Abstract
GARP encoded by the Lrrc32 gene is the cell surface docking receptor for latent TGF-β which is expressed naturally by platelets and regulatory T cells. Although Lrrc32 is amplified frequently in breast cancer, the expression and relevant functions of GARP in cancer have not been explored. Here we report that GARP exerts oncogenic effects, promoting immune tolerance by enriching and activating latent TGF-β in the tumor microenvironment. We found that human breast, lung and colon cancers expressed GARP aberrantly. In genetic studies in normal mammary gland epithelial and carcinoma cells, GARP expression increased TGF-β bioactivity and promoted malignant transformation in immune deficient mice. In breast carcinoma-bearing mice that were immune competent, GARP overexpression promoted Foxp3+ regulatory T cell activity, which in turn contributed to enhancing cancer progression and metastasis. Notably, administration of a panel of GARP-specific monoclonal antibodies limited metastasis in an orthotopic model of human breast cancer. Overall, these results define the oncogenic effects of the GARP-TGF-β axis in the tumor microenvironment and suggest mechanisms that might be exploited for diagnostic and therapeutic purposes.
Introduction
GARP gene Lrrc32 was first discovered in the human chromosomal 11q13-14 region that is frequently amplified in breast cancer (1,2). The biological significance of GARP in cancer, however, is entirely unknown. The renewed interests in GARP were catalyzed by the finding that GARP is expressed by Foxp3+ regulatory T cells (Tregs) (3,4), but not conventional T cells or other immune cells except platelets (5). GARP is a type I transmembrane protein with a large ectodomain composed of 20 leucine-rich repeats, a transmembrane domain and a short cytoplasmic tail without obvious signaling motifs. The predicted configuration of GARP based on its primary amino acid sequence suggests that it is a cell surface acceptor molecule for the purpose of enriching ligands to cell surface, thus increasing the ligand availability. Indeed, GARP was later shown to be the docking receptor for latent transforming growth factor - β (TGF-β) (5–9), and was reported to increase the activation of latent TGF-β in an integrin-dependent fashion (6).
TGF-β is a pleiotropic cytokine expressed by most cells. Aberrance in its signaling has been implicated in multiple diseases, including cancer (10,11). In addition to causing growth arrest, TGF-β induces a variety of malignant phenotypes including invasion, loss of cellular adhesion, epithelial-mesenchymal transition and metastasis (10,12,13). Importantly, the role of TGF-β in shaping the tumor microenvironment is a critical aspect of its function in carcinogenesis. For example, TGF-β1 is a potent inducer of angiogenesis (14), by directly inducing VEGF expression (15), or recruiting other cells such as monocytes which in turn secrete pro-angiogenic molecules (16). TGF-β can also manipulate the tumor microenvironment to favor the evasion of cancer cells from immune surveillance via tampering with the antitumor functions of T cells, NK cells, B cells and other cells (17,18). This activity of TGF-β is mediated through its direct effect on these cells, as well as via its ability to induce Foxp3+ Tregs (19). Both cancer-intrinsic and immune-mediated effect of TGF-β in breast cancers have been described (20–24).
Biochemically, TGF-β exists in at least 4 different forms: 1) freely soluble active TGF-β; 2) soluble TGF-β associated with latency associated peptide (LAP) to form a TGF-β-LAP complex, known as latent TGF-β or LTGF-β); 3) LTGF-β associated covalently with large TGF-β-binding protein (LTBP), thus forming the TGF-β-LAP-LTBP complex; and 4) cell surface TGF-β (19,25), due primarily to its association with GARP (5–9). Only LAP-free TGF-β is known to be biologically active. Therefore, a large pool of TGF-β is sequestered in the extracellular matrix in the latent form before being activated by proteases such as matrix metalloproteinase (MMP)-2, MMP9 and plasmin (26–28), which are in turn secreted by tumor cells and other cells in the tumor microenvironment. Recently, it was reported that GARP-TGF-β can also be shed from the cell surface and that the soluble form of GARP-TGF-β has immunosuppressive roles (9,29,30).
To investigate a potential role of GARP-TGF-β axis in cancer, we examined GARP expression in a variety of epithelial cancer types including breast cancer and found that GARP was aberrantly upregulated compared to normal tissues. Importantly, by both gain- and loss-of-function studies, we found that GARP expression in cancer cells enhanced cancer invasion, epithelial-mesenchymal cell transition, immune tolerance and metastasis. Finally, we generated a panel of GARP-specific antibodies and demonstrated the therapeutic efficacy of GARP antibodies in a pre-clinical model of mammary carcinoma.
Materials and Methods
Cell lines and mice
Pre-B cell line (70Z/3) was a gift from Brian Seed (Harvard University) (31). 4T1, NMuMG and NMuMG* subline with silencing of hnRNP E1 were described previously (32). 70Z/3 was validated by flow cytometry using B cell lineage markers. Cancer cells were authenticated by gene expression analysis, in vivo growth and histology. All the lines were monitored for pathogens as per MUSC regulations and we routinely perform mycoplasma analysis on the lines. 293FT and other cell lines were purchased from ATCC.
BALB/c and NOD–Rag-1−/− mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All mouse experiments were approved by MUSC’s Institutional Animal Care and Use Committee, and the established guidelines were followed.
Human tumor microarrays
All human tumor microarrays (TMAs) were made out of formalin-fixed, paraffin embedded tissues, collected at the Medical University of South Carolina (Charleston, SC). Each patient specimen in these TMAs was represented in two cores on the slide and each core measured 1 mm in diameter. These patient specimens were available in a single core of 2 mm in diameter. Clinical and demographic information were obtained from the Cancer Registry of the Hollings Cancer Center at MUSC or provided by the commercial source. This study was approved by the Institutional Review Board (IRB) at MUSC.
Immunohistochemistry (IHC)
The mouse anti-human GARP (hGARP) antibody (ALX-804-867-C100, Enzo Life Sciences) was first verified by Western blot using hGARP-transfected HEK-293 cells and by IHC with hGARP-transfected 70Z/3 cells. Both analyses demonstrated specificity of the antibody and dilutions used from 1:250 (colon cancer) to 1:60 (all other cancers).
TMA slides were processed and antigen retrieved as described previously (33). For mouse IHC, tissue was either placed into OCT media for fresh frozen sections or fixed in 4% paraformaldehyde overnight. Fixed tissue was incubated in 70% ethanol overnight prior to paraffin embedding, and then cut for hematoxylin and eosin (H&E) staining. For p-Smad-2/3 on fresh frozen tumor sections, 5 μm sections were fixed with 4% paraformaldehyde followed by incubation with 3% H2O2. To minimize nonspecific staining, sections were incubated with the appropriate animal serum for 20 min at room temperature, followed by incubation with primary anti-p-Smad-2/3 antibody (EP823Y; Abcam) overnight at 4°C. Staining with secondary antibodies (Vectastain ABC Kit) was then performed before development using DAB substrate (Vector Labs SK-4100). The staining intensity of GARP and pSmad-2/3 was graded as follows with the sample identity blinded (0: negative; 1: faint; 2: moderate; 3: strong but less intense than 4; and 4: intense).
GARP knockdown by lentivirus-expressed short hairpin RNA
A lentivirus vector-expressing short hairpin RNA (shRNA) targeting the mouse GARP transcript was purchased from Sigma-Aldrich (St. Louis, MO). Ecotropic GARP shRNA and control scrambled lentiviral shRNA particles were produced in HEK293FT cells as described previously (34,35). To knock down GARP in NMuMG* cells, the cells were transduced with lentiviral supernatants targeting GARP and scrambled control. The knockdown efficiency was assessed by RT-PCR (Applied Biosystems Step-One Plus), flow cytometry (BD Verse) and Western blot using an anti-mouse GARP antibody (eBioscience).
Generation of GARP-expression vectors
GARP was amplified by PCR and subcloned between the BglII and HpaI sites in a MigR1 retroviral vector (34). A cDNA construct for expression of the GARP-Fc fusion protein (sGARP) was generated by joining the extracellular domain of GARP to the sequence encoding the Fc portion of murine IgG2a by PCR. Ecotropic GARP and sGARP retroviral particles were packaged into the Phoenix-Ecotropic cells. Virus propagation and transduction of Pre-B cells, 4T1 cells and NMuMG* cells were based on the established protocols (34,36). Cells were stably selected by culturing in presence of blasticidin 48 h post transduction for at least 72 h.
Purification of sGARP
For purification of sGARP, MigR1 vector was transfected into Chinese hamster ovary (CHO) cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Stably transfected clones were selected by blasticidin (5 μg/ml) and protein expression was quantified by SDS-PAGE and Western blot under reducing conditions using anti–mouse GARP and anti–mouse Fc antibody. Recombinant sGARP was purified from cell culture supernatants by protein A affinity chromatography (GE Health).
Generation and characterization of anti-GARP antibody
Four BALB/c mice were immunized with recombinant human GARP (R&D Systems, Minneapolis, MN) with Freund’s complete adjuvant, followed by boosting with SP2/0 cells stably expressing human GARP for 2–3 times. Splenic B cells from mice with high anti-GARP antibody titers were fused to SP2/0 cells in the presence of polyethylene glycol. Hybridomas were selected in HAT medium and cloned by limiting dilution assay. The specificity of antibody was screened and determined by ELISA and flow cytometry using 70Z/3 cells stably transduced with empty vector (70Z/3-EV) and overexpression of human GARP (70Z/3-GARP).
Protein extraction, immunoprecipitation, and Western blot analysis
Cells were harvested by trypsin-EDTA when necessary, washed in PBS, and lysed on ice in radio-immunoprecipitation assay (RIPA) lysis buffer in the presence of a protease inhibitor cocktail (Sigma-Aldrich). Nuclear-free protein lysate was quantified by Bradford assay (Bio-Rad), and an equal amount of lysate was analyzed by SDS-PAGE and Western blot under reducing conditions using anti–mouse GARP (AF6229; R&D system), anti-mouse Vimentin (D21H3; Cell signaling), anti-mouse E-Cadherin (24E10; Cell Signaling) and anti-mouse p-Smad-2/3 (EP823Y; Abcam) antibodies.
Cell proliferation and in vitro wound healing assay
To measure cell proliferation, 2.5 ×104 NMuMG* cells were seeded in a 96-well plate in complete medium (DMEM, 10% FCS, 1% penicillin-streptomycin) and incubated overnight. Proliferation was determined with 3-[4,5 dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), which was added to the cells at the indicated times and incubated for an additional 3 h at 37°C. The medium was then removed and mixed with 100 μl of DMSO for 15 minutes by shaking. Absorbance at 570 nm was then measured using a plate reader. The cell migration was measured by the wound-healing assay: at 100% confluence, two parallel wounds were made using a 1 ml pipette tip. Migration was assessed after 24, 48 and 72 hours and quantification of wound closure was measured using the ImageJ software (NIH).
4T1 Tumor model, CD25+ cell depletion and GARP antibody therapy
Female BALB/c mice, 6–8-week old were inoculated in the fourth mammary fat pad subcutaneously (s.q.) with 5 × 105 cells (4T1-EV, 4T1-GARP, or 4T1-sGARP). Tumor growth was monitored three times per week with a digital vernier caliper and tumor volume was calculated using the following formula: tumor volume (mm3) = [(width)2×length]/2. In GARP antibody therapy experiments, beginning at 3 days post-tumor inoculation, anti-GARP antibody or isotype-controlled antibody (0.1 mg/mouse in 0.1 mL PBS; three times per week) were administered intraperitoneally (i.p.) into mice. For combination therapy with cyclophosphamide (CY) and antibody, mice were treated with one injection of CY (4 mg/mouse) 3 days post-tumor inoculation in addition to the antibody treatment. For CD25+ cell depletion, mice received 500 μg PC61 antibody via i.p. administration every 4 days, beginning 2 days before 4T1 injection. At end-point, mice were sacrificed and the primary tumor, spleen and lungs were isolated. Primary tumors were weighted and IHC performed. Lung macro- and micro- metastases were determined by visual inspection and microscopic analysis respectively. Tumor infiltrated lymphocytes were isolated by Collagenase D (Sigma) digestion followed by Histopaque-1083 (Sigma) mediated density separation.
NMuMG tumor model
Female NOD-Rag-1−/− (n=5 each group; 6–8 week old) mice were inoculated in the fourth and left mammary fat pad using 5 × 105 cells (NMuMG*-EV, GARP knockdown NMuMG*). Animals were weighed and tumors measured weekly. At endpoint, primary tumors, lungs and livers were harvested. In another experiment, female NOD-Rag-1−/− mice (n=4–5 each group; 6–8 week old) were inoculated in the fourth left mammary fat pad with 5 × 105 cells (NMuMG-GARP-Luc, NMuMG-sGARP-Luc or NMuMG-Luc cells). In vivo luciferase imaging was evaluated weekly as follows: mice were intraperitoneally injected with D-luciferin (Perkin Elmer) at a dose of 150 mg/kg per mouse and anesthetized. Bioluminescence images were then acquired using Xenogen IVIS imaging system. Bioluminescence signal was quantified as photon flux (photons/second/cm2/steradian, or p/s/cm2/sr) in defined regions of interest using Living Image software (Xenogen).
Soluble TGF-β1 analysis
Active TGF-β1 and total TGF-β1 levels were measured using TGF-β1 ELISA kits (BioLegend, San Diego, CA) according to the manufacturer’s protocols.
Statistical Analysis
In TMAs where specimens were spotted in duplicate, the average of both cores was used as the representative value. The Student’s t-test was implemented to compare categorical variables such as normal versus cancer or different disease stages or categories. Kaplan-Meier analysis for correlation of GARP with survival was performed using X-tile software (37). Population characteristics were tested for statistically significant differences between low and high GARP expressers using Chi-squared test. Tumor curve analysis was performed using 2-way analysis of variance (ANOVA) or a Wad test. For bioluminescence imaging study, random effects linear regression was used to model tumor volume over time. To adhere to model assumptions, a square-root transform of tumor size was taken before model estimation. Bayesian information criteria was used to determine the best fitting model. The final model included three main effects of time, and interactions between group and time. Wald tests were used to compare coefficients and significance across groups; all other experiments were analyzed using Two-tailed Student’s t-test with GraphPad Prism. All data are presented as mean ± SEM. P values less than 0.05 were considered to be statistically significant.
Results
GARP expression and significance in mammary carcinoma
To examine GARP protein expression in archived formalin-fixed cancer specimens, we developed an IHC assay. The specificity of the anti-human GARP antibody was ascertained by its staining of a mouse mammary gland carcinoma cell line 4T1 stably expressing human GARP (Figure 1A). Given that Lrrc32 was amplified in human breast cancer (1), we evaluated GARP expression in this disease on a human breast cancer microarray. Analysis of uninvolved normal breast tissue (n=16) versus primary breast cancer (n=39) indicated a significant increase of GARP expression on cancer tissues (Figure 1B and 1C). By RT-PCR, Lrrc32 mRNA expression was increased by ≥ 2-fold in 28.5% of patients with breast cancer (n=42) compared with normal breast tissues (data not shown).
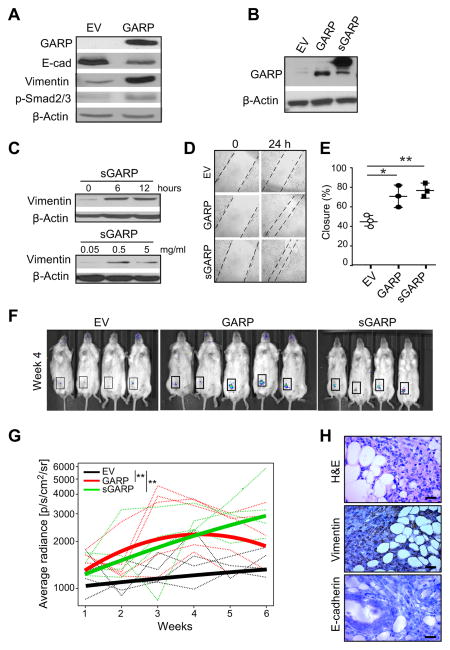
Figure 1. GARP expression in human breast cancer and its oncogenic roles in murine mammary gland epithelial cells.
(A) IHC validation of the human GARP antibody using murine 4T1 cells expressing human GARP (hGARP). Upper panels: staining of 4T1 cells expressing hGARP with anti-hGARP or control antibody. Lower panels: hGARP antibody was pre-adsorbed with 10 μg of recombinant hGARP (rhGARP) or 10 μg of bovine serum albumin (BSA) for 30 minutes at room temperature prior to the application on the slides. Scale bar: 40 μm. (B) Representative images of GARP IHC (brown color) of breast cancer with their matched normal breast tissues. Each patient specimen in these TMAs was represented in two cores on the slide and each core measured 1 mm in diameter. Scale bar: 50 μm. (C) Expression intensity of GARP-positive cells in breast cancer specimens and normal breast tissues. (D) shRNA knockdown of GARP mRNA in NMuMG* cells. Cells treated with scrambled shRNA (SCR) were used as control. Real-time RT-PCR was performed to quantify mRNA of Lrrc32 using β-actin as a control. MNE: mean normalized expression. (E) Flow cytometric analysis of cell surface GARP expression on GARP KD and SCR NMuMG* cells. (F) Immunoblot of GARP and TGF-β level in GARP KD and SCR NMuMG* whole cell lysates. (G) In vitro cell proliferation assay for GARP KD and SCR NMuMG* cells by MTT assay. (H) NMuMG* SCR and NMuMG*-GARP KD cells were injected into NOD-Rag-1−/− mice, followed by weekly monitoring of the tumor growth kinetics. (I) 120 days after tumor injection, lungs gross metastatic nodules were counted visually on the surface of the organ. **P<0.01. Statistical analysis was determined by 2-way ANOVA or two-tailed T-test, where appropriate. Data are representative of at least two independent experiments.
To understand the functional significance of GARP expression in mammary carcinoma, we screened murine mammary carcinoma cell lines for GARP expression by flow cytometry. A variant of the normal murine mammary gland epithelial cell line (NMuMG*), in which an RNA-binding protein hnRNPE1 is knocked down by RNA interference, was recently described as being capable of forming tumors in nude mice (38). Intriguingly, we found these cells expressed a significantly high level of endogenous GARP (Figure 1D–1F), raising the possibility that increased GARP expression, in addition to the silencing of the TGF-β-mediated translation repression complex, drives mammary cancer in this model. To test this hypothesis, we performed short hairpin RNA (shRNA) knock down (KD) of GARP in the NMuMG* cells (Figure 1D–1F). GARP silencing did not affect the in vitro proliferation of NMuMG* cells as determined by MTT assay (Figure 1G). Remarkably, silencing of GARP alone in the NMuMG* cells significantly attenuated their growth in vivo (Figure 1H). Further, the ability of these GARP KD cells to metastasize to the lungs was compromised (Figure 1I).
Enforced GARP expression in normal murine mammary epithelial cells upregulates TGF-β bioactivity and drives oncogenesis
In the parental NMuMG cells, TGF-β exerts both a growth inhibitory response and an epithelial-to-mesenchymal cell transition (EMT) response (39). As such, NMuMG cells have been extensively utilized to study TGF-β signaling and biology (40). We found that stable GARP-expressing NMuMG cells induced Smad-2/3 phosphorylation and expression of vimentin, but downregulated E-cadherin, consistent with increased canonical TGF-β signaling (Figure 2A). Recently it was found that GARP can also be secreted as a soluble form and that soluble GARP enhances the biologically active TGF-β (9,29,30). To further address this point, we generated a soluble GARP (sGARP) by fusing the entire ectodomain of GARP with the Fc portion of murine IgG (Figure 2B). We found indeed that sGARP can drive EMT by inducing time- and dose-dependent upregulation of vimentin (Figure 2C). We next performed a “scratch” assay to gauge the migratory properties of GARP-expressing cells. The closure rate of the scratch gap was significantly increased with GARP-expressing cells, indicating increased acquired migratory ability (Figure 2D and 2E). More importantly, we examined whether enforced GARP expression enabled NMuMG cells to establish tumors in vivo. We injected luciferase-expressing GARP-NMuMG or EV-NMuMG to female immunodeficient NOD-Rag-1−/− mice in the fourth mammary fat pad. By in vivo imaging of the bioluminescence, we found that the significant bioactive mass formed in mice that received GARP+ or sGARP+ NMuMG, but not in mice receiving EV transduced cells (Figure 2F and 2G). This tumor formation by GARP-expressing cells was confirmed by histology (Figure 2H). Collectively, we demonstrate that GARP has a transforming property via upregulation of TGF-β, identifying GARP as a potential novel oncogene.
Figure 2. Recombinant soluble GARP and enforced soluble or membrane-bound GARP expression drives epithelial-mesenchymal cell transition (EMT) and invasion of normal mammary gland epithelial cells.
(A) Immunoblot analysis for GARP, E-cadherin, Vimentin and Smad2/3 phosphorylation in NMuMG cells stably transfected with GARP or empty vector. (B) Immunoblot analysis for GARP in NMuMG cells stably transduced with expression vector for GARP, sGARP or empty vector. (C) NMuMG cells were treated for the indicated time points and with increasing doses of soluble GARP in serum-free culture medium, followed by immunoblot for vimentin and β-actin. (D) In vitro scratch assay to indicate the difference in the gap closure at 24 hours. (E) Summary statistics of three independent scratch assays. (F) In vivo imaging of the luceferin-enhanced bioluminescence in mice after injection of GARP, sGARP and control NMuMG cells, with images from Week 4. (G) Quantification of luciferase signal in the region of interests (boxed); data were weekly quantified by average radiance (photons/s/cm2/steradian). Thin lines represent data from individual mouse, whereas the three thick curves reflect mean values of each group. (H) Week 4 H&E and IHC analysis of Vimentin and E-cadherin expression (brown color) for GARP-expressing NMuMG tumors. Scale bar: 20 μm. *P<0.05; **P<0.01. Statistical significance was determined by the Wald test (panel G) and two-tailed T-test (panel E). Western Blot images are representative of 2 independent experiments and scratch assay are representative of 3 independent experiments.
GARP upregulation in murine mammary cancer cells promotes TGF-β activation, tumor growth, metastasis and immune tolerance
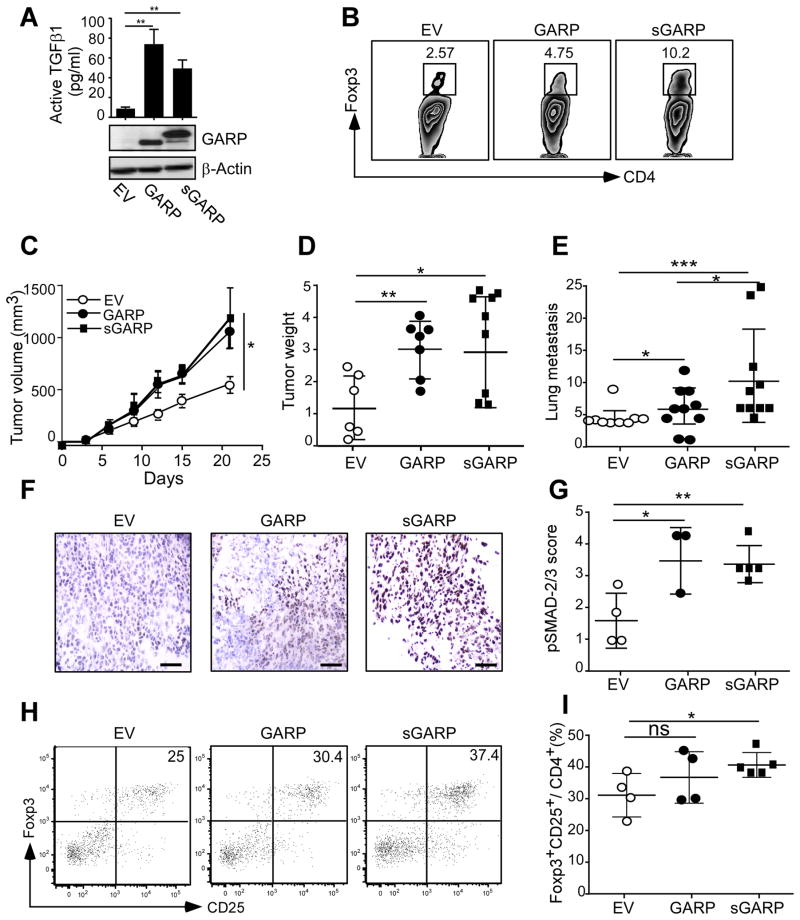
Cancer-intrinsic TGF-β signaling has been shown to promote breast cancer invasion and metastasis (11,20,21). The other aspect of TGF-β biology in cancer is its cancer-extrinsic role via modulating the host immune response (19), which is under-studied. We thus turned our attention next to an examination of how GARP impacts cancer growth and metastasis in a syngeneic immune-sufficient setting. We chose the highly aggressive and metastatic 4T1 mammary carcinoma model in BALB/c mice (41). Similar to the NMuMG system, over-expression of GARP or sGARP in 4T1 cells led to increased production of active TGF-β (Figure 3A). One of the key mechanisms by which TGF-β inhibits tumor-specific immunity is via the induction of Foxp3+ Tregs. To this end, purified naïve CD4+ T cells were cultured in vitro with conditioned media from 4T1-GARP, 4T1-sGARP and empty vector (EV) control cells in the presence of polyclonal T cell activators for 3 days. The conditioned media from GARP-expressing cells was 2–3 fold more efficient at inducing Treg differentiation compared to medium from control cells (Figure 3B). We next injected 4T1-EV, 4T1-GARP and 4T1-sGARP cells orthotopically in the fourth right mammary fat pad of 6–8 weeks old female BALB/c mice. We found that GARP-expressing cells were more aggressive, as indicated by both increased growth kinetics of the primary tumor (Figure 3C and 3D) and increased lung metastasis (Figure 3E). We found that this aggressiveness correlated with enhanced TGF-β signaling in the tumor microenvironment as determined by increased p-Smad-2/3 in cancer cells (Fig 3F and 3G), as well as by expansion of Foxp3+ tolerogenic Treg cells (Figure 3H and 3I).
Figure 3. GARP upregulation in murine mammary cancer cells promotes TGFβ activation, tumor growth, metastasis and immune tolerance.
(A) Concentration of active TGFβ in 3 days conditioned medium measured by ELISA, and immunoblot for GARP in 4T1 cells stably expressing GARP, sGARP or EV. (B) Treg differentiation assay. Naïve splenic CD4+CD25− cells were stimulated with agonistic antibody against CD3 and CD28 for 3 days in the presence of 50% 72-hour conditioned media from 4T1-GARP, 4T1-sGARP, or 4T1-EV cells. Flow cytometry was then performed to define the percentage of Treg conversion from naïve non-Treg cells. (C) Female BALB/c mice were injected in the 4th mammary fat pad with 5×105 tumor cells. Tumor volume was measured every 3 days. (D) Three weeks after tumor injection, mice were sacrificed and the primary tumors were resected and weighted. (E) Lungs were isolated, paraffin embedded, and H&E stained for histological analysis. The number of micro-metastatic tumor nodules in the lungs were enumerated by a pathologist. (F) Portions of the primary tumors were isolated and embedded in OCT and snap frozen. The fresh frozen sections were stained for the presence of p-SMAD-2/3; representative images are shown. Scale bar: 20 μm. (G) Summary statistics for p-SMAD-2/3 staining intensity, defined independently by a pathologist (S.S). (H) Tumor-infiltrating lymphocytes were isolated and the percentages of CD4+CD25+Foxp3+ Tregs in CD4+ cells were determined by flow cytometry with representative flow plots. (I) Summary of the percentage of Tregs in the tumor microenvironment. *P<0.05; **P<0.01; ***P<0.001. Statistical significance was determined by two-tailed Student’s t-test or Two-way ANOVA, where appropriate. Data is representative of at least two independent experiments.
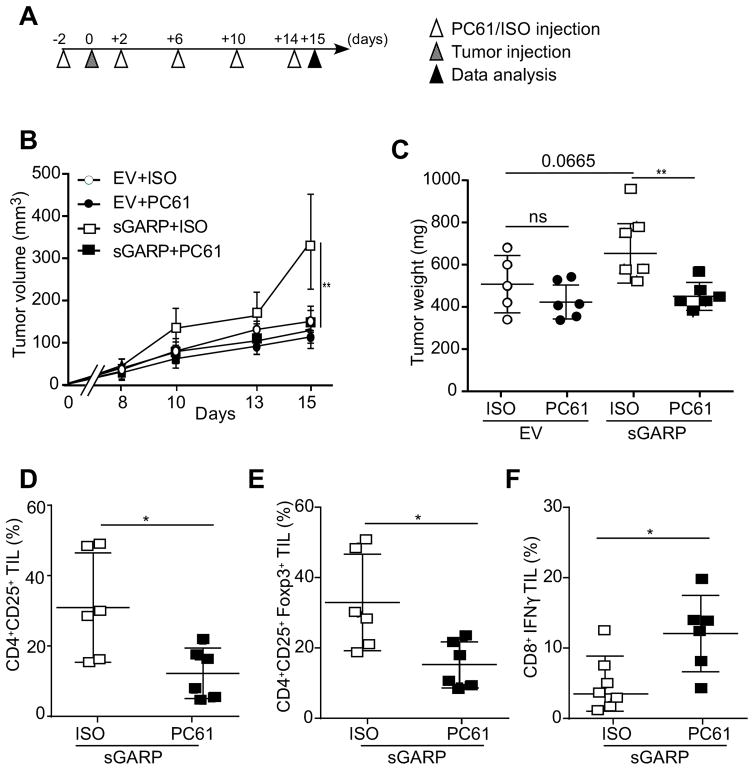
Depletion of CD25+ Treg cells abolished the aggressiveness of soluble GARP expressing mammary tumors
To determine if the increased aggressiveness of 4T1-sGARP cells was mediated by Tregs, we next depleted CD25+ Treg cells with CD25-specific antibody PC61 (42,43), followed by implanting either 4T1-EV or 4T1-sGARP cells (Figure 4A). Indeed, Treg depletion abolished the aggressiveness of 4T1-sGARP measured by tumor growth kinetics (Figure 4B), as well as tumor volume (Figure 4C). We also examined the tumor-infiltrating T cells, and confirmed that PC61 injection resulted in reduction of CD4+CD25+ (Figure 4D) and CD4+CD25+Foxp3+ Tregs (Figure 4E), and increased percentage of IFNγ-producing CD8+ T cells (Figure 4F).
Figure 4. Depletion of CD25+ T cells abolished the aggressiveness of soluble GARP expressing mammary tumors.
(A) Experimental design. Female BALB/c mice received intra peritoneal injection of PC61 anti-CD25 monoclonal antibody (100 μl ascites) every 4 days, starting 2 days before tumor injection. On Day 0 mice were injected in the 4th mammary fat pad with 5×105 tumor cells. Mice were sacrificed on day 15. (B) Tumor volume measured every 3 days (n=5–6). (C) Mice were sacrificed at day 15 after tumor injection and the primary tumors were resected and weighted. (D–E) Tumor-infiltrating lymphocytes were isolated and the percentages of CD4+CD25+ T cells, CD4+CD25+Foxp3+ Tregs in the whole CD4+ population were determined by flow cytometry. (F) Tumor-infiltrating CD8+ T lymphocytes were isolated, ex vivo stimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin. IFNγ production was quantified by flow cytometry. *p<0.05; **P<0.01. Significance was determined by the Wald test (panel B) and two-tailed Student’s t-test (panels C–F).
GARP is a novel therapeutic target in cancer
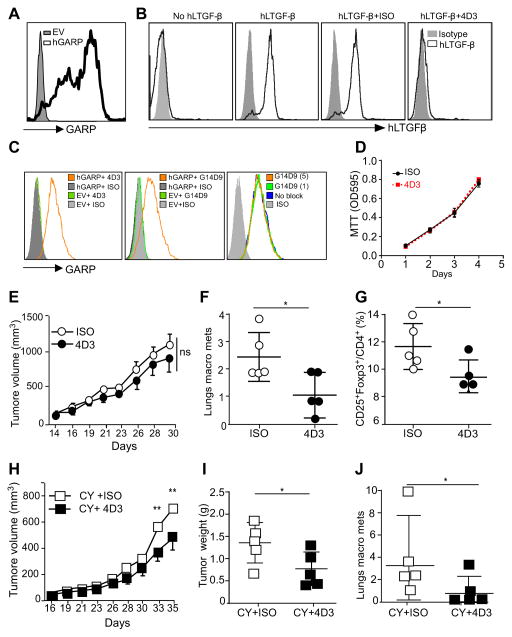
We next determined whether GARP could serve as a novel therapeutic target in cancer, using an antibody-based strategy. For the generation of anti-GARP monoclonal antibodies (mAbs), mice were immunized with recombinant human GARP, followed by boosting with irradiated whole myeloma SP2/0 cells stably expressing human GARP, with the aim of generating mAbs against GARP that were conformation-specific. More than 20 mAbs were generated that specifically recognize human GARP as determined by flow cytometry. All of these clones were specific for human GARP (hGARP) including clone 4D3 (Figure 5A). Importantly, 4D3 but none of the other clones, was able to block the binding of exogenous human LTGFβ-1 (huLTGFβ-1) to surface GARP (Figure 5B). By sequential staining of hGARP-expressing cells, we found that 4D3 recognize a different epitope from the one reactive to a commonly used GARP antibody from commercial sources (clone G14D9) (Figure 5C). The mere presence of 4D3 did not inhibit tumor cell growth in vitro (Figure 5D). To examine if GARP antibody had any direct anti-tumor activities in vivo against the primary tumor or metastasis, BALB/c mice were inoculated orthotopically with 4T1-GARP cells. Mice were then treated with either 4D3 (IgG1) or isotype control antibody (ISO), with or without a single dose of cyclosphosphamide (CY). This regimen was chosen because it was shown previously that a TGF-β-neutralizing antibody 1D11 was able to potentiate the ability of CY to control 4T1 (44). We found that 4D3 did not inhibit the primary tumor growth (Figure 5E), but it significantly blunted lung metastasis compared with the isotype-treated group (Figure 5F). The reduction of lung metastasis was associated with decreased Tregs in the blood (Figure 5G). Concomitant treatment with chemotherapy and 4D3 resulted in significant better control of primary tumors (Figure 5H and 5I), as well as the lung metastasis (Figure 5J).
Figure 5. GARP-specific antibody has therapeutic value against preclinical model of breast cancer.
(A) Surface staining of pre-B cells stably expressing human GARP (pre-B-hGARP) by 4D3 GARP antibody. Grey histogram represents staining with isotype control antibody. (B) pre-B-hGARP cells were incubated without or with human LTGFβ (huLTGF-β), in the presence of GARP antibodies or Isotype control antibody. Cells were then stained for cell surface hLTGF-β, in order to determine the activity of the antibody to block the binding of hLTGF-β to GARP. (C) 4T1-EV (empty vector) cells and 4T1-GARP (human GARP overexpression cells) was stained with 4D3 or a commercial anti-GARP antibody (G14D9), followed by flow cytometry. To determine if G14D9 blocks staining by 4D3 antibody, cells were pre-incubated with 1 or 5 μg/ml G14D9 antibody, followed by 4D3 antibody (1 μg/ml) and appropriate fluorochrome-labeled secondary antibody, and flow cytometric analysis. (D) 4T1-GARP cells were seeded in triplicates in a 96-well plate (2,000 cells per well) with 10 μg/ml 4D3 or mouse IgG followed by MTT assay kinetically. (E) BALB/c mice were injected with 5×105 4T1-hGARP mammary tumors orthotopically in the 4th mammary fat pad, followed by IP injection of 200 μg/mouse of 4D3 GARP antibody every three days. The primary tumor growth kinetics was measured three times a week. Significance is indicated at various time points along the tumor curves. (F) Four weeks after tumor injection, mice were sacrificed. Lungs were isolated and the numbers of visible metastatic nodules were counted. (G) Spleens were harvested and percentage of Treg (FoxP3+CD25+CD4+) cells in the antibody treated mice versus Isotype antibody groups were determined by flow cytometry. (H) BALB/c mice were injected with 5×105 4T1-hGARP mammary tumors orthotopically in the 4th mammary fat pad, followed by one dose of cyclophosphamide (CY) and IP injection of 200 μg/mouse 4D3 GARP antibody every three days. The primary tumor growth kinetics was measured three times a week. Significance is indicated at various time points along the tumor curves. (I) Five weeks after tumor injection, mice were sacrificed, tumors were excised and weighted. (J) Lungs were isolated and numbers of visible metastatic nodules were counted. *P<0.05; **P<0.01; ***P<0.001. Significance was determined by two-tailed Student’s t-test. Results are representative of two independent experiments.
Broad GARP expression in human cancers
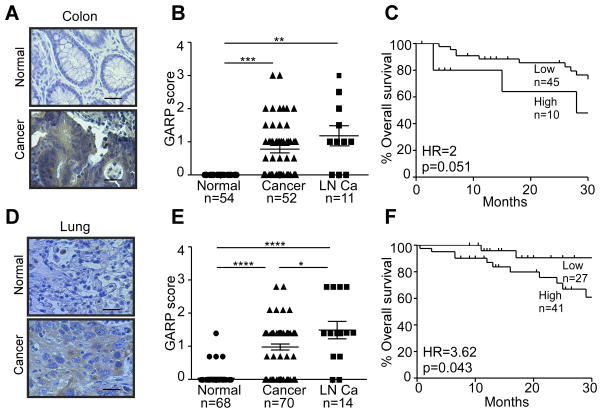
Finally, we reasoned that if GARP expression is an important mechanism for immune tolerance and oncogenesis, aberrant GARP expression shall not be restricted to the breast cancer. The recent cancer genomic revolution, including The Cancer Genome Atlas (TCGA) effort, indeed unveiled that Lrrc32 is amplified in up to 30% of patients with many human cancer types, including ovarian, lung, breast, and head and neck cancers (data not shown). By IHC, normal colonic epithelial cells showed no significant GARP positivity (Figure 6A). However, the primary colon cancers and lymph node (LN) metastatic lesions stained variably positive for GARP (uniformly negative with isotype control antibody, data not shown) (Figure 6A). On a scale of 0 to 4, GARP intensity score ranged between 0 and 3, averaging at 0.78 (p=1.1×10−8) in primary colon cancers and 1.18 (p=0.003) in LN metastasis (Figure 6B). Higher GARP expression is associated with the trend of worsening overall survival (Figure 6C). Similarly, we found significantly increased GARP levels in primary cancers of the lung and lymph node metastasis (Figure 6D and 6E). More importantly, patients with higher GARP expression (staining intensity ≥ 1) showed significant worse survival comparing with patients with no or lower GARP expression (staining intensity < 1) (Figure 6F). Overall, we demonstrated for the first time that GARP is widely expressed in human cancers, suggesting that it is a general mechanism in oncogenesis and immune tolerance.
Figure 6. GARP upregulation in human colon and lung cancer correlates with poor prognosis.
(A) Representative images of GARP IHC (brown color) of colon cancers with their respective patient matched normal tissue. Each patient specimen in these TMAs was represented in two cores on the slide and each core measured 1 mm in diameter. Scale bar: 20 μm. (B) Expression intensity of GARP-positive cells in colon cancer. (C) Correlation between GARP expression and overall survival of colon cancer. (D) Representative images of GARP IHC (brown color) of lung cancers with their respective patient matched normal tissue. Each patient specimen in these TMAs was represented in two cores on the slide and each core measured 1 mm in diameter. Scale bar: 20 μm. (E) Expression intensity of GARP-positive cells in lung cancer and normal lungs. (F) Correlation between GARP expression and overall survival of lung cancer. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. Statistical significance was analyzed by two-tailed Student’s t-test or Kaplan Meier analysis; the numbers of samples (n) are indicated.
Discussion
Surface expression of TGF-β in cancer has been recognized (45,46), but its biological significance has remained elusive. This is in part due to lack of understanding of the molecular basis for membrane-bound TGF-β. The discovery of GARP to be the sole cell surface docking receptor for latent TGF-β finally created an experimental opportunity to manipulate the level of surface TGF-β through altering GARP. In this study, we demonstrated that the GARP-TGF-β axis plays both cancer-intrinsic and extrinsic roles in promoting oncogenesis.
Using both human and mouse systems with a combination of genetic and immunological tools, we have made several fundamental discoveries in this study: (i) GARP is widely expressed by human cancer cells, but less so by normal epithelial cells, and GARP expression correlates with advanced stage of cancer and poor prognosis; (ii) GARP itself has a transformation potential, which renders normal mammary gland epithelial cells tumorigenic; (iii) GARP expression in cancer cells leads to increased TGF-β activity, likely due to its ability to concentrate LTGF-β in cis as well as in trans, contributing to cancer aggressiveness and metastasis; (iv) GARP expression in the tumor microenvironment promotes the induction of Treg cells and thus blunts the function of effector T cells against cancers; and (v) neutralizing GARP by blocking its ability to bind to TGF-β reduces tumor metastasis, even in the absence of chemotherapy or shrinkage of primary tumors.
Mechanistically, we discovered that GARP expression enhances TGF-β activation. We demonstrated this in both NMuMG and 4T1 that enhanced GARP expression translated into increased canonical signaling, such as phosphorylation of Smad-2/3. Importantly, the accumulation of active TGF-β within the tumor microenvironment impairs anti-tumor immunity through multiple mechanisms including the induction of Treg cells (47,48). We investigated the latter utilizing a syngeneic tumor model, namely 4T1 mammary carcinoma (BALB/c). Our data indicate that GARP expression in 4T1 induces Treg cells which blunts the ability of effector T cells to control cancer. Thus, by positively regulating TGF-β in the tumor microenvironment, GARP promotes oncogenesis through cancer-intrinsic as well as cancer-extrinsic mechanism.
The oncogenic roles of TGF-β span from promoting invasion, metastasis and angiogenesis, to maintaining stemness and inducing immune tolerance (11). Thus, TGF-β remains an attractive target for the treatment of cancer. However, the development of therapeutics that target TGF-β has thus far been hampered by multiple factors, not the least of which is the presence of multiple ligands in multiple forms and the context-dependent function of this pleotropic cytokine (49). Surface expression of GARP provides cancer cells a means to concentrate TGF-β locally and thus influence TGF-β-dependent growth, transformation and invasion through activation of both integrins and the TGF-β receptors. GARP therefore represents a unique alternative target for blocking the TGF-β pathway in the tumor microenvironment. In this study, we generated a panel of highly specific mAbs against human GARP, and demonstrated that 1 of these mAbs, 4D3, with or without cyclophosphamide, successfully treated 4T1-hGARP tumors by inhibiting lung metastasis. We believe that GARP-targeted mAbs may exert their therapeutic benefits via multiple mechanisms, including: 1) inhibition of LTGF-β binding to GARP and thus blockade of TGF-β activation, 2) direct tumor-killing through antibody-dependent cell cytotoxicity (ADCC) and complement-dependent cell cytotoxicity (CDCC) and, 3) Ablation of immunosuppressive Treg cells. Interestingly, 4D3 clone is murine IgG1 isotype which has very limited ADCC and CDCC activities (data not shown). Thus, the therapeutic effect of 4D3 is likely through its ability to block the binding between LTGF-β and GARP, a speculation that is in part supported by the in vitro binding assay and the reduction of Tregs in the tumor microenvironment. Moreover, although the effect of 4D3 alone on the primary tumor growth appeared to be marginal, it however did potentiate the effect of cyclophosphamide. Thus, anti- GARP antibody may be beneficial therapeutically against both primary and metastatic cancers in conjunction with other modalities.
Finally, we found that GARP expression may serve as a novel biomarker for cancer. GARP is aberrantly expressed in breast, colon and lung cancers. Although there was a limited sample size for breast and colon cancer, we demonstrated that higher expression of GARP by lung cancer correlates with poorer prognosis. The clinical significance of this finding warrants further investigation to determine particularly if GARP expression defines a subgroup of patients with immune evasive signature and if they thus may benefit the most from immunotherapy.
In conclusion, we have discovered that GARP is a novel oncogene due to its cancer-intrinsic roles in promoting invasion and metastasis, as well as its cancer-extrinsic roles in inducing immune tolerance. It may serve as a novel diagnostic and therapeutic target for cancer.
Acknowledgments
Grant support: This work was supported in part by NIH grants: P01CA186866, R01CA188419, R01AI070603 and P30CA138313 (Z. Li), TL1 TR001451 and UL1 TR001450 (C.W. Fugle), as well as P30 CA138313 (Z. Li., P. Howe and E. Garrett-Mayer).
We thank Drs. Feng Hong, Ephraim Ansa-Addo, Eric Meissner and Jordan Morreall for critical reading of the manuscript, MUSC Center for Biomedical Imaging and Pathology Core Facilities for Technical Assistant.
Footnotes
Conflict of interests: A provisional patent has been filed for developing GARP and GARP-based antibodies in oncology for diagnostic and therapeutic purpose.
References
- 1.Szepetowski P, Ollendorff V, Grosgeorge J, Courseaux A, Birnbaum D, Theillet C, et al. DNA amplification at 11q13. 5-q14 in human breast cancer. Oncogene. 1992;7(12):2513–7. [PubMed] [Google Scholar]
- 2.Ollendorff V, Noguchi T, deLapeyriere O, Birnbaum D. The GARP gene encodes a new member of the family of leucine-rich repeat-containing proteins. Cell growth & differentiation: the molecular biology journal of the American Association for Cancer Research. 1994;5(2):213–9. [PubMed] [Google Scholar]
- 3.Wang R, Kozhaya L, Mercer F, Khaitan A, Fujii H, Unutmaz D. Expression of GARP selectively identifies activated human FOXP3+ regulatory T cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(32):13439–44. doi: 10.1073/pnas.0901965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R, Wan Q, Kozhaya L, Fujii H, Unutmaz D. Identification of a regulatory T cell specific cell surface molecule that mediates suppressive signals and induces Foxp3 expression. PloS one. 2008;3(7):e2705. doi: 10.1371/journal.pone.0002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM. GARP (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated FOXP3+ regulatory T cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(32):13445–50. doi: 10.1073/pnas.0901944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R, Zhu J, Dong X, Shi M, Lu C, Springer TA. GARP regulates the bioavailability and activation of TGFbeta. Molecular biology of the cell. 2012;23(6):1129–39. doi: 10.1091/mbc.E11-12-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stockis J, Colau D, Coulie PG, Lucas S. Membrane protein GARP is a receptor for latent TGF-beta on the surface of activated human Treg. European journal of immunology. 2009;39(12):3315–22. doi: 10.1002/eji.200939684. [DOI] [PubMed] [Google Scholar]
- 8.Edwards JP, Fujii H, Zhou AX, Creemers J, Unutmaz D, Shevach EM. Regulation of the expression of GARP/latent TGF-beta1 complexes on mouse T cells and their role in regulatory T cell and Th17 differentiation. Journal of immunology. 2013;190(11):5506–15. doi: 10.4049/jimmunol.1300199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn SA, Stahl HF, Becker C, Correll A, Schneider FJ, Tuettenberg A, et al. Soluble GARP has potent antiinflammatory and immunomodulatory impact on human CD4(+) T cells. Blood. 2013;122(7):1182–91. doi: 10.1182/blood-2012-12-474478. [DOI] [PubMed] [Google Scholar]
- 10.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nature genetics. 2001;29(2):117–29. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 11.Massague J. TGFbeta in Cancer. Cell. 2008;134(2):215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oft M, Heider KH, Beug H. TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8(23):1243–52. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 13.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, et al. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Molecular biology of the cell. 2001;12(1):27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, et al. Transforming growth factor type beta: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(12):4167–71. doi: 10.1073/pnas.83.12.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pertovaara L, Kaipainen A, Mustonen T, Orpana A, Ferrara N, Saksela O, et al. Vascular endothelial growth factor is induced in response to transforming growth factor-beta in fibroblastic and epithelial cells. J Biol Chem. 1994;269(9):6271–4. [PubMed] [Google Scholar]
- 16.Sunderkotter C, Goebeler M, Schulze-Osthoff K, Bhardwaj R, Sorg C. Macrophage-derived angiogenesis factors. Pharmacol Ther. 1991;51(2):195–216. doi: 10.1016/0163-7258(91)90077-y. [DOI] [PubMed] [Google Scholar]
- 17.Kehrl JH, Wakefield LM, Roberts AB, Jakowlew S, Alvarez-Mon M, Derynck R, et al. Production of transforming growth factor beta by human T lymphocytes and its potential role in the regulation of T cell growth. The Journal of experimental medicine. 1986;163(5):1037–50. doi: 10.1084/jem.163.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopp HG, Placke T, Salih HR. Platelet-derived transforming growth factor-beta down-regulates NKG2D thereby inhibiting natural killer cell antitumor reactivity. Cancer research. 2009;69(19):7775–83. doi: 10.1158/0008-5472.CAN-09-2123. [DOI] [PubMed] [Google Scholar]
- 19.Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134(3):392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegel PM, Shu W, Cardiff RD, Muller WJ, Massague J. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(14):8430–5. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133(1):66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanks BA, Holtzhausen A, Evans KS, Jamieson R, Gimpel P, Campbell OM, et al. Type III TGF-beta receptor downregulation generates an immunotolerant tumor microenvironment. The Journal of clinical investigation. 2013;123(9):3925–40. doi: 10.1172/JCI65745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nature reviews Cancer. 2006;6(7):506–20. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 24.Bos PD, Plitas G, Rudra D, Lee SY, Rudensky AY. Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. The Journal of experimental medicine. 2013;210(11):2435–66. doi: 10.1084/jem.20130762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran DQ. TGF-beta: the sword, the wand, and the shield of FOXP3(+) regulatory T cells. J Mol Cell Biol. 2012;4(1):29–37. doi: 10.1093/jmcb/mjr033. [DOI] [PubMed] [Google Scholar]
- 26.Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes & development. 2000;14(2):163–76. [PMC free article] [PubMed] [Google Scholar]
- 27.Sato Y, Rifkin DB. Inhibition of endothelial cell movement by pericytes and smooth muscle cells: activation of a latent transforming growth factor-beta 1-like molecule by plasmin during co-culture. J Cell Biol. 1989;109(1):309–15. doi: 10.1083/jcb.109.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyons RM, Gentry LE, Purchio AF, Moses HL. Mechanism of activation of latent recombinant transforming growth factor beta 1 by plasmin. J Cell Biol. 1990;110(4):1361–7. doi: 10.1083/jcb.110.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fridrich S, Hahn SA, Linzmaier M, Felten M, Zwarg J, Lennerz V, et al. How Soluble GARP Enhances TGFbeta Activation. PloS one. 2016;11(4):e0153290. doi: 10.1371/journal.pone.0153290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gauthy E, Cuende J, Stockis J, Huygens C, Lethe B, Collet JF, et al. GARP is regulated by miRNAs and controls latent TGF-beta1 production by human regulatory T cells. PloS one. 2013;8(9):e76186. doi: 10.1371/journal.pone.0076186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randow F, Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nature cell biology. 2001;3(10):891–6. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- 32.Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MC, et al. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Molecular cell. 2011;41(4):419–31. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rachidi SM, Qin T, Sun S, Zheng WJ, Li Z. Molecular profiling of multiple human cancers defines an inflammatory cancer-associated molecular pattern and uncovers KPNA2 as a uniform poor prognostic cancer marker. PloS one. 2013;8(3):e57911. doi: 10.1371/journal.pone.0057911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S, Hong F, Gewirth D, Guo B, Liu B, Li Z. The molecular chaperone gp96/GRP94 interacts with Toll-like receptors and integrins via its C-terminal hydrophobic domain. The Journal of biological chemistry. 2012;287(9):6735–42. doi: 10.1074/jbc.M111.309526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong F, Liu B, Chiosis G, Gewirth DT, Li Z. alpha7 Helix Region of alphaI Domain Is Crucial for Integrin Binding to Endoplasmic Reticulum Chaperone gp96: A POTENTIAL THERAPEUTIC TARGET FOR CANCER METASTASIS. The Journal of biological chemistry. 2013;288(25):18243–8. doi: 10.1074/jbc.M113.468850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Wu BX, Metelli A, Thaxton JE, Hong F, Rachidi S, et al. GP96 is a GARP chaperone and controls regulatory T cell functions. The Journal of clinical investigation. 2015;125(2):859–69. doi: 10.1172/JCI79014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10(21):7252–9. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 38.Howley BV, Hussey GS, Link LA, Howe PH. Translational regulation of inhibin betaA by TGFbeta via the RNA-binding protein hnRNP E1 enhances the invasiveness of epithelial-to-mesenchymal transitioned cells. Oncogene. 2015 doi: 10.1038/onc.2015.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie L, Law BK, Aakre ME, Edgerton M, Shyr Y, Bhowmick NA, et al. Transforming growth factor beta-regulated gene expression in a mouse mammary gland epithelial cell line. Breast cancer research: BCR. 2003;5(6):R187–98. doi: 10.1186/bcr640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell research. 2009;19(2):156–72. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pulaski BA, Ostrand-Rosenberg S. Mouse 4T1 breast tumor model. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 20. Chapter 20. 2001. p. 2. [DOI] [PubMed] [Google Scholar]
- 42.McHugh RS, Shevach EM. Cutting edge: depletion of CD4+CD25+ regulatory T cells is necessary, but not sufficient, for induction of organ-specific autoimmune disease. Journal of immunology. 2002;168(12):5979–83. doi: 10.4049/jimmunol.168.12.5979. [DOI] [PubMed] [Google Scholar]
- 43.Dai J, Liu B, Ngoi SM, Sun S, Vella AT, Li Z. TLR4 Hyperresponsiveness via Cell Surface Expression of Heat Shock Protein gp96 Potentiates Suppressive Function of Regulatory T Cells. Journal of immunology. 2007;178(5):3219–25. doi: 10.4049/jimmunol.178.5.3219. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Yang Y, Zhou Q, Weiss JM, Howard OZ, McPherson JM, et al. Effective chemoimmunotherapy with anti-TGFbeta antibody and cyclophosphamide in a mouse model of breast cancer. PloS one. 2014;9(1):e85398. doi: 10.1371/journal.pone.0085398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker K, Raut P, Jass JR. Colorectal cancer cells express functional cell surface-bound TGFbeta. International journal of cancer Journal international du cancer. 2008;122(8):1695–700. doi: 10.1002/ijc.23312. [DOI] [PubMed] [Google Scholar]
- 46.Ahn YO, Lee JC, Sung MW, Heo DS. Presence of membrane-bound TGF-beta1 and its regulation by IL-2-activated immune cell-derived IFN-gamma in head and neck squamous cell carcinoma cell lines. Journal of immunology. 2009;182(10):6114–20. doi: 10.4049/jimmunol.0803725. [DOI] [PubMed] [Google Scholar]
- 47.Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limon P. The polarization of immune cells in the tumour environment by TGFbeta. Nature reviews Immunology. 2010;10(8):554–67. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beyer M, Schultze JL. Regulatory T cells in cancer. Blood. 2006;108(3):804–11. doi: 10.1182/blood-2006-02-002774. [DOI] [PubMed] [Google Scholar]
- 49.Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nature reviews Drug discovery. 2012;11(10):790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]