of differentiation.21, 22 Several studies have suggested that the suppression of transcriptional repressors can induce mesenchymal stem cell differentiation into IPCs.23, 24 Accordingly, we hypothesize that silencing the expression of transcriptional repressors expressed in bmMSCs can improve differentiation into IPCs.
Chicken ovalbumin upstream promoter transcriptional factors (COUP-TFs) belong to the steroid/thyroid hormone receptor superfamily.25 There are two major homologs, COUP-TFI and COUP-TFII, which bind to a similar DNA binding domain.26 COUP-TFI is essential for neural,27, 28 optical,29 and cardiac development.30 It is also as an important regulator in differentiation of oligodendrocytes,31 retinal progenitor cells,32 and hair cells.33 COUP-TFI directly binds to the DR1 (direct repeats with 1 spacer) elements to repress gene expression.34 It is unclear whether the DR1 element is present in the mouse insulin promoter and/or involved in transcriptional regulation.
In this study, we have identified a DR1 element in the Ins2 promoter. COUP-TFI binds to that site to negatively regulate Ins2 expression in bmMSCs. This leads to the reprogramming of bmMSCs into IPCs by overexpressing MafA and silencing COUP-TFI.
Results
Characterization of bmMSCs
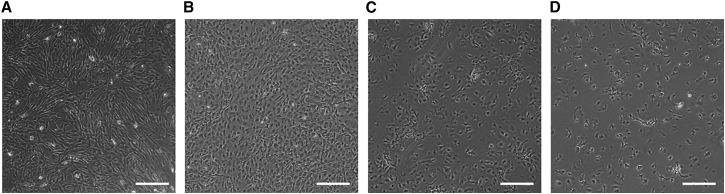
bmMSCs grew with fibroblast-like morphology (Figure 1A). To further characterize bmMSCs, flow cytometry analysis revealed that these cells expressed Sca-1, CD44, and CD29 and did not express the hematopoietic markers CD117, CD31, and CD45 (see Figure S1A). To confirm multipotency, bmMSCs were induced to differentiate into chondrocytes, osteoblasts, and adipocytes (see Figures S1B–S1D, respectively). The bmMSCs could not be induced in the absence of differentiation medium (Figures S1E–S1G). Cultured for 30 days, 60 days, and 90 days (Figures 1B–1D, respectively), the bmMSCs atrophied and changed to a polygonal shape.
Figure 1.
Growth of bmMSCs In Vitro
(A) The morphology of bmMSCs at the third passage. (B) bmMSCs were cultured in normal medium for 30 days. (C) bmMSCs were cultured in normal medium for 60 days. (D) bmMSCs were cultured in normal medium for 90 days. Scale bars represent 100 μm.
Identification of Transcription Factors in the Ins2 Promoter
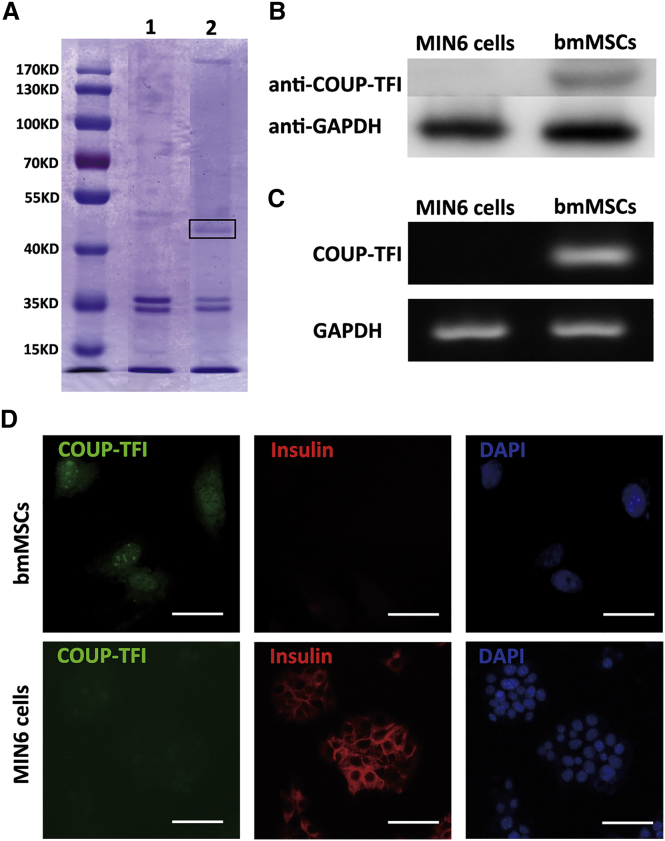
To identify novel transcription factors in the Ins2 promoter, we performed a DNA affinity precipitation assay. After precipitation, the specific band (shown inside the black box in Figure 2A) was selected and liquid chromatography (LC)-mass spectrometry (MS) analysis was performed. The data suggested that COUP-TFI was enriched by the Ins2 promoter sequence in bmMSCs (Figure 2A).
Figure 2.
COUP-TFI Is Expressed in bmMSCs and Binds to the Ins2 Promoter
(A) The DNA affinity precipitation assay was carried out with the nuclear extracts and biotinylated PCR products. The protein-DNA complexes were separated with streptavidin-labeled beads. The numbers 1 and 2 indicate nuclear extracts from MIN6 cells and from bmMSCs, respectively. (B and C) Western blotting (B) and semiquantitative PCR analysis (C) for COUP-TFI expression levels in MIN6 cells and bmMSCs. (D) MIN6 cells and bmMSCs were stained for COUP-TFI (left) and insulin (middle). Nuclei were stained with DAPI (right). Scale bars represent 50 μm.
To determine COUP-TFI expression levels, we used western blotting and semiquantitative PCR. COUP-TFI was highly expressed in bmMSCs, but not in MIN6 cells (Figures 2B and 2C). Immunofluorescence revealed that COUP-TFI was primarily localized in the nucleus of bmMSCs (Figure 2D). Considering the lack of expression of Ins2 in bmMSCs, we hypothesize that COUP-TFI act as a repressor for Ins2.
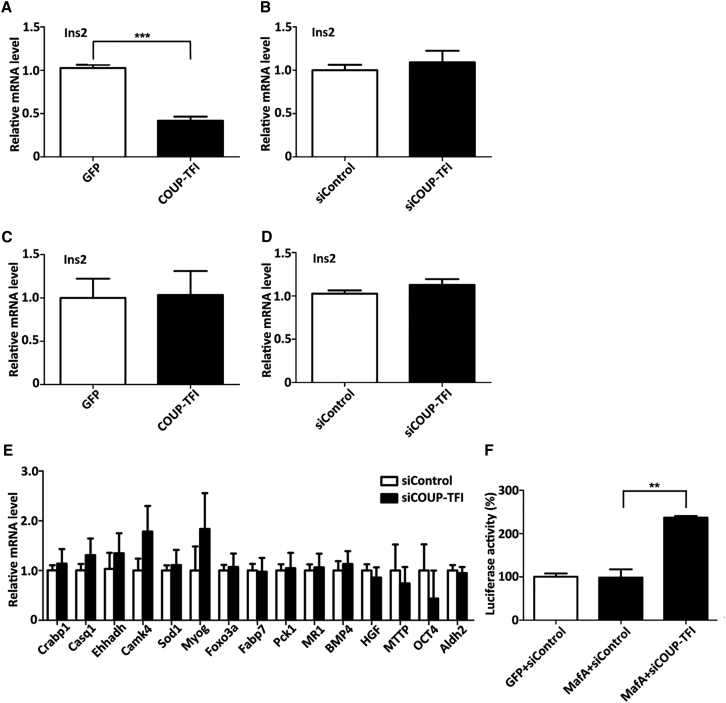
When COUP-TFI in MIN6 cells was overexpressed, we observed a robust knockdown of the endogenous Ins2 mRNA expression by qPCR (Figure 3A). COUP-TFI silencing could not affect Ins2 mRNA expression (Figure 3B). These observations indicate that COUP-TFI represses Ins2 expression in MIN6 cells. We hypothesize that bmMSCs could be induced to IPCs by repression of COUP-TFI.
Figure 3.
COUP-TFI-Mediated Transcriptional Regulation of the Ins2 Gene
(A and B) MIN6 cells were transfected with the COUP-TFI overexpression vector (A) or siCOUP-TFI (B). (C and D) bmMSCs were transfected with the COUP-TFI overexpression vector (C) or siCOUP-TFI (D). After 72 hr, Ins2 mRNA expression levels were determined by qPCR assay. (E) Expression levels of targets of COUP-TFI were determined by qPCR assay after siRNA knockdown of COUP-TFI in bmMSCs. (F) 3T3 cells were co-transfected with pGL3-Ins2 and phRL-TK in the presence or absence of MafA or siCOUP-TFI, as indicated. The results are expressed relative to the activity observed in the presence of GFP and siControl. Data are presented as means ± SD. Asterisks indicate statistical significance (n = 3, **p < 0.01, ***p < 0.001).
Differentiation of bmMSCs into IPCs
When COUP-TFI in bmMSCs was overexpressed, the expression of Ins2 was not altered by qPCR (Figure 3C). To test whether COUP-TFI silencing triggers differentiation of bmMSCs into IPCs, we knocked down the expression of COUP-TFI in bmMSCs. However, the expression of Ins2 was not elevated significantly (Figure 3D). To determine the other targets of COUP-TF1 (including Crabp1,34 Casq1,34 Ehhadh,35 Camk4,36 Sod1,34 Myog,37 Foxo3a,34 Fabp7,34 Pck1,38 MR1,39 BMP4,40 HGF,41 MTTP,42 OCT4,43 and Aldh244), we detected the change in expression levels after small interfering RNA (siRNA) knockdown of COUP-TFI in bmMSCs. The results showed that disruption of COUP-TFI did not induce global effects (Figure 3E). Thus, we hypothesize that knocking down COUP-TFI led to a state where the response to an activator of Ins2 expression might be improved.
It was previously shown that MafA can activate endogenous Ins2 in αTC cells. MafA improves the differentiation efficiency of mouse embryonic stem cells into IPCs.17 However, overexpression of MafA has no additional effect on insulin promoter activity.19 We found that when expression of COUP-TFI was knocked down in bmMSCs, Ins2 promoter activity was significantly enhanced in response to overexpression of MafA (Figure 3F).
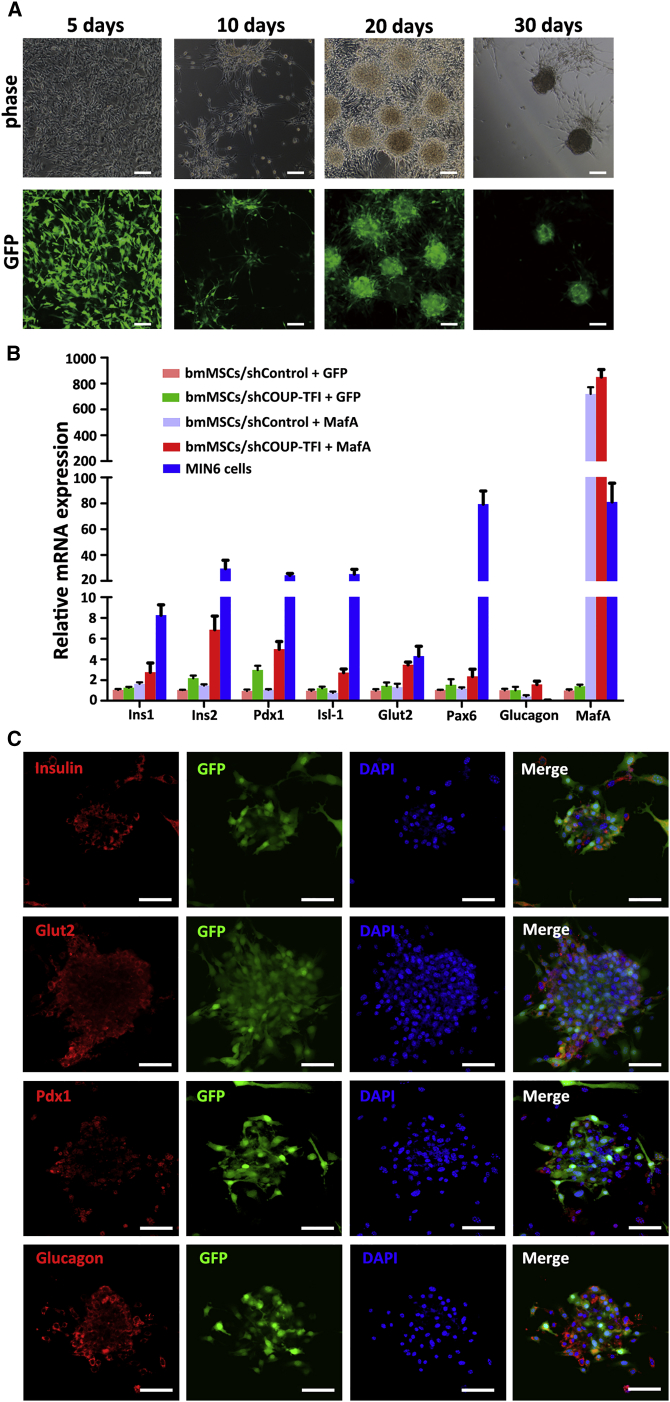
To improve the differentiation, we infected bmMSCs with lentiviral vector-mediated (Lv)-shCOUP-TFI and Lv-MafA. Along with differentiation, the shCOUP-TFI plus MafA group’s morphology was islet-like in structure (Figure 4A). The expression of several genes specific to islet endocrine cells was detected by qPCR on day 30 of differentiation. As shown in Figure 4B, the expression level of endocrine cell markers (including Ins1, Ins2 Pdx1, Isl-1, Glut2, and Pax6) in the shCOUP-TFI plus MafA group was higher than that in any other group, but lower than that in MIN6 cells. To further determine the differentiation, we performed immunofluorescence staining on the differentiated cells. The results showed that differentiated bmMSCs infected with shCOUP-TFI plus MafA expressed insulin, Glut2, Pdx1, and glucagon (Figure 4C). Together, these data show that differentiated bmMSCs can express endocrine cell markers.
Figure 4.
Differentiation of bmMSCs into IPCs
(A) Cell clusters were observed at various stages (5–30 days) during the differentiation process. The morphology of cells was observed in a light microscope (top panel) and a fluorescence microscope (bottom panel). (B) qPCR was used to detect mRNA expression levels of Ins1, Ins2, Pdx1, Isl-1, Glut2, Pax6, glucagon, and MafA. The level of mRNA in cells infected with controlled shRNA (shControl) plus GFP was defined as 1. MIN6 cells were used as a positive control. Data are presented as means ± SD from three independent experiments. (C) Immunofluorescence analysis for insulin, Glut2, Pdx1, and glucagon in differentiated bmMSCs infected with shRNA targeting COUP (shCOUP)-TFI plus MafA, which was visualized using confocal microscopy on day 30. DAPI was used for nuclear staining. Scale bars represent 50 μm.
Functional Analysis of Differentiated bmMSCs In Vitro
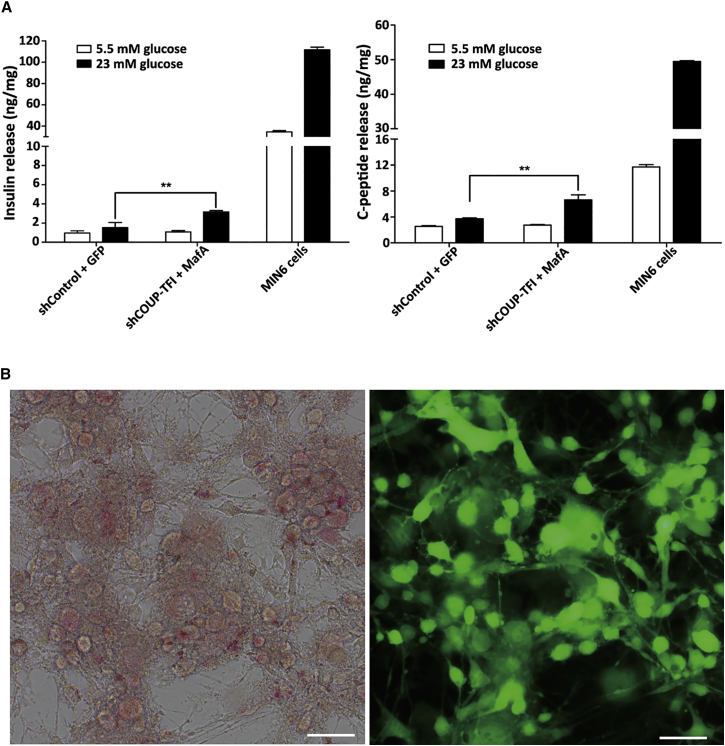
To determine the glucose responsiveness of differentiated cells, insulin and C-peptide levels were measured using ultrasensitive ELISA kits. After a 23 mmol/L glucose challenge, the cells infected with shCOUP-TFI plus MafA displayed significantly elevated levels of insulin and C-peptide compared to the control group (Figure 5A). However, the quantity of insulin and C-peptide secreted by the differentiated bmMSCs was significantly lower than that secreted by MIN6 cells.
Figure 5.
Functional Analysis of Differentiated bmMSCs In Vitro
(A) Insulin and C-peptide were determined in response to 5.5 mM and 23 mM glucose in differentiated bmMSCs infected with indicated lentivirus by ELISA. MIN6 cells were used as a positive control. Data are presented as means ± SD. Asterisks indicate statistical significance (n = 3, **p < 0.01). (B) Zinc staining in differentiated bmMSCs with dithizone. Cell clusters stained positively for zinc, as shown by distinct red staining. Cell morphology was observed under a light microscope (left) and a fluorescence microscope (right). Scale bars represent 50 μm.
Because they are known to contain zinc, pancreatic β cells can be stained with dithizone, a zinc-chelating agent.45 To verify whether the differentiated cells contained zinc, dithizone staining was performed. As shown in Figure 5B, the islet-like cell clusters were distinctly stained crimson red by dithizone. These findings suggest that differentiated bmMSCs can produce and secrete insulin and C-peptide in vitro.
Functional Analysis of Differentiated bmMSCs In Vivo
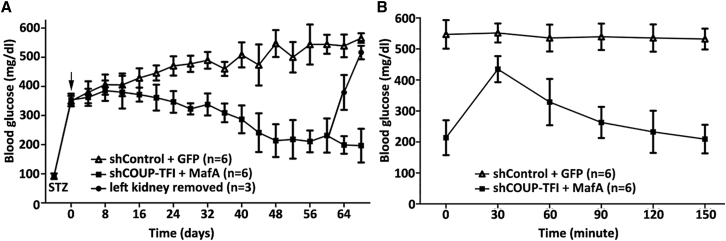
Transplantation of shCOUP-TFI- and MafA-infected bmMSCs under the left renal capsule of streptozotocin (STZ)-treated diabetic C57/bl mice resulted in a reduction of blood glucose levels, from greater than 350 mg/dL to approximately 210 mg/dL at 48 days post-transplantation. In contrast, blood glucose levels of mice that received transplanted control virus-infected bmMSCs remained elevated to greater than 500 mg/dL (Figure 6A). Glucose tolerance testing performed after 48 days post-transplantation revealed that differentiated cells were able to respond to the glucose challenge (Figure 6B). In addition, after removal of the left kidney transplanted with differentiated bmMSCs on day 60, the blood glucose levels rapidly rose to greater than 500 mg/dL (Figure 6A). These results demonstrate that IPCs can reverse hyperglycemia in STZ-treated diabetic mice.
Figure 6.
Functional Analysis of Differentiated bmMSCs In Vivo
(A) bmMSCs infected with lentivirus were transplanted into STZ-treated diabetic mice whose glucose levels had reached ≥350 mg/dL. The arrow shows the day of implantation (day 0). Glucose levels were monitored every 4 days. On day 60, the left kidney implanted with differentiated bmMSCs was removed (n = 3). Blood glucose levels are presented as means ± SD. (B) Glucose tolerance was tested on mice in 48 days after transplantation. Blood glucose levels are presented as means ± SD.
COUP-TFI Acts as a Repressor of Ins2 Expression
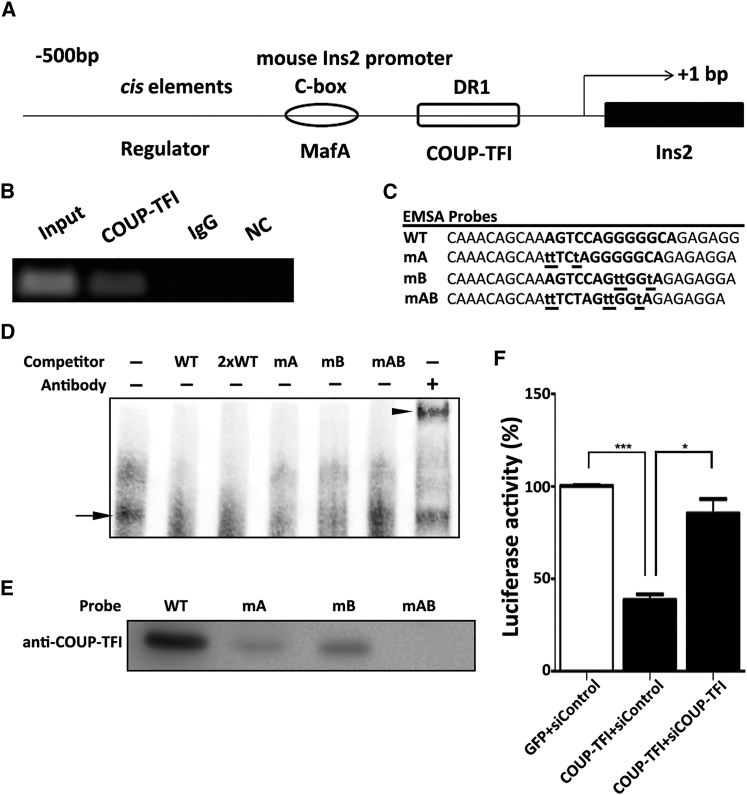
COUP-TFI recognizes consensus sequences [5′-(A/G) G (G/T) TCA-3′] and represses gene expression by directly binding to the DR1 site of the target genes. Through Internet-based sequence alignment (“Patch,” http://gene-regulation.com), we found a DR1 element located within −53 bp to −40 bp (AGGTCAgGGGGCA) of the Ins2 promoter (Figure 7A). To determine whether COUP-TFI binds to the DR1 element in the Ins2 promoter of bmMSCs, we carried out electrophoretic mobility shift assays (EMSAs) and ChIP assays.
Figure 7.
COUP-TFI Directly Binds to the Ins2 Gene Promoter
(A) Illustration of MafA, COUP-TFI, and the DR1 element in a region 500 bp downstream of the Ins2 gene promoter. (B) The ChIP assay was carried out in bmMSCs using anti-COUP-TFI antibody and non-specific IgG. The immunoprecipitated DNA fragments were amplified and the appropriately sized product was determined by electrophoresis in an agarose gel. Input indicates 1% of total DNA, IgG is normal goat serum IgG, and NC is the negative control. (C) Probe sequences used for EMSA analysis. (D) EMSA analysis was performed using nuclear extracts from bmMSCs with biotinylated double-stranded oligonucleotide probes. EMSA probes used for competition include mutant cold probes (mA, mB, and mAB) and wild-type cold probes (50-fold [WT] and 100-fold [2 × WT]) molar excess of unlabeled oligonucleotide. The supershift was performed by adding COUP-TFI antibody to the binding reactions. (E) Nuclear extracts from bmMSCs were subjected to a DNA affinity precipitation assay using biotinylated double-stranded oligonucleotide probes carrying either a WT or mutant (mA, mB, or mAB) sequence. COUP-TFI binding to the sequences was confirmed by western blotting. (F) MIN6 cells were co-transfected with pGL3-Ins2 and phRL-TK in the presence or absence of COUP-TFI and siCOUP-TFI, as indicated. The results are expressed relative to the activity observed in the presence of GFP and controlled siRNA (siControl). Data are presented as means ± SD. Asterisks indicate statistical significance (n = 3, *p < 0.05, ***p < 0.001).
Our data showed that the DR1 element was amplified in COUP-TFI-precipitated DNA samples (Figure 7B). The probe sequences for the EMSA assay are listed in Figure 7C. The specific shift complex was formed when nuclear extracts from bmMSCs were incubated with the DR1 probe alone (as indicated by a black arrow in Figure 7D). The complex was effectively competed away by either a wild-type (WT) cold probe or the half-site mutated probes (mA or mB). This competition was lost with mutation of both half-sites (mAB). These results indicated that both half-sites are necessary for binding to the DR1 site. In addition, the complex was entirely supershifted (as indicated by a black arrowhead) by the addition of COUP-TFI antibody (Figure 7D). Binding of COUP-TFI to the DR1 element was further confirmed by western blotting in the DNA affinity precipitation assay (Figure 7E). Luciferase experiments showed that the overexpression of COUP-TFI decreased Ins2 promoter activity in MIN6 cells, whereby co-transfected siRNA targeting COUP-TFI (siCOUP-TFI) could be recruited (Figure 7F).
Data from multiple assays above strongly suggest that COUP-TFI acts as a repressor by negatively regulating the Ins2 promoter activity through binding to the DR1 element.
Discussion
Notably, in our studies, COUP-TFI is identified as a negative regulator that binds to the mouse Ins2 promoter. Positive regulators have been extensively investigated, although there are few known negative regulators that repress insulin transcription in β cells. Although recent evidence indicates that COUP-TFII can bind an unrelated imperfect repeat on the rat Ins2 promoter between −55 and −38,46 it is unclear whether COUP-TFII represses rat Ins2 gene promoter transcription. To our knowledge, whether COUP-TFs bind to the mouse Ins2 promoter as a negative regulator has not yet been reported. First, we found that COUP-TFI could be enriched by using the Ins2 promoter sequence in a DNA affinity precipitation assay. Second, and importantly, we identified a DR1 element located between 53 bp and 40 bp downstream of the mouse Ins2 promoter and found that COUP-TFI bound to that site, as indicated by EMSA and ChIP experiments. Mechanistic studies revealed that COUP-TFI suppressed Ins2 expression by binding to the DR1 element in its promoter. Therefore, COUP-TFI acts as a negative regulator involved in Ins2 transcription.
MafA is known as a special activator of Ins2, but its ability to induce Ins2 expression in bmMSCs by itself is weak. We previously showed that the suppression of negative regulators and overexpression of positive regulators can generate islet-like cells from bmMSCs.24 Luciferase assays indicated that COUP-TFI overexpression could repress Ins2 promoter activity in MIN6 cells. However, in our COUP-TFI knockdown experiments, Ins2 expression was not significantly elevated in bmMSCs. Therefore, an activating factor may be necessary to stimulate Ins2 expression in bmMSCs. In our study, the Ins2 promoter was significantly activated by co-transfection with MafA and siCOUP-TFI. Our data suggest that removal of a repressor is important for the differentiation of bmMSCs into IPCs. To improve the generation of IPCs, COUP-TFI may be a novel target.
siRNAs are chemically synthesized in vitro to silence expression of endogenous target genes.47 siRNA-based therapeutics has been studied in clinical trials for treatment of cancers, viral infections, and age-related macular degeneration.48 In the current study, a siRNA targeting COUP-TFI could be a potential strategy with clinical implications. Overexpression of activators is still needed for β cell regeneration, although delivery of siRNAs in humans shows better promise than delivery of overexpression vectors, especially considering the future of siRNAs. Future studies of repressors may provide more efficient siRNAs that can be designed for T1D treatment.
Our findings demonstrate that the differentiated bmMSCs infected with shCOUP-TFI plus MafA expressed crucial β cell transcription factors and functional marker genes. The cells stained positive for insulin, Glut2, Pdx1, and glucagon. Pax6 expression was repressed in the nucleus of COUP-TFI-overexpressing ARPE-19 cells.49 Consistent with the previous study, we found that increased expression of Pax6 was present when COUP-TFI was silenced. Hepatocyte nuclear factor 4 alpha (HNF4α) has been shown to regulate the expression of pancreatic β cell genes through hepatocyte nuclear factor 1 alpha (HNF1α).50 Overexpression of HNF4α can induce reprogramming of pancreatic alpha cells to beta-like cells.51 COUP-TFI was previously shown to suppress the ability of HNF4α to activate the ALDH2 promoter.44 Dai and Hussain demonstrated that COUP-TFI suppresses synergistic activation of the MTTP promoter by HNF-4α/HNF-1α by binding to the DR1 element.42 We show that disruption of COUP-TFI did not upregulate the ALDH2 and MTTP expression in bmMSCs. It is possible that HNF-4α is absent. Abolishment of COUP-TFI could enhance the activation of HNF-4α to trigger insulin expression.
However, the secretion of insulin and C-peptide was much lower in differentiated bmMSCs than in MIN6 cells, as confirmed by ELISA. A similar situation also occurred in the expression levels of β cell transcription factors and functional marker genes between differentiated cells and MIN6 cells (Figure 4B). It is likely that these cells are not fully mature in terms of their secretory capacity in vitro. The levels of secreted insulin and C-peptide must be enhanced by suppression of not only COUP-TFI but also other negative regulators, which is a future direction of our studies. Many cell factors, including activin-A, exendin-4, and nicotinamide, have been used to stimulate differentiation and maturation in tissue-specific MSCs18, 52; however, none of these were used in our study. Interestingly, in the transplantation experiment, the differentiated bmMSCs significantly reduced the blood glucose levels of STZ-treated C57/bl mice. Our finding is consistent with previous studies,11 and this may be caused by further differentiation in vivo.
Transcriptional co-repressors that are recruited by repression domain of COUP-TFs have been shown to regulate gene silencing.29, 53 Further studies are therefore necessary to determine co-repressors that act with COUP-TFI to repress the differentiation of bmMSCs.
In conclusion, our studies indicate that COUP-TFI represses Ins2 expression in bmMSCs by binding to the DR1 element in the promoter region of Ins2. Therefore, reduced COUP-TFI expression, coupled with overexpression of MafA, can trigger differentiation of bmMSCs into IPCs. Our data indicate that repressor removal is important for the differentiation process. COUP-TFI is presented as a novel target to enhance the differentiation of bmMSCs into IPCs.
Materials and Methods
Cell Culture and Transfection
MIN6 cells were maintained in a monolayer culture on tissue culture-treated plates (Thermo Scientific) in DMEM, supplemented with GlutaMAX, glucose (4.5 g/L; Thermo Scientific), 15% (v/v) fetal bovine serum (FBS; GE Healthcare Bio-Sciences), and 70 μM β-mercaptoethanol (Thermo Scientific).
Mouse bmMSCs derived from bone marrow (Cyagen) were cultured as follows.54 For adipogenic, chondrogenic, or osteogenic differentiation, bmMSCs were grown in adipogenic, chondrogenic, or osteogenic differentiation medium (Cyagen). After the appropriate number of days for adipogenic or osteogenic differentiation, the following procedure was used according to the manufacturer’s instructions. For chondrogenic differentiation, the chondrogenic pellet was fixed in 4% paraformaldehyde for 1 hr at room temperature, embedded with optimal cutting temperature medium (Leica), and 8-μm sections were made using a cryostat microtome (Leica). The slides were rinsed once with distilled water, followed by staining with alcian blue solution (Cyagen). 30 min later, the slides were rinsed three times in water. Images were obtained using a light microscope.
3T3 or 293FT cells were cultured in DMEM supplemented with 4.5 g/L glucose and 10% FBS. The cells were passaged using standard trypsinization technique on tissue culture-treated plates.
For transient transfection, cells were seeded at 70% confluence on tissue culture-treated plates the day before transfection. siRNA (Santa Cruz) or a control non-targeting siRNA (Santa Cruz) was transfected using Lipofectamine RNAiMAX reagent (Thermo Scientific). The plasmid DNA for overexpression was transfected using Lipofectamine LTX and Plus reagent (Thermo Scientific).
Plasmid Construction
Sequences 993 bp upstream and 50 bp downstream of the transcription initiation site of the mouse Ins2 were obtained from genomic DNA using PCR. The PCR products were cloned into a pMD18-T vector (Takara) to generate pMD-1000. The mouse COUP-TFI coding sequence (CDS) was amplified by PCR from pCMV6-Entry-COUP-TFI (Origene), digested with EcoRI and NotI (NEB), and inserted into the pCDH-EF1-T2A-GFP vector (a generous gift from Dr. Sally Temple) to generate pCDH-COUP-TFI. The mouse MafA CDS was amplified from pENTR223.1-MafA (DNASU), digested with XbaI and BamHI (NEB), and inserted into the pCDH-EF1-T2A-GFP to generate pCDH-MafA.
Sequences 475 bp upstream and 25 bp downstream of the transcription initiation site of the mouse Ins2 were obtained by PCR from pMD-1000. The promoter fragments were cloned into pGL3-Basic (Promega) between MluI and XhoI sites to generate pGL3-Ins2.
The primers used for vector construction are listed in Table S1. All plasmids were verified by sequencing (Sangon Biotech).
Flow Cytometry Analysis
The bmMSCs at passage three were released by trypsinization. The cells were incubated with antibodies conjugated to phycoerythrin (PE) for CD29, CD44, Sca-1, CD117, CD31, and CD45 (BioLegend). Flow cytometry analysis was performed as previously described.55 Rat immunoglobulin G2a (IgG2a) or IgG2b conjugated to PE was used as a negative control.
RNA Analyses
Total RNA was isolated using RNAiso reagent (Takara). cDNA was synthesized using a GoScript Reverse Transcription Kit (Promega) as described by the manufacturer. Real-time qPCR was performed using SYBR Premix Ex TaqII (Takara) according to the manufacturer’s instructions. For qPCR analysis, changes in gene expression were calculated using the ΔΔCT method for relative quantification of each target gene, normalized to the housekeeper control gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Semiquantitative PCR was analyzed using DreamTaq Green PCR Master Mix (Thermo Scientific) according to the manufacturer’s protocol. PCR products were separated by agarose gel electrophoresis. Primers are listed in Table S1.
Western Blotting
Cell pellets were lysed in radio immunoprecipitation assay (RIPA) buffer. Immunoblotting procedures were performed as described previously.56 Detection was performed with an LAS-3000 imaging system (Fujifilm). Dilutions and sources of antibodies were as follows: anti-COUP-TFI (1:500; Abcam) and anti-GAPDH (1:2,000; Santa Cruz).
Preparation of Nuclear Extracts and DNA Affinity Precipitation Assay
The 5′-biotinylated primers (see Table S1) corresponding to positions 475 bp downstream and 25 bp upstream in the Ins2 promoter were synthesized by Sangon Biotech. The template used for cloning was pMD-1000. PCR products were purified using a DNA clean-up kit (Omega Bio-Tek). The nuclear extracts from cells were prepared by using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) according to the manufacturer’s instructions. The nuclear extract (200 μg) was incubated at 4°C for 4 hr with biotinylated PCR products previously coupled to streptavidin-coated magnetic beads (Dynabeads M-280; Thermo Scientific). The protein-DNA complexes were separated with a Magna GrIP Rack (Millipore), denatured in SDS buffer, and subjected to SDS-PAGE using a 10% polyacrylamide gel, followed by Coomassie blue (Sigma-Aldrich) staining.
ChIP Assays
ChIP assays were performed according to the manufacturer’s instructions (EZ-Magna ChIP A/G; Millipore). Cells were grown to 80% confluence in 15-cm tissue culture plates and were then fixed with 1% formaldehyde for 10 min. Glycine was added to a final concentration of 0.125 M. After 5 min at room temperature, cells were washed twice with PBS, collected by scraping and centrifugation, and lysed with 0.5 mL lysis buffer containing protease inhibitor cocktail (Thermo Scientific). Cell lysates were sonicated on ice using 15 cycles of 5-s pulses with 60 s between each pulse, at an amplitude of 30% using an UP50H (Dr. Hielscher) with a microtip set at 1.0 power. Following sonication, 10 μg anti-COUP-TFI antibody (Santa Cruz) and 20 μL fully suspended protein A/G magnetic beads were added to each supernatant, and the samples were incubated overnight at 4°C with rotation. As a negative control, chromatin was precipitated with 10 μg normal goat IgG (Santa Cruz). Protein A/G magnetic beads were pelleted with a magnetic separator. Protein-DNA complexes were eluted from the beads and reverse cross-linked for 2 hr at 62°C. DNA was purified using spin columns (Millipore). Purified DNA was amplified using primers targeting the DR1 element and a region 2 kb downstream of the DR1 element as a negative control. PCR reactions were carried out and the appropriately size product was determined by electrophoresis in an agarose gel. Primers are listed in Table S1.
EMSA and Supershift Analyses
The electrophoretic mobility shift assay (EMSA) was carried out using double-stranded DNA probes pre-labeled with biotin (Sangon Biotech). Probes were incubated at room temperature for 30 min with 10 μg nuclear extract from cells in binding buffer with 1 μg poly(dI/dC). Supershift analyses were performed by adding 1 μg of the appropriate antibody after the initial incubation period for 45 min on ice before electrophoresis. Binding reactions were loaded onto 5% native polyacrylamide gels and run for 2 hr in 0.5× Tris-borate-EDTA (TBE). Binding reactions were transferred to a nylon membrane and cross-linked at a distance of approximately 0.5 cm from the membrane for 5–10 min with a hand-held UV lamp equipped with 254-nm bulbs. Blots were developed using enhanced chemiluminescence (ECL) SuperSignal West Pico chemiluminescent reagents (Thermo Scientific) and were visualized with an LAS-3000 imaging system (Fujifilm).
Immunofluorescent Staining
Cells were fixed with 4% formaldehyde at room temperature for 30 min and then treated with 0.25% Triton X-100 in PBS for 10 min. After blocking with 10% normal donkey serum containing 1% BSA in PBS for 30 min, cells were incubated with primary antibodies against COUP-TFI (1:200; Abcam), insulin (1:200; Cell Signaling Technology), Glut2 (1:200; Abcam) Pdx1 (1:1,000; Abcam), or glucagon (1:200; Santa Cruz) at 4°C overnight. Suitable fluorescent-labeled secondary antibodies were used for detection (Thermo Scientific). Nuclei were stained with DAPI (1 μg/mL; Roche). Images were acquired by fluorescence microscopy.
Luciferase Assays
Cells were seeded in 24-well plates and co-transfected the following day with 0.1 μg siRNA and 0.2 μg plasmid DNA using TransMessenger Transfection Reagent (QIAGEN). The medium was changed to normal growth medium 24 hr after the transfection. For the luciferase reporter assay, 500 ng pGL3-Ins2 vector and 20 ng phRL-TK vector (Promega) were added to each well for all transfections. The relative luciferase activities (ratios of firefly and Renilla luciferase activity) of lysates were measured using the Dual Luciferase Reporter Assay System (Promega).
Lentivirus Generation
Lentivirus was generated as previously described.57 Briefly, constructs of pCDH-MafA were co-transfected with pCMV-VSVG and pCMV-dvpr (a generous gift from Dr. Sally Temple) into 293FT cells. The supernatants were collected at 48 hr and 72 hr after transfection. Lentivirus was further concentrated by ultra-centrifugation (Beckman Coulter). Lentivirus containing either MafA CDS or the empty vector was termed Lv-MafA or Lv-GFP, respectively. The lentivirus for small hairpin RNA (shRNA) knockdown against COUP-TFI was designed and produced by GeneChem. The target sequence was 5′-GCAGTTTCAACTGGCCTTA-3′. Subsequently, the viral titer was determined using fluorescence-activated cell sorting. bmMSCs infected with lentivirus were seeded in six-well plates at a density of 1 × 104 cells/cm2 and cultured with DMEM containing 2% FBS.
Measurement of Insulin and C-Peptide Secretion by ELISA
Cells were switched to serum-free medium containing 0.5% BSA for 12 hr, washed twice with PBS, and then preincubated with Krebs-Ringer bicarbonate (KRB) buffer for 1 hr, followed by KRB buffer containing 5.5 or 23 mM glucose for 1 hr at 37°C. The supernatant was collected for ELISA (R&D Systems) according to the manufacturer’s instructions. The total protein content was measured by using a bicinchoninic acid assay kit (Thermo Scientific).
Dithizone Staining
Dithizone (Sigma-Aldrich) staining was performed by adding 10 μL stock dithizone (10 mg/mL) to 1 mL culture medium. The staining solution was filtered using a 0.22-μm filter and then added to culture dishes. Following the addition of dithizone, the dishes were incubated at 37°C for 30 min. The dishes were then washed three times with PBS. The stained cells were examined microscopically.
Cell Transplantation
All animal experiments were approved by the Institutional Animal Care and Use Committee of China Medical University in accordance with protocols. Male C57/bl mice received a single intraperitoneal injection of 200 mg STZ/kg body weight according to published procedures, with minor modifications.58 Blood glucose levels were monitored using an AccuChek glucometer (Roche Diagnostics). Within 12 days of injection, all C57/bl mice became hyperglycemic, with blood glucose levels >350 mg/dL, and then 3 × 106 cells were transplanted under the left renal capsule of diabetic mice. Blood glucose levels were monitored every 4 days following transplantation. For the glucose tolerance test, mice were fasted for 6–10 hr and then injected with 1 mg glucose in saline/g body weight. Blood glucose levels were detected at the indicated time points in samples obtained from the tail vein.
Statistical Analysis
All data are presented as means ± SD. Statistical significance was assessed with Student’s t test using GraphPad Prism 5.0 software (GraphPad Software).
Author Contributions
T.Z. and X.-H.L. performed the experiments and wrote the paper; D.-B.Z. provided study materials; X.-Y.L. and F.Z. contributed to data analysis and interpretation; X.-W.L., R.W., and H.-X.L. contributed to collection and assembly of data; X.-N. P. designed the study and provided final approval of manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (2012CB518103), the National Natural Science Foundation of China (81370883), the Doctoral Fund of the Ministry of Education of China (20132104110020), and the Shenyang Key Laboratory Project (F15-157-1-00). We thank Dr. Sally Temple for providing pCDH-EF1-T2A-GFP, pCMV-VSVG, and pCMV-dvpr vectors.
Footnotes
Supplemental Information includes one figure and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.06.016.
Supplemental Information
References
- 1.Atkinson M.A., Eisenbarth G.S. Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet. 2001;358:221–229. doi: 10.1016/S0140-6736(01)05415-0. [DOI] [PubMed] [Google Scholar]
- 2.Bluestone J.A., Herold K., Eisenbarth G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464:1293–1300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mineo D., Pileggi A., Alejandro R., Ricordi C. Point: steady progress and current challenges in clinical islet transplantation. Diabetes Care. 2009;32:1563–1569. doi: 10.2337/dc09-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Androutsellis-Theotokis A., Rueger M.A., Park D.M., Mkhikian H., Korb E., Poser S.W., Walbridge S., Munasinghe J., Koretsky A.P., Lonser R.R., McKay R.D. Targeting neural precursors in the adult brain rescues injured dopamine neurons. Proc. Natl. Acad. Sci. USA. 2009;106:13570–13575. doi: 10.1073/pnas.0905125106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salewski R.P., Mitchell R.A., Shen C., Fehlings M.G. Transplantation of neural stem cells clonally derived from embryonic stem cells promotes recovery after murine spinal cord injury. Stem Cells Dev. 2015;24:36–50. doi: 10.1089/scd.2014.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gómez-Villafuertes R., Rodríguez-Jiménez F.J., Alastrue-Agudo A., Stojkovic M., Miras-Portugal M.T., Moreno-Manzano V. Purinergic receptors in spinal cord-derived ependymal stem/progenitor cells and their potential role in cell-based therapy for spinal cord injury. Cell Transplant. 2015;24:1493–1509. doi: 10.3727/096368914X682828. [DOI] [PubMed] [Google Scholar]
- 7.Miura Y. Regulation of hematopoiesis by mesenchymal stem cells. Rinsho Ketsueki. 2013;54:431–435. [PubMed] [Google Scholar]
- 8.Laurenti E., Doulatov S., Zandi S., Plumb I., Chen J., April C., Fan J.B., Dick J.E. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat. Immunol. 2013;14:756–763. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanini C., Bruno S., Mandili G., Baci D., Cerutti F., Cenacchi G., Izzi L., Camussi G., Forni M. Differentiation of mesenchymal stem cells derived from pancreatic islets and bone marrow into islet-like cell phenotype. PLoS ONE. 2011;6:e28175. doi: 10.1371/journal.pone.0028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Limbert C., Päth G., Ebert R., Rothhammer V., Kassem M., Jakob F., Seufert J. PDX1- and NGN3-mediated in vitro reprogramming of human bone marrow-derived mesenchymal stromal cells into pancreatic endocrine lineages. Cytotherapy. 2011;13:802–813. doi: 10.3109/14653249.2011.571248. [DOI] [PubMed] [Google Scholar]
- 11.Karnieli O., Izhar-Prato Y., Bulvik S., Efrat S. Generation of insulin-producing cells from human bone marrow mesenchymal stem cells by genetic manipulation. Stem Cells. 2007;25:2837–2844. doi: 10.1634/stemcells.2007-0164. [DOI] [PubMed] [Google Scholar]
- 12.Docherty H.M., Hay C.W., Ferguson L.A., Barrow J., Durward E., Docherty K. Relative contribution of PDX-1, MafA and E47/beta2 to the regulation of the human insulin promoter. Biochem. J. 2005;389:813–820. doi: 10.1042/BJ20041891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glick E., Leshkowitz D., Walker M.D. Transcription factor BETA2 acts cooperatively with E2A and PDX1 to activate the insulin gene promoter. J. Biol. Chem. 2000;275:2199–2204. doi: 10.1074/jbc.275.3.2199. [DOI] [PubMed] [Google Scholar]
- 14.Kaneto H., Matsuoka T.A., Kawashima S., Yamamoto K., Kato K., Miyatsuka T., Katakami N., Matsuhisa M. Role of MafA in pancreatic beta-cells. Adv. Drug Deliv. Rev. 2009;61:489–496. doi: 10.1016/j.addr.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 15.Hay C.W., Docherty K. Comparative analysis of insulin gene promoters: implications for diabetes research. Diabetes. 2006;55:3201–3213. doi: 10.2337/db06-0788. [DOI] [PubMed] [Google Scholar]
- 16.Yuan H., Li J., Xin N., Zhao Z., Qin G. Expression of Pdx1 mediates differentiation from mesenchymal stem cells into insulin-producing cells. Mol. Biol. Rep. 2010;37:4023–4031. doi: 10.1007/s11033-010-0061-y. [DOI] [PubMed] [Google Scholar]
- 17.Xu H., Tsang K.S., Chan J.C., Yuan P., Fan R., Kaneto H., Xu G. The combined expression of Pdx1 and MafA with either Ngn3 or NeuroD improves the differentiation efficiency of mouse embryonic stem cells into insulin-producing cells. Cell Transplant. 2013;22:147–158. doi: 10.3727/096368912X653057. [DOI] [PubMed] [Google Scholar]
- 18.Li L., Li F., Qi H., Feng G., Yuan K., Deng H., Zhou H. Coexpression of Pdx1 and betacellulin in mesenchymal stem cells could promote the differentiation of nestin-positive epithelium-like progenitors and pancreatic islet-like spheroids. Stem Cells Dev. 2008;17:815–823. doi: 10.1089/scd.2008.0060. [DOI] [PubMed] [Google Scholar]
- 19.Matsuoka T.A., Kaneto H., Miyatsuka T., Yamamoto T., Yamamoto K., Kato K., Shimomura I., Stein R., Matsuhisa M. Regulation of MafA expression in pancreatic beta-cells in db/db mice with diabetes. Diabetes. 2010;59:1709–1720. doi: 10.2337/db08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishizawa M., Kataoka K., Vogt P.K. MafA has strong cell transforming ability but is a weak transactivator. Oncogene. 2003;22:7882–7890. doi: 10.1038/sj.onc.1206526. [DOI] [PubMed] [Google Scholar]
- 21.Nechiporuk T., McGann J., Mullendorff K., Hsieh J., Wurst W., Floss T., Mandel G. The REST remodeling complex protects genomic integrity during embryonic neurogenesis. eLife. 2016;5:e09584. doi: 10.7554/eLife.09584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaarenstroom T., Hill C.S. TGF-β signaling to chromatin: how Smads regulate transcription during self-renewal and differentiation. Semin. Cell Dev. Biol. 2014;32:107–118. doi: 10.1016/j.semcdb.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Li B., Wang S., Liu H., Liu D., Zhang J., Zhang B., Yao H., Lv Y., Wang R., Chen L. Neuronal restrictive silencing factor silencing induces human amniotic fluid-derived stem cells differentiation into insulin-producing cells. Stem Cells Dev. 2011;20:1223–1231. doi: 10.1089/scd.2010.0195. [DOI] [PubMed] [Google Scholar]
- 24.Li H.T., Jiang F.X., Shi P., Zhang T., Liu X.Y., Lin X.W., Pang X.N. In vitro reprogramming of rat bone marrow-derived mesenchymal stem cells into insulin-producing cells by genetically manipulating negative and positive regulators. Biochem. Biophys. Res. Commun. 2012;420:793–798. doi: 10.1016/j.bbrc.2012.03.076. [DOI] [PubMed] [Google Scholar]
- 25.Wang L.H., Tsai S.Y., Cook R.G., Beattie W.G., Tsai M.J., O’Malley B.W. COUP transcription factor is a member of the steroid receptor superfamily. Nature. 1989;340:163–166. doi: 10.1038/340163a0. [DOI] [PubMed] [Google Scholar]
- 26.Qiu Y., Tsai S.Y., Tsai M.J. COUP-TF an orphan member of the steroid/thyroid hormone receptor superfamily. Trends Endocrinol. Metab. 1994;5:234–239. doi: 10.1016/1043-2760(94)p3081-h. [DOI] [PubMed] [Google Scholar]
- 27.Zhou C., Qiu Y., Pereira F.A., Crair M.C., Tsai S.Y., Tsai M.J. The nuclear orphan receptor COUP-TFI is required for differentiation of subplate neurons and guidance of thalamocortical axons. Neuron. 1999;24:847–859. doi: 10.1016/s0896-6273(00)81032-6. [DOI] [PubMed] [Google Scholar]
- 28.Tomassy G.S., De Leonibus E., Jabaudon D., Lodato S., Alfano C., Mele A., Macklis J.D., Studer M. Area-specific temporal control of corticospinal motor neuron differentiation by COUP-TFI. Proc. Natl. Acad. Sci. USA. 2010;107:3576–3581. doi: 10.1073/pnas.0911792107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang K., Tsai S.Y., Tsai M.J. COUP-TFs and eye development. Biochim. Biophys. Acta. 2015;1849:201–209. doi: 10.1016/j.bbagrm.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo L., Lynch J., Nakamura K., Fliegel L., Kasahara H., Izumo S., Komuro I., Agellon L.B., Michalak M. COUP-TF1 antagonizes Nkx2.5-mediated activation of the calreticulin gene during cardiac development. J. Biol. Chem. 2001;276:2797–2801. doi: 10.1074/jbc.C000822200. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi H., Zhou C., Lin S.C., Durand B., Tsai S.Y., Tsai M.J. The nuclear orphan receptor COUP-TFI is important for differentiation of oligodendrocytes. Dev. Biol. 2004;266:238–251. doi: 10.1016/j.ydbio.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 32.Inoue M., Iida A., Satoh S., Kodama T., Watanabe S. COUP-TFI and -TFII nuclear receptors are expressed in amacrine cells and play roles in regulating the differentiation of retinal progenitor cells. Exp. Eye Res. 2010;90:49–56. doi: 10.1016/j.exer.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 33.Tang L.S., Alger H.M., Pereira F.A. COUP-TFI controls Notch regulation of hair cell and support cell differentiation. Development. 2006;133:3683–3693. doi: 10.1242/dev.02536. [DOI] [PubMed] [Google Scholar]
- 34.Montemayor C., Montemayor O.A., Ridgeway A., Lin F., Wheeler D.A., Pletcher S.D., Pereira F.A. Genome-wide analysis of binding sites and direct target genes of the orphan nuclear receptor NR2F1/COUP-TFI. PLoS ONE. 2010;5:e8910. doi: 10.1371/journal.pone.0008910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcus S.L., Capone J.P., Rachubinski R.A. Identification of COUP-TFII as a peroxisome proliferator response element binding factor using genetic selection in yeast: COUP-TFII activates transcription in yeast but antagonizes PPAR signaling in mammalian cells. Mol. Cell. Endocrinol. 1996;120:31–39. doi: 10.1016/0303-7207(96)03813-0. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y.Y., Brent G.A. A complex deoxyribonucleic acid response element in the rat Ca(2+)/calmodulin-dependent protein kinase IV gene 5′-flanking region mediates thyroid hormone induction and chicken ovalbumin upstream promoter transcription factor 1 repression. Mol. Endocrinol. 2002;16:2439–2451. doi: 10.1210/me.2001-0324. [DOI] [PubMed] [Google Scholar]
- 37.Muscat G.E., Rea S., Downes M. Identification of a regulatory function for an orphan receptor in muscle: COUP-TF II affects the expression of the myoD gene family during myogenesis. Nucleic Acids Res. 1995;23:1311–1318. doi: 10.1093/nar/23.8.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beale E.G., Forest C., Hammer R.E. Regulation of cytosolic phosphoenolpyruvate carboxykinase gene expression in adipocytes. Biochimie. 2003;85:1207–1211. doi: 10.1016/j.biochi.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Zhao B., Hou S., Ricciardi R.P. Chromatin repression by COUP-TFII and HDAC dominates activation by NF-kappaB in regulating major histocompatibility complex class I transcription in adenovirus tumorigenic cells. Virology. 2003;306:68–76. doi: 10.1016/s0042-6822(02)00079-x. [DOI] [PubMed] [Google Scholar]
- 40.Feng J.Q., Chen D., Cooney A.J., Tsai M.J., Harris M.A., Tsai S.Y., Feng M., Mundy G.R., Harris S.E. The mouse bone morphogenetic protein-4 gene. Analysis of promoter utilization in fetal rat calvarial osteoblasts and regulation by COUP-TFI orphan receptor. J. Biol. Chem. 1995;270:28364–28373. doi: 10.1074/jbc.270.47.28364. [DOI] [PubMed] [Google Scholar]
- 41.Jiang J.G., Bell A., Liu Y., Zarnegar R. Transcriptional regulation of the hepatocyte growth factor gene by the nuclear receptors chicken ovalbumin upstream promoter transcription factor and estrogen receptor. J. Biol. Chem. 1997;272:3928–3934. doi: 10.1074/jbc.272.7.3928. [DOI] [PubMed] [Google Scholar]
- 42.Dai K., Hussain M.M. NR2F1 disrupts synergistic activation of the MTTP gene transcription by HNF-4α and HNF-1α. J. Lipid Res. 2012;53:901–908. doi: 10.1194/jlr.M025130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben-Shushan E., Sharir H., Pikarsky E., Bergman Y. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct-3/4 expression in embryonal carcinoma cells. Mol. Cell. Biol. 1995;15:1034–1048. doi: 10.1128/mcb.15.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.You M., Fischer M., Cho W.K., Crabb D. Transcriptional control of the human aldehyde dehydrogenase 2 promoter by hepatocyte nuclear factor 4: inhibition by cyclic AMP and COUP transcription factors. Arch. Biochem. Biophys. 2002;398:79–86. doi: 10.1006/abbi.2001.2713. [DOI] [PubMed] [Google Scholar]
- 45.Shiroi A., Yoshikawa M., Yokota H., Fukui H., Ishizaka S., Tatsumi K., Takahashi Y. Identification of insulin-producing cells derived from embryonic stem cells by zinc-chelating dithizone. Stem Cells. 2002;20:284–292. doi: 10.1634/stemcells.20-4-284. [DOI] [PubMed] [Google Scholar]
- 46.Hwung Y.P., Crowe D.T., Wang L.H., Tsai S.Y., Tsai M.J. The COUP transcription factor binds to an upstream promoter element of the rat insulin II gene. Mol. Cell. Biol. 1988;8:2070–2077. doi: 10.1128/mcb.8.5.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novina C.D., Sharp P.A. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 48.Ozcan G., Ozpolat B., Coleman R.L., Sood A.K., Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv. Drug Deliv. Rev. 2015;87:108–119. doi: 10.1016/j.addr.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang K., Xie X., Park J.I., Jamrich M., Tsai S., Tsai M.J. COUP-TFs regulate eye development by controlling factors essential for optic vesicle morphogenesis. Development. 2010;137:725–734. doi: 10.1242/dev.040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H., Maechler P., Antinozzi P.A., Hagenfeldt K.A., Wollheim C.B. Hepatocyte nuclear factor 4alpha regulates the expression of pancreatic beta -cell genes implicated in glucose metabolism and nutrient-induced insulin secretion. J. Biol. Chem. 2000;275:35953–35959. doi: 10.1074/jbc.M006612200. [DOI] [PubMed] [Google Scholar]
- 51.Sangan C.B., Jover R., Heimberg H., Tosh D. In vitro reprogramming of pancreatic alpha cells towards a beta cell phenotype following ectopic HNF4α expression. Mol. Cell. Endocrinol. 2015;399:50–59. doi: 10.1016/j.mce.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 52.Kassem D.H., Kamal M.M., El-Kholy Ael.-L., El-Mesallamy H.O. Exendin-4 enhances the differentiation of Wharton’s jelly mesenchymal stem cells into insulin-producing cells through activation of various β-cell markers. Stem Cell Res. Ther. 2016;7:108. doi: 10.1186/s13287-016-0374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie X., Tang K., Yu C.T., Tsai S.Y., Tsai M.J. Regulatory potential of COUP-TFs in development: stem/progenitor cells. Semin. Cell Dev. Biol. 2013;24:687–693. doi: 10.1016/j.semcdb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin X., Wu L., Zhang Z., Yang R., Guan Q., Hou X., Wu Q. MiR-335-5p promotes chondrogenesis in mouse mesenchymal stem cells and is regulated through two positive feedback loops. J. Bone Miner. Res. 2014;29:1575–1585. doi: 10.1002/jbmr.2163. [DOI] [PubMed] [Google Scholar]
- 55.Li S.W., Tang D., Ahrens K.P., She J.X., Braylan R.C., Yang L. All-trans-retinoic acid induces CD52 expression in acute promyelocytic leukemia. Blood. 2003;101:1977–1980. doi: 10.1182/blood-2002-05-1426. [DOI] [PubMed] [Google Scholar]
- 56.Karbiener M., Pisani D.F., Frontini A., Oberreiter L.M., Lang E., Vegiopoulos A., Mössenböck K., Bernhardt G.A., Mayr T., Hildner F. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells. 2014;32:1578–1590. doi: 10.1002/stem.1603. [DOI] [PubMed] [Google Scholar]
- 57.Phoenix T.N., Temple S. Spred1, a negative regulator of Ras-MAPK-ERK, is enriched in CNS germinal zones, dampens NSC proliferation, and maintains ventricular zone structure. Genes Dev. 2010;24:45–56. doi: 10.1101/gad.1839510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soria B., Roche E., Berná G., León-Quinto T., Reig J.A., Martín F. Insulin-secreting cells derived from embryonic stem cells normalize glycemia in streptozotocin-induced diabetic mice. Diabetes. 2000;49:157–162. doi: 10.2337/diabetes.49.2.157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.