Abstract
Aeromonas spp. are ubiquitous bacteria primarily recovered from aquatic ecosystems. They are found in fresh water as well as estuarine and marine waters, and in association with numerous autochthonous aquatic organisms in these environments. However, aeromonads are also etiologic agents of fish diseases and are now recognized as emerging pathogens in humans. The estuary is therefore a key environment, harboring autochthonous aeromonads, and aeromonads originating from humans and animals, mainly released by treated WWTP effluent or watershed run-off via tributaries. The present study compares the abundance and the diversity of Aeromonas populations. Over 2 years of monitoring (eight campaigns from February 2013 to November 2015), the occurrence of Aeromonas was investigated within the water column (water and fluid mud) and in association with copepods. Moreover, the diversity of Aeromonas populations was ascertained by analyzing gyrB and radA sequences, and the antibiotic-resistance phenotypes were determined using the disk diffusion method. This study shows, for the first time, the presence of Aeromonas spp. in water (1.1 × 102 to 1.2 ± 0.3 × 103 CFU.100 mL-1), fluid mud (2.6 ± 2.6 × 102 to 9.8 ± 0.9 × 103 CFU.g-1) and in association with living copepods (1.9 ± 0.7 × 102 to >1.1 × 104 CFU.g-1) in the Seine estuary. Moreover, the diversity study, conducted on 36 strains isolated from the water column and 47 strains isolated from copepods, indicates distinct populations within these two compartments. Strains distributed in five clusters corresponding to A. bestiarum (n = 6; 5.45%), A. encheleia (n = 1; 0.91%), A. media (n = 22; 20.0%), A. rivipollensis (n = 34; 30.91%) and A. salmonicida (n = 47; 42.73%). A. salmonicida is the most abundant species associated with Eurytemora affinis (n = 35; 74.47%). In contrast, A. salmonicida accounts for only 30.56% (n = 11) of isolates in the water column. This study shows the coexistence of distinct populations of Aeromonas in the oligohaline area of an anthropized estuary. Moreover, A. media, a putative human pathogen, present in the water column and abundant in the WWTP samples, was not detected in association with living copepods.
Keywords: Aeromonas, diversity, copepods, estuary, antibiotic resistance
Introduction
Aeromonas spp. are ubiquitous bacteria primarily recovered from aquatic ecosystems (Janda and Abbott, 2010). They are found in fresh water as well as estuarine and marine waters, and in association with numerous autochthonous aquatic organisms in these environments (Gugliandolo et al., 2008; Janda and Abbott, 2010; Khor et al., 2015; Laviad and Halpern, 2016; Chenia and Duma, 2017). They are also isolated from various habitats such as the gastrointestinal tracts of healthy animals and as transient flora in foods such as vegetables, dairy products, meat, seafood, and drinking water.
Several species are involved in pathologic interaction with numerous animals from the aquatic environment, mainly fishes but also corals, for example (Janda and Abbott, 2010; Hamid et al., 2016; Chenia and Duma, 2017). Some of these species have a major economic impact in aquaculture: the species A. hydrophila, A. caviae, and A. veronii are frequent etiologic agents of fish diseases, e.g., motile Aeromonas septicemia and ulcerative syndrome; A. salmonicida is responsible for fish furunculosis, which can cause death within hours (Menanteau-Ledouble et al., 2016). In addition, the hypervirulent pathotype of A. hydrophila is considered an emerging pathogen responsible for outstanding epidemic outbreaks in farmed warm-water fishes (Rasmussen-Ivey et al., 2016).
Moreover, aeromonads are recognized as emerging pathogens in humans (Janda and Abbott, 2010). The severity of the disease varies from diarrhea to septicemia, depending on individual’s susceptibility, mainly the immune status (Aujoulat et al., 2012; Figueras and Beaz-Hidalgo, 2015; Teunis and Figueras, 2016). The species A. hydrophila, A. veronii, A. caviae, A. dhakensis, and A. media are responsible for more than 85% of human cases of aeromonosis (Figueras and Beaz-Hidalgo, 2015; Teunis and Figueras, 2016). Transmission of pathogenic strains of Aeromonas, including the fecal-oral route, is often due to direct or indirect contact with water (Lamy et al., 2009; Janda and Abbott, 2010; Li et al., 2015; Pal et al., 2016).
To date, widespread of antibiotic-resistant bacteria such as Aeromonas is a major public health issue related to the One Health concept, considering that aeromonads circulate within the major ecosystems: human, animals and water. Occurrence of antibiotic-resistant Aeromonas has been reported in these three ecosystems (Figueira et al., 2011; Piotrowska and Popowska, 2014; Esteve et al., 2015; Li et al., 2015; Patil et al., 2016; Baron et al., 2017). In this context, Aeromonas has been recently proposed as an indicator to assess the spread of antibiotic resistance in the aquatic environment (Berendonk et al., 2015).
Among water environments, estuaries are of particular interest for Aeromonas ecology. They are a transitional zone between a freshwater river and the seawater, characterized by a salinity gradient, where the level of contamination by chemicals and fecal bacteria reflect the land use of the watershed. In this environment, Aeromonas has been isolated in both water and sediments; some isolates exhibited resistance to multiple antibiotics (Henriques et al., 2006; Silva et al., 2014). Thus, in a lagoon estuarine area (Abidjan, Africa), the maximum abundance of Aeromonas was linked to both discharges from urban areas and the river-flow period when the water salinity was low (Marcel et al., 2002). In estuaries, aeromonads were reported to be associated with copepods, the most abundant zooplankton living in the salinity gradient zone and a key component in estuarine food chains. Interestingly, in coastal water in Italy, Gugliandolo et al. (2008) showed that abundance of Aeromonas spp. associated with the copepods is higher than in the water column, suggesting that copepods may function as an aeromonad reservoir. However, today little is known about the putative role of copepods as a vector of pathogenic strains for humans, and the population diversity of Aeromonas associated with copepods remains poorly documented. The topic is complicated by the fact that (i) there are still no simple culturable methods to recover Aeromonas from the environment (Latif-Eugenín et al., 2016) and (ii) the aeromonad population structure is a complex of species, making taxonomic classification difficult. Multilocus phylogenetic analysis (MLPA) and/or whole-genome sequencing are now used for a more precise delineation of the Aeromonas species (Alperi et al., 2010a; Roger et al., 2012; Colston et al., 2014; Talagrand-Reboul et al., 2017), with new species of Aeromonas autochthonous in water environments regularly described (e.g., Marti and Balcázar, 2015; Latif-Eugenín et al., 2016).
The Seine estuary is the largest macrotidal estuary opening into the English Channel, which is characterized by strong anthropic pressure exerted on its watershed (76,650 km2): 30% of the French population is located mainly in its urban areas, with 40% of the country’s economic activity (mostly the chemical industry) and 30% of the national agricultural activity. The estuarine water quality has been extensively studied within the framework of the French multidisciplinary scientific program1. The microbiological quality of the Seine estuary water is poor, mainly impacted by wastewater treatment plants (WWTPs) treating wastewater from Paris and its suburbs during high flow-periods, while the input of Seine tributaries are predominant during low-flow periods (Garcia-Armisen et al., 2005; Servais et al., 2007; Touron et al., 2007). In addition, high contamination by trace metals, mainly cadmium and lead, and organic compounds such as polycyclic aromatic hydrocarbons, (PAHs), polychlorinated biphenyls (PCBs) and pesticides make the Seine estuary one of the most contaminated in Europe (Carpentier et al., 2002).
This macrotidal estuary is characterized by the presence of a high turbidity zone (HTZ) in the lower estuary (in the mouth of the estuary) where suspended particulate matter (SPM) and the associated contaminants are concentrated. Therefore, in this area the behavior of microorganisms such as fecal bacteria, mainly associated with organomineral particles, is strongly influenced by hydrosedimentary processes (Guézennec et al., 1999; Pachepsky and Shelton, 2011; Malham et al., 2014). During a semidiurnal tidal cycle, bacteria associated with particles suspended in the water column settle during slack high water and then are concentrated in fluid mud at the water–sediment interface. In contrast, at the beginning of a flood, when current velocities increase, bacteria trapped within the fluid mud can be resuspended in the water column (Berthe et al., 2008). Another characteristic of this area is the abundance of Eurytemora affinis, a distinctly dominant copepod species (crustacean, calanoid). In this oligohaline zone, Eurytemora affinis accounts for up to more than 90% of the zooplankton (Mouny and Dauvin, 2002; Devreker et al., 2010). The dynamics of population of Eurytemora affinis was driven by environmental parameters (SPM, salinity and temperature) and consequently – as bacteria dynamic-closely linked to the tidal cycle (Devreker et al., 2010).
Thus, the mouth of Seine estuary is a key environment and provides an outstanding model for studying autochthonous aeromonads in the estuary, and those from human and animal origin, mainly released by treated effluent from WWTPs or watershed run-off via tributaries. The present study aims to compare the abundance and the diversity of Aeromonas populations (i) in the water column, i.e., water and fluid mud, (ii) in association with copepods and (iii) from treated effluent from WWTP released in the same area. For this purpose, a 2-year monitoring campaign was carried out in the Seine estuary, and the diversity of the Aeromonas population was investigated based on a combined culturable and molecular approach, and the phenotypic antibiotic-resistance profile was determined.
Materials and Methods
Sampling Strategy
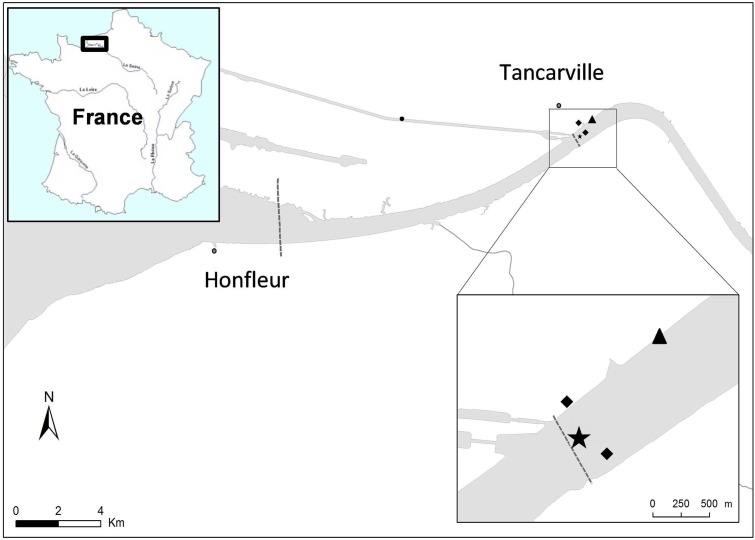
Copepods, water, and fluid mud were collected in the mouth of Seine estuary (France N 49° 28′ 30.26″ E 0° 27′ 48.65″) (Figure 1). This site is located in the mesohaline zone where salinity can range from 0 to 15 during a semidiurnal cycle (twice a day). In this area the microbiological quality based on Escherichia coli and Enterococci abundance ranged from 3.0 × 101 to 2.5 × 103 CFU.100 mL-1 and 1.0 × 101 to 2.3 × 103 CFU.100 mL-1, respectively (Touron et al., 2007), resulting in (i) intra-estuarine inputs (WWTP and seven main tributaries) and (ii) inputs of the estuarine entrance mainly dominated by one of the largest WWTPs in Europe, which treats the wastewater of Paris and its suburbs (6.5 million inhabitants). Moreover, a Waste Water Treatment Plan (Tancarville WWTP, 1800 inhabitants) is located 1 km upstream from the sampling site. Between February 2012 and November 2015, eight sampling campaigns were carried out at 0 and 2 h after the high water slack for various hydrological periods: in the high-flow period (>800 m3.s-1) and low-flow period (<500 m3.s-1, Table 1). The HTZ was located in this area for five of the eight campaigns (February 2012, August 2014, June 2015, October 2015, and November 2015). Surface water (-50 cm depth) and bottom water (+50 cm from the bottom) were sampled with a 3-L Niskin bottle and were transferred to sterile bottles before being analyzed. Fluid mud was sampled (i) on the intertidal mudflat (August 2014, February, June and November 2015) and directly collected with sterile plastic tubes at three equidistant points 50 cm apart; (ii) on the subtidal mudflat, closest to the copepod sample site (March and May 2013, and October 2015) with an Ekman sediment grab sampler (15 cm × 15 cm area). All the samples were immediately stored at 4–6°C after sampling and microbiological analysis was carried out within 4 h. Treated WWTP effluent (500 mL) (Figure 1) was collected every hour for 24 h using an ISCO 6700 portable sampler (Teledyne Isco, Inc., Lincoln, NE, United States). The mean daily sample consists of a subsamples mixture (1 flask of 1 L every hour) of identical volume (250 mL) collected during the sampling period (24 h).
FIGURE 1.
Study area and sampling point. Water, fluid mud and copepods were sampled from the mouth of Seine estuary (France N 49° 28′ 30.26″ E 0° 27′ 48.65″) located at 1.2 km downstream of a wastewater treatment plant (WWTP; 1800 inhabitants). Surface water (–50 cm) was sampled in the middle of the channel, fluid muds were sampled from the subtidal mudflat (February 2012, March, May 2013 and October 2015) and from the intertidal mudflat (August 2014, February, June and November 2015) and copepods were collected from the right bank (February 2012, March 2013 and October 2015).  , Water sampling point;
, Water sampling point;  , Fluid mud sampling points;
, Fluid mud sampling points;  , WWTP.
, WWTP.
Table 1.
Hydrological, physicochemical, microbial, and biotic characteristics of the sampling site.
| February 2012 | March 2013 | May 2013 | August 2014 | February 2015 | June 2015 | October 2015 | November 2015 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hydrological and physico-chemical parameters | ||||||||||||||||
| Flow ratea | 385 m3.s-1 | 877 m3.s-1 | 1090 m3.s-1 | 366 m3.s-1 | 1144 m3.s-1 | 226 m3.s-1 | 217 m3.s-1 | 199 m3.s-1 | ||||||||
| Pluviometryb | 2.8 mm | 6.4 mm | 35.4 mm | 37 mm | 20.7 mm | 16.9 mm | 13.4 mm | 6.6 mm | ||||||||
| Salinity | 1.3 | 0.2 | 0 | 0.6 | 0 | 8.3 | 5.2 | 2.8 | ||||||||
| Temperature | 7°C | 7.2°C | 15°C | 19.5°C | 5.4°C | 19.8°C | 15.4°C | 12°C | ||||||||
| Conductivity | 1784 μS.cm-1 | 784 μS.cm-1 | NA | 1400 μS.cm-1 | 477 μS.cm-1 | 13086 μS.cm-1 | 8896 μS.cm-1 | 4012 μS.cm-1 | ||||||||
| SPMc | 100 mg.L-1 | 50 mg.L-1 | 10 mg.L-1 | 68 mg.L-1 | 88 mg.L-1 | 76 mg.L-1 | 81 mg.L-1 | 185 mg.L-1 | ||||||||
| Microbiological quality | ||||||||||||||||
| Waterd | Fluid mude | Water | Fluid mud | Water | Fluid mud | Water | Fluid mud | Water | Fluid mud | Water | Fluid mud | Water | Fluid mud | Water | Fluid mud | |
| E. coli | NA | NA | 7.0 ± 0.2 × 101 | 6.1 ± 0.7 × 100 | NA | 5.5 ± 0.3 × 101 | 1.1 ± 0.2 × 102 | 2.1 ± 1.2 × 101 | 3.0 ± 1.4 × 101 | 3.5 ± 0.9 × 102 | 6.2 ± 0.1 × 101 | <5.0 101f | 2.3 ± 0.4 × 102 | 3.9 ± 1.6 × 101 | 5.0 ± 1.0 × 101 | 6.6 ± 2.0 × 100 |
| Enterococci | 2.0 × 101 | NA | 5.0 ± 0.5 × 101 | 6.5 ± 0.3 × 100 | NA | 5.7 ± 1.5 × 101 | 2.5 ± 0.7 × 101 | 5.0 ± 5.0 × 100 | 2.6 ± 0.4 × 102 | 5.2 ± 0.9 × 102 | <5.0 101f | 2.6 ± 0.5 × 101 | 7.5 ± 3.5 × 101 | 3.6 ± 0.4 × 101 | <5.0 × 101f | 8.0 ± 3.0 × 101 |
| Copepods | ||||||||||||||||
| Eurytemora affinis | + | + | - | + | NS | NS | + | NS | ||||||||
Surface water (-50 cm below the surface) and fluid mud were sampled in the mouth of the Seine Estuary at Tancarville site (N 49° 28′ 30.26″ E 0° 27′ 48.65″). aAverage daily flow rate (GIP Seine-Aval database); bcumulated rainfall 7 days before the sampling (Météo France database); cSuspended Particulate Matter (wet weight); dCFU.100 mL-1; eCFU.g-1 (wet weight); fcount below the detection threshold; NA: not analyzed; NS: not sampled.
Eurytemora affinis Sampling
Living copepods were collected in the subsurface using a WP2 plankton net (200-mm mesh size; 1 m in diameter) as previously described (Cailleaud et al., 2007). Immediately after sampling, copepods were sorted using two sieves (500- and 100-μm mesh size) in order to eliminate particles and predators such as Mysidacea and Gammaridae. Copepods were then stored in insulated containers filled with estuarine water and microbiological analysis was carried out within 4 h.
Chemical and Physical Parameters
Temperature, salinity, and conductivity were measured using a in situ multi-parameter probe (TetraCon 325, WTW, Germany). To determine the SPM concentration, the water was filtered through preweighed 0.45-μm pore-size filters (Whatman GF/F, Sigma–Aldrich). After filtration, the filters were rinsed with distilled water (10% filtered volume) to remove the salt and dried for 48 h at 50°C before being weighed again to determine the total SPM concentration. Rainfall was extracted from Méteo-France database2. River flow was extracted from the GIP-SA database3, as was the flow rate.
Enumeration and Isolation of Culturable E. coli, Enterococcus, and Presumptive Aeromonas
Escherichia coli, Enterococcus, and presumptive Aeromonas were enumerated using membrane filtration methods (ACN, 0.45 μm, Sartorius). β-D-galactosidase- and β-D-glucuronidase-positive E. coli were isolated from the water samples with selective chromogenic media specific for E. coli, with the addition of a selective supplement for water samples (RAPID’E. coli 2 Medium and Supplement; Biorad, United States); plates were incubated for 24 h at 37°C. Enterococcus was isolated from the water samples with selective chromogenic media specific for Enterococcus (RAPID’Enterococcus Medium; Biorad, France); the plates were incubated for 48 h at 44°C. Presumptive Aeromonas were isolated on Aeromonas isolation agar based on Ryan’s formulation (Sigma–Aldrich, United States) supplemented with ampicillin (5 mg.L-1) (Sigma–Aldrich, United States). Plates were incubated for 48 h at 22°C. Dark green colonies (sulfide production and no acid formation) were considered as presumptive Aeromonas until molecular characterization, as described in Section Antimicrobial Susceptibility Testing. The sediment was analyzed with the following modifications: 3 g (w/w) were added to 27 mL of NaCl 0.85% (w/v) supplemented with Na4P2O7 (1 mM, final concentration) and mixed vigorously for 3 min to dissociate bacteria from organic mineral particles. Ten-milliliter volumes of appropriate dilutions were then filtered (ACN, 0.45 μm, Sartorius) before plating. Further identification of presumptive Aeromonas (radA-) was carried out using mass spectrometry (matrix-assisted laser desorption ionization mass spectrometry-time of flight, MALDI-TOF MS); Biotyper (Bruker Daltonics, Germany).
Copepods previously collected in estuarine water were separated from suspended particles by phototropism (915 lumens) for 15 min, then sorted using a 200-μm sieve, resuspended in 5 L of artificial brackish water (15 PSU), and filtered on a 1.2-mm filter (Sartorius, France) before being weighed. Finally, copepods were suspended in 30 mL of NaCl 0.9% (w/v) and mixed with a blender (Ultra Turrax T10, Imlab, France) for 1 min at 4°C before being analyzed. Ten-milliliter volumes of appropriate dilutions were then filtered before plating (0.45 μm HA047, Millipore). The threshold values for the enumeration of Enterococcus, E. coli, and presumptive Aeromonas in water was 5 CFUs per 100 mL. For each sample, non-confluent colonies of presumptive Aeromonas were selected on the filter and then streaked on Luria Broth agar (Thermo Fisher Scientific). Finally, a total of 476 presumptive Aeromonas strains (dark green colonies) were isolated and stored on a cryo-bead system (Dutscher) at -80°C.
Antimicrobial Susceptibility Testing
Aeromonas resistance to antibiotics was tested using the disk diffusion method according to the recommendations of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines V1.0 2015. The categorical interpretations (susceptible, S; intermediate, I; resistant, R) were based on the EUCAST interpretative criteria for Enterobacteriaceae after incubation at 22 and 35°C for 24 h. E. coli CIP 7624 (ATCC 25922) was used as a control. The antibiotics tested (16) included the most commonly used in France for treatment of Aeromonas infections in human and veterinary medicine: ampicillin (AM, 10 μg), amoxicillin + clavulanic acid (AMC, 20 + 10 μg), ticarcillin (TIC, 75 μg), ticarcillin + clavulanic acid (TCC, 75 + 10 μg), piperacillin (PRL, 30 μg), piperacillin + tazobactam (TBZ, 30 + 6 μg), cefotaxime (CTX, 5 μg), cefoxitin (FOX, 30 μg), cefepime (FEP, 30 μg), ertapenem (ETP, 10 μg), imipenem (IPM, 10 μg), gentamicin (CN, 10 μg), tobramycin (TOB, 10 μg), norfloxacin (NOR, 10 μg), ciprofloxacin (CIP, 5 μg), trimethoprim + sulfamethoxazole (SXT, 23.75 + 1.25 μg). As recommended by Magiorakos et al. (2012), Aeromonas strains resistant to at least one antibiotic in three or more antimicrobial classes (penicillins not included) were considered a multidrug-resistant strain.
DNA Extraction and PCR Amplification
Cell suspensions were prepared with two or three colonies in 200 μL of sterile water and total bacterial DNA was extracted by boiling (10 min at 94°C). All presumptive Aeromonas strains were tested and identified at the species level by amplification and sequencing of housekeeping genes. PCRs were performed with specific primers targeting the gyrB gene (Yáñez et al., 2003) and the radA gene (Roger et al., 2012), as previously described.
The PCR products were separated in 1.5% agarose gel in 0.5× TBE buffer. The products were sequenced using forward amplification primers in an ABI 3730XL automatic sequencer (Beckman Coulter Genomics). Membership in the genus Aeromonas was checked by comparison with the NCBI database using the Basic Local Alignment Search Tool (BLAST4). All the sequencing data were submitted to the GenBank database: the accession numbers of the sequences are KX898587 to KX898810.
Phylogenetic Analysis
Phylogenetic analysis was performed as previously described by Roger et al. (2012). Briefly, gene sequences were codon-aligned using the ClustalW application within the Bioedit Sequence Alignment Editor. Phylogenetic analyses were performed for each of the two gene sequences and for a manually concatenated sequence. Gaps in concatenated sequences were deleted with Bioedit. The sequences were converted to Phylip format using the EMBOSS Seqret online program5. A maximum likelihood (ML) method-based phylogenetic tree was reconstructed using evolutionary distance analyzed with the PhyML v3.1 software using GTR, with a gamma distribution parameter estimated from the dataset and invariant sites as a substitution model. ML bootstrap support was calculated after 100 reiterations. Type strain sequences were downloaded from the NCBI database.
Statistical Analysis
The chi-squared test of the Fisher exact test was performed to compare the antimicrobial profiles; the Pearson coefficient was used to measure the degree of linear correlation between abundance of Aeromonas in water and fluid mud; the Student’s t-test was used to compare the abundance of Aeromonas inside the water column. All data analyses were performed with XLSTAT (XLSTAT, Boston, MA, United States V2016.3).
Results
Abundance of Aeromonas in the Water Column and Copepods in the Estuarine Environment
In the oligohaline area of the Seine estuary, Aeromonas was detected in the water column, (i.e., surface water and fluid mud of the estuary mouth) for all flow rates and for water temperatures ranging from 5.4 to 19.8°C (Tables 1, 2A). Copepods were collected in water characterized by a temperature varying between 7 and 19.5°C and a salinity between 0.6 and 5.2 (Table 1). Copepods were collected during campaigns (February 2012, August 2014, and October 2015) corresponding to low-flow periods when the HTZ was located in this area. Copepods were also detected during an increase of the river flow (March 2013), but not when the river flow reached 1000 m3s-1 (May 2013). For all of these campaigns, Aeromonas was always bound to copepods (Table 2B).
Table 2A.
Occurrence of presumptive Aeromonas in water column.
| February 2012 | March 2013 | May 2013 | August 2014 | February 2015 | June 2015 | October 2015 | November 2015 | |
|---|---|---|---|---|---|---|---|---|
| Surface water (CFU.100 mL-1) | ||||||||
| Presumptive Aeromonas spp. | 1.1 × 102 | 6.0 ± 0.2 × 102 | 6.1 ± 1.3 × 102 | 1.4 ± 0.5 × 102 | 3.0 ± 2.8 × 102 | 4.6 ± 0.3 × 102 | 1.2 ± 0.3 × 103 | 1.3 ± 0.6 × 102 |
| Fluid mud (CFU.g-1) | ||||||||
| Presumptive Aeromonas spp. | NA | 1.2 ± 0.3 × 103 | 5.7 ± 0.7 × 103 | 9.0 ± 2 × 102 | 5.3 ± 0.9 × 102 | 9.1 ± 0.3 × 102 | 9.8 ± 0.9 × 103 | 2.6 ± 2.6 × 102 |
NA, not analyzed.
Table 2B.
Occurrence of presumptive Aeromonas and fecal indicator bacteria associated with Eurytemora affinis in Seine estuary.
| February 2012 | March 2013 | October 2015 | |
|---|---|---|---|
| Presumptive Aeromonasa (CFU.g-1∗) | 1.9 ± 0.7 × 102 | >1.1 × 104 | 4.3 ± 0.6 × 103 |
| Fecal indicator bacteria (CFU.g-1∗) | |||
| E. coli | 3.6 ± 0.3 × 101 | 7.9 ± 0.5 × 101 | 1.0 ± 0.1 × 102 |
| Enterococci | 6.4 ± 1.6 × 101 | 2.9 ± 0.1 × 102 | 1.8 ± 0.4 × 102 |
∗Wet weight copepods. aPresumptive abundance based on phenotypic feature of Aeromonas on Aeromonas Isolation Agar (48 h at 22°C, sulfide production and no acid formation).
In surface water, the abundance of presumptive Aeromonas (i.e., dark green colonies with sulfide formation and no acid formation) ranged from 1.1 × 102 to 1.2 ± 0.3 × 103 CFU.100 mL-1 and was not significantly different from the abundance in the bottom of the water column (P-value = 0.24) (Table 2A). In fluid mud, the abundance ranged from 2.6 ± 2.6 × 102 to 9.8 ± 0.9 × 103 CFU.g-1 (w/w). For each campaign, the abundance of presumptive Aeromonas in water and fluid mud were positively correlated (R2 = 0.8, P-value = 0.006). The abundance of presumptive Aeromonas associated with living copepods (see Materials and Methods) ranged from 1.9 ± 0.7 × 102 to >1.1 × 104 CFU.g-1 (w/w) (Table 2B).
During all sampling campaigns, the microbiological quality of the estuarine water estimated by the abundance of E. coli and Enterococci was of good to average quality according to the French water index (SEQ values, 2 × 102 CFU 100 mL-1 to 2 × 103 CFU 100 mL-1, for E. coli, and 2 × 102 CFU 100 mL-1 to 103 CFU 100 mL-1 for Enterococcus) established by the French Ministry of the Environment and Regional Water Agencies as well the WHO recommendations (WHO, 2011; Table 1). However, no correlation was observed between the abundance of presumptive Aeromonas and indicators of fecal bacteria in water, fluid mud, and copepods. The abundance of presumptive Aeromonas was always about one or two orders of magnitude higher than the abundance of E. coli and Enterococci.
Among the 476 isolates of presumptive Aeromonas that were collected, 213 strains were confirmed as belonging to the Aeromonas genus through partial sequencing of gyrB (169 strains) and/or radA (173 strains). Interestingly, the occurrence of Aeromonas (gyr B+ and/or radA +) seems higher in sediment and associated with copepods than in water (Table 3). Identification of a random sample of 50 isolates of presumptive Aeromonas (radA-) was further analyzed based on matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). Among them, 29 isolates belonging to Pseudomonas spp., 11 isolates of Serratia marcescens, and eight isolates could not be identified by this approach. Interestingly, four isolates were detected as Aeromonas spp. by MALDI-TOF MS.
Table 3.
Occurrence of Aeromonas among presumptive Aeromonas isolated in Seine estuary.
| Water | Fluid mud | Copepods | WWTP | |||||
|---|---|---|---|---|---|---|---|---|
| Presumptive Aeromonas abundancea | ||||||||
| (CFU.100 mL-1 or CFU.g-1) | 6.5 ± 5.3 × 102 | 4.0 ± 5.0 × 103 | 7.6 ± 4.7 × 103 | 5.3 ± 0.3 × 104 | ||||
| Isolated number | 136 | 130 | 125 | 100 | ||||
| Aeromonas gyrB+ and/or radA+ b | 33% | 47% | 50% | 46% | ||||
| (CFU.100 mL-1 or CFU. g-1)c | 2.1 ± 1.7 × 102 | 1.9 ± 2.4 × 103 | 3.8 ± 2.3 × 103 | 2.4 ± 0.1 × 104 | ||||
aPresumptive abundance based on phenotypic feature of Aeromonas on Aeromonas Isolation Agar (48 h at 22°C, sulfide production and no acid formation); bnumber of Aeromonas (gyrB+radA+)/total number of presumptive Aeromonas; cestimated abundance taking into account % of Aeromonas (gyrB+ radA+).
Diversity of Aeromonas Populations from the Water Column and Copepods in the Seine Estuary
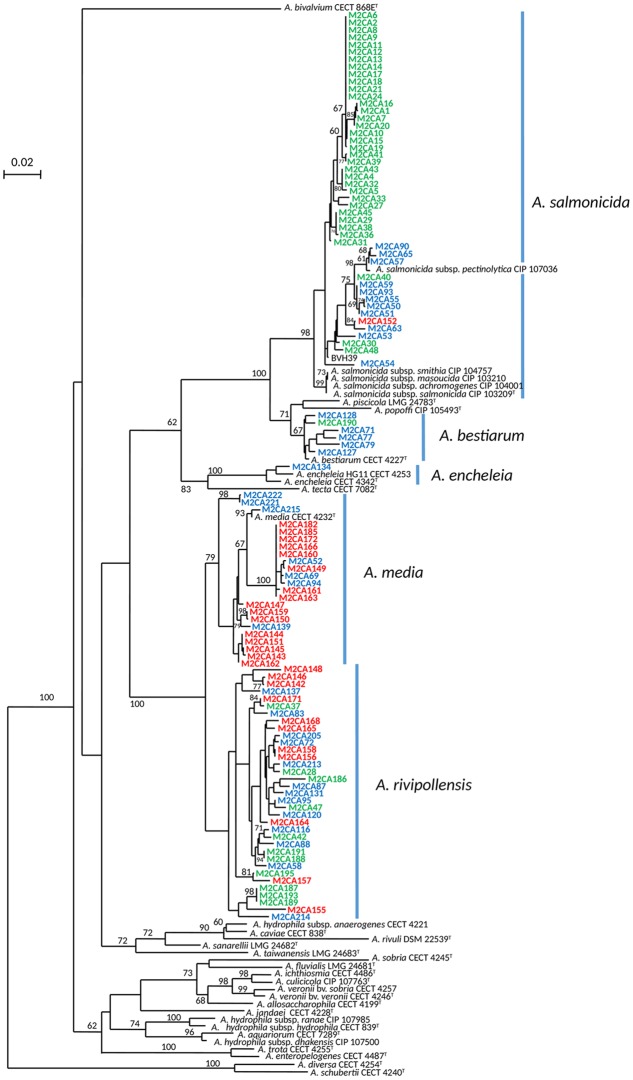
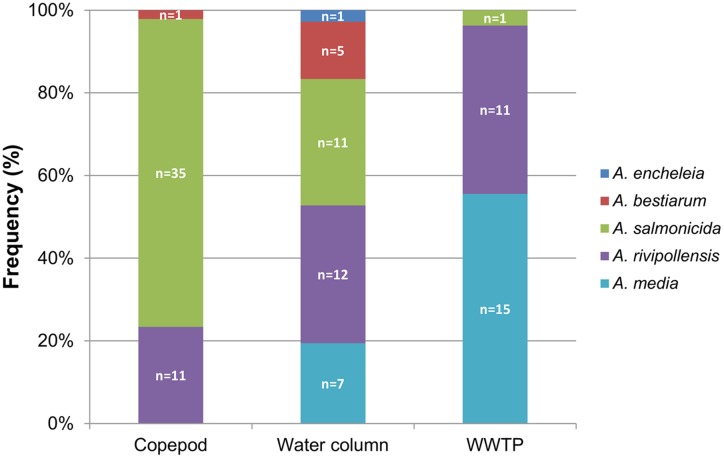
To compare the diversity of the Aeromonas population sampled in the water column (surface water and fluid mud) with the Aeromonas population bound to the copepods, we studied all isolated Aeromonas gyrB+ and radA+, which comprised 47 isolates from copepods and 36 isolates from the water column. Phylogenetic analysis combined with phenotypic antibiotic-resistance profiles were carried out on all these isolates. In addition, 27 isolates of Aeromonas from WWTP-treated effluent (5.3 ± 0.3 × 104 CFU.100 mL-1) located 1 km upstream from the sampling site were analyzed as a control corresponding to an input of allochthonous Aeromonas (i.e., not from an estuarine habitat). The phylogenetic tree reconstructed on the basis of concatenated sequences of gyrB+ radA (1182 nt) made it possible to discriminate the different Aeromonas species that were collected (Figure 2). Strains distributed in five clusters corresponding to A. bestiarum (n = 6; 5.45%), A. encheleia (n = 1; 0.91%), A. media (n = 22; 20.00%), and A. salmonicida (n = 47; 42.73%). The last cluster identified is phylogenetically very close to the A. media cluster and is probably affiliated with the recently described species A. rivipollensis (n = 34; 30.91%) (Marti and Balcázar, 2015) (Figure 2). In the water column, the five species were isolated as follows: A. rivipollensis (n = 12; 33.33%), A. salmonicida (n = 11; 30.56%), A. media (n = 7; 19.44%), A. bestiarum (n = 5; 13.89%), and A. encheleia (n = 1; 2.78%). Among the less common species, A. bestiarum was mainly isolated from the water column. While A. salmonicida is the major species that colonizes copepods, like more than 74.47% (n = 35) of Aeromonas strains, other species recovered include A. rivipollensis (n = 11; 23.40%) and A. bestiarum (only one strain). In contrast, in treated WWTP effluent, A. media (n = 15; 55.56%) and A. rivipollensis (n = 11; 40.74%) were mainly observed while only one strain of A. salmonicida was isolated (3.7%). It should be noted that A. media, a putative human pathogen, present in the water column and abundant in the WWTP samples, was not detected in association with living copepods (Figures 2, 3).
FIGURE 2.
Unrooted maximum-likelihood tree based on concatenated sequences of the two housekeeping gene fragments (1182 nt). The tree shows the structure of the studied Aeromonas spp. population (110 strains). The origin of the trains is indicated by the font color, WWTP (red font), column water (blue font) and copepods (green font). The horizontal lines represent genetic distance, with the scale bar indicating the number of substitutions per nucleotide position. The numbers at the nodes are support values estimated with 100 bootstrap replicates. Only bootstrap values > 60 are indicated on the tree. Species names corresponding to the five clusters are indicated close to the blue bar.
FIGURE 3.
Distribution of Aeromonas species in the water column, associated with copepods, and in the effluent of WWTP. n = number of isolates.
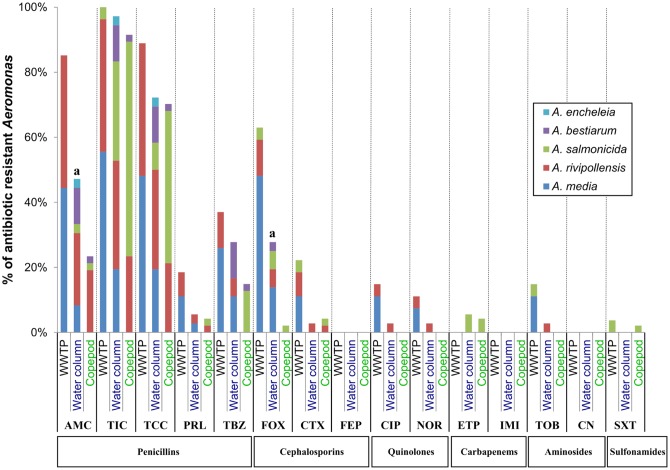
Considering the profiles of antibiotic resistance phenotypes, no significant difference between Aeromonas (gyrB+ radA+) populations from the water column and copepods was observed, except for AMC (P-value = 0.02) and FOX (P-value = 0.001) resistance, which is higher in the Aeromonas population from the water column than in the Aeromonas population from copepods (Figure 4). However, at the species level, 62.9% of A. salmonicida associated with copepods were resistant to TCC versus 27.3% in the Aeromonas population from the water column (P-value = 0.04; Supplementary Table S1). No multiresistant Aeromonas was isolated from copepods. Only two multiresistant Aeromonas were isolated in the water column (A. rivipollensis) and in the WWTP effluent (A. media) (Table 4).
FIGURE 4.
Comparative antibiotic resistance phenotypes of Aeromonas populations from water column, copepods and WWTP. The number (n) of isolates resistant to each of the 16 antibiotics was estimated using the agar diffusion method at 22°C, for Aeromonas populations sampled from the water column (N = 36); copepods (N = 47) and the effluent of WWTP (N = 27). Percentages of strains resistant to each antibiotic within Aeromonas population corresponded n/N. aSignificant difference between percentages of resistance to an antibiotic between the Aeromonas population isolated from the water column and copepods by the chi-squared test of Fischer (P-value = 0.02 for AMC and 0.001 for FOX). AM, Ampicillin; AMC, Amoxicillin + clavulanic acid; TIC, Ticarcillin; TCC, Ticarcillin + clavulanic acid; PRL, Piperacillin; TBZ, Piperacillin + tazobactam; CTX, Cefotaxime; FOX, Cefoxitin; FEP, Cefepime; ETP, Ertapenem; IMI, Imipenem; CN, Gentamicin; TOB, Tobramycin; NOR, Norfloxacin; CIP, Ciprofloxacin; SXT, Trimetropin + sulfamethoxazole.
Table 4.
Antibiotic-resistant phenotypes of Aeromonas (gyrB+ radA+) from Seine Estuary.
| Origin of sample | Species | No. of isolates | No. of resistance | Pattern |
|---|---|---|---|---|
| Copepod | A. salmonicida | 3 | 1 | AM |
| 1 | 2 | AM, TCC | ||
| 6 | 2 | AM, TIC | ||
| 19 | 3 | AM, TIC, TCC | ||
| 1 | 4 | AM, TIC, TCC, ETP | ||
| 1 | 4 | AM, AMC, TIC, TCC | ||
| 1 | 4 | AM, TIC, TCC, TBZ | ||
| 1 | 4 | AM, TIC, TCC, FOX | ||
| 1 | 5 | AM, TIC, TCC, CTX, SXT | ||
| 1 | 6 | AM, TIC, TCC, PRL, TBZ, ETP | ||
| A. rivipollensis | 1 | 3 | AM, TIC, TCC | |
| 9 | 4 | AM, AMC, TIC, TCC | ||
| 1 | 4 | AM, TIC, PRL, CTX | ||
| A. bestiarum | 1 | 5 | AM, AMC, TIC, TCC, TBZ | |
| Water columna | A. salmonicida | 7 | 2 | AM, TIC |
| 1 | 3 | AM, TIC, FOX | ||
| 1 | 4 | AM, AMC, TIC, TCC | ||
| 1 | 4 | AM, TIC, TCC, ETP | ||
| 1 | 5 | AM, TIC, TCC, FOX, ETP | ||
| A. rivipollensis | 1 | 2 | AM, TIC | |
| 2 | 3 | AM, TIC, TCC | ||
| 6 | 4 | AM, AMC, TIC, TCC | ||
| 1 | 5 | AM, AMC, TIC, TCC, TBZ | ||
| 1 | 5 | AM, AMC, TIC, TCC, FOX | ||
| 1b | 10 | AM, TIC, TCC, PRL, TBZ, CTX, FOX, TOB, NOR, CIP | ||
| A. bestiarum | 1 | 1 | AM | |
| 3 | 5 | AM, AMC, TIC, TCC, TBZ | ||
| 1 | 6 | AM, AMC, TIC, TCC, TBZ, FOX | ||
| A. media | 1 | 3 | AM, TIC, TCC | |
| 1 | 4 | AM, TIC, TCC, FOX | ||
| 1 | 4 | AM, TIC, TCC, TBZ | ||
| 1 | 5 | AM, AMC, TIC, TCC, FOX | ||
| 2 | 6 | AM, AMC, TIC, TCC, TBZ, FOX | ||
| 1 | 6 | AM, TIC, TCC, PRL, TBZ, FOX | ||
| A. encheleia | 1 | 4 | AM, AMC, TIC, TCC | |
| WWTP | A. salmonicida | 1 | 6 | AM, TIC, CTX, FOX, TOB, SXT |
| A. rivipollensis | 4 | 4 | AM, AMC, TIC, TCC | |
| 3 | 5 | AM, AMC, TIC, TCC, FOX | ||
| 1 | 5 | AM, AMC, TIC, TCC, TBZ | ||
| 1 | 6 | AM, AMC, TIC, TCC, NOR, CIP | ||
| 2 | 7 | AM, AMC, TIC, TCC, PRL, TBZ, CTX | ||
| A. media | 1 | 2 | AM, TIC | |
| 1 | 4 | AM, TIC, TCC, FOX | ||
| 5 | 5 | AM, AMC, TIC, TCC, FOX | ||
| 1 | 6 | AM, AMC, TIC, TCC, TBZ, FOX | ||
| 1 | 7 | AM, AMC, TIC, TCC, PRL, TBZ, FOX | ||
| 1 | 7 | AM, AMC, TIC, TCC, TBZ, CTX, FOX | ||
| 3 | 8 | AM, AMC, TIC, TCC, PRL, TBZ, CTX, FOX | ||
| 2b | 9 | AM, AMC, TIC, TCC, TBZ, FOX, TOB, NOR, CIP | ||
ai.e., water + fluid mud; b strain resistant to at least three antibiotic classes (Penicillin group not included, as it is considered as an intrinsic resistance for Aeromonas, Baron et al., 2017); AM, Ampicillin; TIC, Ticarcillin; TCC, Ticarcillin + clavulanic acid; PRL, Piperacillin; AMC, Amoxicillin + clavulanic acid; TBZ, Piperacillin + tazobactam; CTX, Cefotaxime; FEP, Cefepime; FOX, Cefoxitin; CIP, Ciprofloxacin; NOR, Norfloxacin; ETP, Ertapenem; IMI, Imipenem; TOB, Tobramycin; CN, Gentamicin; SXT, Trimetropin + sulfamethoxazol.
Discussion
Aquatic environments including aquatic organisms are considered Aeromonad’s primary habitat (Janda and Abbott, 2010). Over the last decade, molecular approaches have greatly enhanced the knowledge of Aeromonas diversity in coastal or estuarine waters (Gugliandolo et al., 2008; Silva et al., 2014). Here, we confirm that Aeromonas should be identified using molecular methods for a better understanding of the diversity of Aeromonas in the oligohaline zone of a highly anthropized estuary (Seine). In this environment, only 44.7% of the presumptive Aeromonas, i.e., able to grow on a selective medium, were identified as Aeromonas based on the sequence of radA and/or gyrB genes. Further identification using mass spectrometry (MALDI-TOF) shows that presumptive Aeromonas (radA-) could be mainly related to Pseudomonas and Serratia. These strains have the same culturable characteristics than Aeromonas on the selective media used in this study (Aeromonas Isolation Agar). However, mass spectrometry is not yet the most accurate method to identify all Aeromonas species or environmental bacteria (Shin et al., 2015). In this environment contaminated by metals, it has been shown that prevalence of Pseudomonas and Enterobacteriaceae as well as Aeromonas is high (Henriques et al., 2006). These results confirmed that housekeeping gene sequencing is the most accurate method to identify Aeromonas strains at the species level for strains recovered from aquatic environments, as described for aeromonads from other sources (Alperi et al., 2010b; Roger et al., 2012; Colston et al., 2014; Talagrand-Reboul et al., 2017).
Here we show that in the mouth of the macrotidal Seine estuary, Aeromonas spp. are detected in water and fluid mud during all hydrological periods. In contrast, in estuary water as well as in the coastal environment, a seasonal variation of Aeromonas abundance was reported. The maximum abundance of Aeromonas was observed in the urban area, during the high river flow period in a eutrophic tropical estuary (Abidjan, Senegal) and in spring in coastal waters (Messina, Italy) (Marcel et al., 2002; Silva et al., 2014). Moreover, in this study, the abundance of Aeromonas in water never exceeded 2 ± 1.7 × 102 CFU.100 mL-1 and were lower than those reported in Italian and Brazilian anthropized estuaries (101 to 106 UFC 100 mL-1) (Gugliandolo et al., 2008; Silva et al., 2014). These discrepancies could be explained by (i) the methods used for counting Aeromonas, (ii) the impact of hydrosedimentary processes on Aeromonas behavior in a macrotidal estuary, (iii) the land use of the catchment basin that controls inputs of Aeromonas from soils, humans, and animals by surface runoff and waste waters, and (iv) the temperature of the Seine estuary water that is lower than the aeromonad optimal growth temperature of 22–25°C (Janda and Abbott, 2010). We show here that Aeromonas spp. are detected at the sediment–water interface of the fluid mud, which corresponds to the suspended particulate that settles at the bottom during slack water at high tide. Indeed, these results suggest that the behavior of Aeromonas in the mouth of the Seine estuary was strongly influenced by both the hydrology and particle dynamics (Guézennec et al., 1999; Malham et al., 2014). In the mouth of the Seine estuary, occurrence of Aeromonas is mainly related to the upstream input – i.e., treated WWTP effluent or watershed run off – as well as autochthonous Aeromonas able to grow in the oligohaline area of this highly anthropized environment. In addition, a secondary input of Aeromonas was related to the resuspension of surficial sediment, which previously settled on the intertidal mudflats located in this area (Berthe et al., 2008).
In the mouth of Seine estuary, Aeromonas spp. are also associated with Eurytemora affinis whose abundance depends on both their lifecycle and – as bacteria – the hydrosedimentological processes (Mouny and Dauvin, 2002; Devreker et al., 2010). Indeed, here we show that copepods are present when there is a HTZ in the oligohaline area of the Estuary. These results are consistent with a study reported by Devreker et al. (2010) that shows link between both salinity and the dynamic of SPM and those of copepods in the mouth of the Seine estuary. In similar estuarine water (e.g., the Adriatic Sea, Italy), it has also been shown that Aeromonas spp. were associated with copepods (Temora stylifera, Acartia clausii, Centropages typicus, and Paracalanus parvus) (Dumontet et al., 1996; Gugliandolo et al., 2008).
In this study, we show that the diversity of Aeromonas populations from the water column (water and fluid mud) is different from that associated with copepods. In the Seine estuary water column, five species of Aeromonas co-exist: A. salmonicida, A. bestiarum, A. encheleia, A. media, and A. rivipollensis. Indeed, such Aeromonas species could be well adapted to a estuarine environment having a cooler surface water with mean temperature of 13.4°C (min: 5.3°C/ max 21.3°C). Except for A. salmonicida (Henriques et al., 2006), these species, previously observed in farmed fishes (water and fish) (Esteve et al., 1995; Schmidt et al., 2001; Hatha et al., 2005; Behbahani et al., 2014; Bartkova et al., 2016) and in waste water (Figueira et al., 2011; Marti and Balcázar, 2015; Popovic et al., 2015; Vaz-Moreira et al., 2015; Varela et al., 2016), have never been, as far as we know, described in coastal and anthropized estuary waters. Only the species A. hydrophila, A. caviae, A. sobria, and A. veronii have been reported in coastal or estuarine environments (the Adriatic Sea, Italy) (Dumontet et al., 1996; Fiorentini et al., 1998; Gugliandolo et al., 2008). Underestimation of Aeromonas diversity could mainly stem from the identification methodology. However, both the sampling strategy and the water temperature could also explain the differences observed between the diversities of the Aeromonas population in these similar aquatic environments. Consequently, these factors deserve to be precisely reported in further studies. In this study, an input of A. media and A. rivipollensis from the treated influent of the closest WWTP cannot be excluded even if no correlation between abundance of Aeromonas and fecal bacterial indicators was observed. Indeed, in this anthropized estuary, the diversity of Aeromonas populations in the water column probably reflects the coexistence of autochthonous Aeromonas (A. salmonicida, A. bestiarum, A. encheleia), for which the estuary is the primary habitat, and Aeromonas previously released from wastewater. Interestingly, one of the dominant Aeromonas species from WWTP outflow in Monfort and Baleux’s (1990) study was A. caviae, which could possibly be wrongly identified because (i) this species is known to be difficult to phenotypically distinguish from A. media and A. rivipollensis, and (ii) A. rivipollensis was an unrecognized species at time of study.
In the Seine estuary, the Aeromonas population associated with Eurytemora affinis was composed of three species described today as non-human pathogens and was dominated by A. salmonicida. None of these species (A. rivipollensis, A. salmonicida, or A. bestiarum) has been previously described as being associated with copepods. To date, only the species A. hydrophila has been reported in association with copepods, probably due to the lack of resolution of the biochemical methods used in these studies (Dumontet et al., 1996; Gugliandolo et al., 2008). The higher aeromonad density associated with copepods may be due to the known aeromonad ability to produce biofilm and favor copepod colonization, and this should require further study. Interestingly, higher resistance to TCC was observed within the A. salmonicida population associated with copepods. As aeromonads are naturally resistant to Penicillins (Baron et al., 2017), these results suggest the existence of a specific population of A. salmonicida belonging to the microbiota of Eurytemora affinis, although further study is warranted to confirm this assumption.
Conclusion
In the oligohaline zone of a highly anthropized estuary (oceanic climate), Aeromonas spp. (gyrB+ radA+) are present in the water column (water and fluid mud) and associated with living copepods (Eurytemora affinis). However, the diversity of the Aeromonas populations in the water column (water and fluid mud) is different from those associated with copepods. In the water column, the Aeromonas species autochthonous of the estuary, i.e., A. salmonicida, A. bestiarum, A. encheleia, co-exist with Aeromonas species (A. media, A. rivipollensis) originating from human and animal sources discharged by the WWTP effluent. A. salmonicida are the major species bound to the copepods (Eurytemora affinis), even if A. rivipollensis and A. bestiarum are also detected, while no human pathogenic species were associated with copepods. The proportion of A. salmonicida resistant to the antibiotic (TCC) was higher than in the Aeromonas population in the water column. These results underlined the key role played by this type of anthropized estuary, i.e., as an environment where autochthonous aeromonads and those originating from human and animal sources coexist.
Author Contributions
FP, BL, and EJ-B defined the research theme. GC, TB, RL, JF-L, and FP defined sampling strategy and designed methods and experiments. GC carried out the laboratory experiments, strains isolation and antibiotic resistance analysis, FR carried out the phylogeny of aeromonads. GC, FR, TB, and FP analyzed and interpreted the data. FP and GC drafted the manuscript. BL, TB, RL, and JF-L revised the paper critically. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank: Florian Van Dooren and Michel Simon for their technical assistances, the SAUR company that gave us access to the WWTP of Tancarville, Pr. A. Andremont and N. Grall (INSERM, IAME, UMR 1137) for the identification of presumptive Aeromonas using MALDI-TOF mass spectrometry and Pr Ingrid Padilla (Puerto Rico University, United States) for the critical review of the manuscript.
Funding. This work was supported by the scientific programs DYNAPAT and COBAC – Seine Aval (http://www.seine-aval.fr/) and the research federation (FR CNRS SCALE). GC held a research grant (FR SCALE) from the Haute-Normandie Regional Council (France).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01259/full#supplementary-material
References
- Alperi A., Martinez-Murcia A. J., Ko W.-C., Monera A., Saavedra M. J., Figueras M. J. (2010a). Aeromonas taiwanensis sp. nov. and Aeromonas sanarellii sp. nov., clinical species from Taiwan. Int. J. Syst. Evol. Microbiol. 60 2048–2055. 10.1099/ijs.0.014621-0 [DOI] [PubMed] [Google Scholar]
- Alperi A., Martinez-Murcia A. J., Monera A., Saavedra M. J., Figueras M. J. (2010b). Aeromonas fluvialis sp. nov., isolated from a Spanish river. Int. J. Syst. Evol. Microbiol. 60 72–77. 10.1099/ijs.0.011643-0 [DOI] [PubMed] [Google Scholar]
- Aujoulat F., Roger F., Bourdier A., Lotthé A., Lamy B., Marchandin H., et al. (2012). From environment to man: genome evolution and adaptation of human opportunistic bacterial pathogens. Genes 3 191–232. 10.3390/genes3020191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S., Granier S. A., Larvor E., Jouy E., Cineux M., Wilhelm A., et al. (2017). Aeromonas diversity and antimicrobial susceptibility in freshwater—an attempt to set generic epidemiological cut-off values. Front. Microbiol. 8:503 10.3389/fmicb.2017.00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova S., Kokotovic B., Skall H. F., Lorenzen N., Dalsgaard I. (2016). Detection and quantification of Aeromonas salmonicida in fish tissue by real-time PCR. J. Fish Dis. 40 231–242. 10.1111/jfd.12505 [DOI] [PubMed] [Google Scholar]
- Behbahani S. M. M. M., Akhlaghi M., Sharifiyazdi H. (2014). Phenotypic and genetic diversity of motile aeromonads isolated from diseased fish and fish farms. Iran. J. Vet. Res. 15 238–243. 10.22099/IJVR.2014.2533 [DOI] [Google Scholar]
- Berendonk T. U., Manaia C. M., Merlin C., Fatta-Kassinos D., Cytryn E., Walsh F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13 310–317. 10.1038/nrmicro3439 [DOI] [PubMed] [Google Scholar]
- Berthe T., Touron A., Leloup J., Deloffre J., Petit F. (2008). Faecal-indicator bacteria and sedimentary processes in estuarine mudflats (Seine, France). Mar. Pollut. Bull. 57 59–67. 10.1016/j.marpolbul.2007.10.013 [DOI] [PubMed] [Google Scholar]
- Cailleaud K., Forget-Leray J., Souissi S., Lardy S., Augagneur S., Budzinski H. (2007). Seasonal variation of hydrophobic organic contaminant concentrations in the water-column of the Seine Estuary and their transfer to a planktonic species Eurytemora affinis (Calanoïd, copepod). Part 2: alkylphenol-polyethoxylates. Chemosphere 70 281–287. 10.1016/j.chemosphere.2007.06.012 [DOI] [PubMed] [Google Scholar]
- Carpentier S., Moilleron R., Beltran C., Hervé D., Thévenot D. (2002). Quality of dredged material in the river Seine basin (France). II. Micropollutants. Sci. Total Environ. 299 57–72. 10.1016/S0048-9697(02)00278-4 [DOI] [PubMed] [Google Scholar]
- Chenia H. Y., Duma S. (2017). Characterization of virulence, cell surface characteristics and biofilm-forming ability of Aeromonas spp. isolates from fish and sea water. J. Fish Dis. 40 339–350. 10.1111/jfd.12516 [DOI] [PubMed] [Google Scholar]
- Colston S. M., Fullmer M. S., Beka L., Lamy B., Gogarten J. P., Graf J. (2014). Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. mBio 5:e2136–14 10.1128/mBio.02136-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreker D., Souissi S., Molinero J. C., Beyrend-Dur D., Gomez F., Forget-Leray J. (2010). Tidal and annual variability of the population structure of Eurytemora affinis in the middle part of the Seine Estuary during 2005. Estuar. Coast. Shelf Sci. 89 245–255. 10.1016/j.ecss.2010.07.010 [DOI] [Google Scholar]
- Dumontet S., Krovacek K., Baloda S. B., Grottoli R., Pasquale V., Vanucci S. (1996). Ecological relationship between Aeromonas and Vibrio spp. and planktonic copepods in the coastal marine environment in Southern Italy. Comp. Immunol. Microbiol. Infect. Dis. 19 245–254. 10.1016/0147-9571(96)00012-4 [DOI] [PubMed] [Google Scholar]
- Esteve C., Alcaide E., Giménez M. J. (2015). Multidrug-resistant (MDR) Aeromonas recovered from the metropolitan area of Valencia (Spain): diseases spectrum and prevalence in the environment. Eur. J. Clin. Microbiol. Infect. Dis. 34 137–145. 10.1007/s10096-014-2210-z [DOI] [PubMed] [Google Scholar]
- Esteve C., Gutierrez M. C., Ventosa A. (1995). Aeromonas encheleia sp. nov., isolated from European eels. Int. J. Syst. Evol. Microbiol. 45 462–466. 10.1099/00207713-45-3-462 [DOI] [PubMed] [Google Scholar]
- Figueira V., Vaz-Moreira I., Silva M., Manaia C. M. (2011). Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 45 5599–5611. 10.1016/j.watres.2011.08.021 [DOI] [PubMed] [Google Scholar]
- Figueras M. J., Beaz-Hidalgo R. (2015). “Aeromonas infections in humans,” in Aeromonas, ed. Graf J. (Norfolk: Caister Academic Press; ), 65–108. [Google Scholar]
- Fiorentini C., Barbieri E., Falzano L., Matarrese P., Baffone W., Pianetti A., et al. (1998). Occurrence, diversity and pathogenicity of mesophilic Aeromonas in estuarine waters of the Italian coast of the Adriatic Sea. J. Appl. Microbiol. 85 501–511. 10.1046/j.1365-2672.1998.853517.x [DOI] [PubMed] [Google Scholar]
- Garcia-Armisen T., Touron A., Petit F., Servais P. (2005). Sources of faecal contamination in the Seine estuary (France). Estuaries 28 627–633. 10.1007/BF02696073 [DOI] [Google Scholar]
- Guézennec L., Lafite R., Dupont J.-P., Meyer R., Boust D. (1999). Hydrodynamics of suspended particulate matter in the tidal freshwater zone of a macrotidal estuary (the Seine Estuary, France). Estuaries 22 717–727. 10.2307/1353058 [DOI] [Google Scholar]
- Gugliandolo C., Irrera G. P., Lentini V., Maugeri T. L. (2008). Pathogenic Vibrio, Aeromonas and Arcobacter spp. associated with copepods in the Straits of Messina (Italy). Mar. Pollut. Bull. 56 600–606. 10.1016/j.marpolbul.2007.12.001 [DOI] [PubMed] [Google Scholar]
- Hamid R., Ahmad A., Usup G. (2016). Pathogenicity of Aeromonas hydrophila isolated from the Malaysian Sea against coral (Turbinaria sp.) and sea bass (Lates calcarifer). Environ. Sci. Pollut. Res. 23 17269–17276. 10.1007/s11356-016-6655-8 [DOI] [PubMed] [Google Scholar]
- Hatha M., Vivekanandhan A. A., Julie Joice G., Christol (2005). Antibiotic resistance pattern of motile aeromonads from farm raised fresh water fish. Int. J. Food Microbiol. 98 131–134. 10.1016/j.ijfoodmicro.2004.05.017 [DOI] [PubMed] [Google Scholar]
- Henriques I. S., Fonseca F., Alves A., Saavedra M. J., Correia A. (2006). Occurrence and diversity of integrons and beta-lactamase genes among ampicillin-resistant isolates from estuarine waters. Res. Microbiol. 157 938–947. 10.1016/j.resmic.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Janda J. M., Abbott S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23 35–73. 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor W. C., Puah S. M., Tan J. A. M. A., Puthucheary S., Chua K. H. (2015). Phenotypic and genetic diversity of Aeromonas species isolated from fresh water lakes in Malaysia. PLoS ONE 10:e0145933 10.1371/journal.pone.0145933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamy B., Kodjo A. the colBVH Study Group Laurent F. (2009). Prospective nationwide study of Aeromonas infections in France. J. Clin. Microbiol. 47 1234–1237. 10.1128/JCM.00155-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif-Eugenín F., Beaz-Hidalgo R., Figueras M. J. (2016). Evaluation of different conditions and culture media for the recovery of Aeromonas spp. from water and shellfish samples. J. Appl. Microbiol. 121 883–891. 10.1007/s00203-016-1189-510.1111/jam.13210 [DOI] [PubMed] [Google Scholar]
- Laviad S., Halpern M. (2016). Chironomids’ relationship with Aeromonas species. Front. Microbiol. 7:736 10.3389/fmicb.2016.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wang W., Zhu Z., Chen A., Du P., Wang R., et al. (2015). Distribution, virulence-associated genes and antimicrobial resistance of Aeromonas isolates from diarrheal patients and water, China. J. Infect. 70 600–608. 10.1016/j.jinf.2014.11.004 [DOI] [PubMed] [Google Scholar]
- Magiorakos A.-P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Malham S. K., Rajko-Nenow P., Howlett E., Tuson K. E., Perkins T. L., Pallett D. W., et al. (2014). The interaction of human microbial pathogens, particulate material and nutrients in estuarine environments and their impacts on recreational and shellfish waters. Environ. Sci. Process. Impacts 16 2145–2155. 10.1039/c4em00031e [DOI] [PubMed] [Google Scholar]
- Marcel K. A., Antoinette A. A., Mireille D. (2002). Isolation and characterization of Aeromonas species from an eutrophic tropical estuary. Mar. Poll. Bull. 44 1341–1344. 10.1016/S0025-326X(02)00143-1 [DOI] [PubMed] [Google Scholar]
- Marti E., Balcázar J. L. (2015). Aeromonas rivipollensis sp. nov., a novel species isolated from aquatic samples. J. Basic Microbiol. 55 1435–1439. 10.1002/jobm.201500264 [DOI] [PubMed] [Google Scholar]
- Menanteau-Ledouble S., Kumar G., Saleh M., El-Matbouli M. (2016). Aeromonas salmonicida: updates on an old acquaintance. Dis. Aquat. Organ. 120 49–68. 10.3354/dao03006 [DOI] [PubMed] [Google Scholar]
- Monfort P., Baleux B. (1990). Dynamics of Aeromonas hydrophila, Aeromonas sobria, and Aeromonas caviae in a sewage treatment pond. Appl. Environ. Microbiol. 56 1999–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouny P., Dauvin J.-C. (2002). Environmental control of mesozooplankton community structure in the Seine estuary (English Channel). Oceanol. Acta 25 13–22. 10.1016/S0399-1784(01)01177-X [DOI] [Google Scholar]
- Pachepsky Y. A., Shelton D. R. (2011). Escherichia coli and fecal coliforms in freshwater and estuarine sediments. Crit. Rev. Environ. Sci. Technol. 41 1067–1110. 10.1080/10643380903392718 [DOI] [Google Scholar]
- Pal B. B., Pattnaik S. K., Mohanty A., Samal S. K., Khuntia H. K., Nayak S. K. (2016). Incidence of Aeromonas species isolated from Diarrhoea patients and water samples from coastal districts of Odisha, India. Int. J. Curr. Microbiol. Appl. Sci. 5 990–999. 10.20546/ijcmas.2016.507.111 [DOI] [Google Scholar]
- Patil H. J., Benet-Perelberg A., Naor A., Smirnov M., Ofek T., Nasser A., et al. (2016). Evidence of increased antibiotic resistance in phylogenetically-diverse Aeromonas isolates from semi-intensive fish ponds treated with antibiotics. Front. Microbiol. 7:1875 10.3389/fmicb.2016.01875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska M., Popowska M. (2014). The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann. Microbiol. 64 921–934. 10.1007/s13213-014-0911-2 [DOI] [Google Scholar]
- Popovic N., Kazazic S. P., Strunjak-Perovic I., Barisic J., Sauerborn Klobucar R., Kepec S., et al. (2015). Detection and diversity of aeromonads from treated wastewater and fish inhabiting effluent and downstream waters. Ecotoxicol. Environ. Saf. 120 235–242. 10.1016/j.ecoenv.2015.06.011 [DOI] [PubMed] [Google Scholar]
- Rasmussen-Ivey C. R., Hossain M. J., Odom S. E., Terhune J. S., Hemstreet W. G., Shoemaker C. A., et al. (2016). Classification of a hypervirulent Aeromonas hydrophila pathotype responsible for epidemic outbreaks in warm-water fishes. Front. Microbiol. 7:1615 10.3389/fmicb.2016.01615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger F., Marchandin H., Jumas-Bilak E., Kodjo A. colBVH study group Lamy B. (2012). Multilocus genetics to reconstruct aeromonad evolution. BMC Microbiol. 12:62 10.1186/1471-2180-12-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A. S., Bruun M. S., Dalsgaard I., Larsen J. L. (2001). Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl. Environ. Microbiol. 67 5675–5682. 10.1128/AEM.67.12.5675-5682.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servais P., Garcia-Armisen T., George I., Billen G. (2007). Fecal bacteria in the rivers of the Seine drainage network (France): sources, fate and modelling. Sci. Total Environ. 375 152–167. 10.1016/j.scitotenv.2006.12.010 [DOI] [PubMed] [Google Scholar]
- Shin H. B., Yoon J., Lee Y., Kim M. S., Lee K. (2015). Comparison of MALDI-TOF MS, housekeeping gene sequencing, and 16S rRNA gene sequencing for identification of Aeromonas clinical isolates. Yonsei Med. J. 56 550–555. 10.3349/ymj.2015.56.2.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva C. M., Evangelista-Barreto N. S., dos Fernandes Vieira R. H. S., Mendonça K. V., de Sousa O. V. (2014). Population dynamics and antimicrobial susceptibility of Aeromonas spp. along a salinity gradient in an urban estuary in Northeastern Brazil. Mar. Pollut. Bull. 89 96–101. 10.1016/j.marpolbul.2014.10.031 [DOI] [PubMed] [Google Scholar]
- Talagrand-Reboul E., Roger F., Kimper J.-L., Colston S. M., Graf J., Latif-Eugenín F., et al. (2017). Delineation of taxonomic species within complex of species: Aeromonas media and related species as a test case. Front. Microbiol. 8:621 10.3389/fmicb.2017.00621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunis P., Figueras M. J. (2016). Reassessment of the enteropathogenicity of mesophilic Aeromonas species. Front. Microbiol. 7:1395 10.3389/fmicb.2016.01395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touron A., Berthe T., Gargala G., Fournier M., Ratajczak M., Servais P., et al. (2007). Assessment of faecal contamination and the relationship between pathogens and faecal bacterial indicators in an estuarine environment (Seine, France). Mar. Pollut. Bull. 54 1441–1450. 10.1016/j.marpolbul.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Varela A. R., Nunes O. C., Manaia C. M. (2016). Quinolone resistant Aeromonas spp. as carriers and potential tracers of acquired antibiotic resistance in hospital and municipal wastewater. Sci. Total Environ. 542 665–671. 10.1016/j.scitotenv.2015.10.124 [DOI] [PubMed] [Google Scholar]
- Vaz-Moreira I., Varela A. R., Pereira T. V., Fochat R. C., Manaia C. M. (2015). Multidrug resistance in quinolone-resistant gram-negative bacteria isolated from hospital effluent and the municipal wastewater treatment plant. Microb. Drug Resist. 22 155–163. 10.1089/mdr.2015.0118 [DOI] [PubMed] [Google Scholar]
- WHO (2011). Technical Guidance on Water-Related Disease Surveillance. Available at: http://www.euro.who.int/_data/assets/pdf_file/0009/149184/e95620.pdf [Google Scholar]
- Yáñez M. A., Catalán V., Apráiz D., Figueras M. J., Martínez-Murcia A. J. (2003). Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 53 875–883. 10.1099/ijs.0.02443-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.