Abstract
Chronic alcohol exposure produces widespread neuroadaptations and alterations in gene expression in human alcoholics and animal models. Technological advances in the past decade have increasingly highlighted the role of non-protein-coding RNAs (ncRNAs) in the regulation of gene expression and function. These recently characterized molecules were discovered to mediate diverse processes in the central nervous system, from normal development and physiology to regulation of disease, including alcoholism and other psychiatric disorders. This review will investigate the recent studies in human alcoholics and rodent models that have profiled different classes of ncRNAs and their dynamic alcohol-dependent regulation in brain.
Keywords: alcohol, non-coding RNA, microRNA, long noncoding RNA, transcriptome, gene regulation, next generation sequencing
II. Introduction
Non-coding RNA
Protein-coding genes have traditionally been the most well studied sequences in the human genome; however, these genes account for less than 2% of known structural and regulatory molecular elements (Alexander, Fang, Rozowsky, Snyder, & Gerstein, 2010). In recent years, it has become increasingly clear that the non-protein-coding portion of the genome is functionally important and is required for normal development and physiology, and is also linked with a number of diseases (Cech & Steitz, 2014; Mercer, Dinger, & Mattick, 2009). This fundamental change in our understanding of the complexity of the transcriptome has been the result of improved RNA sequencing (RNA-seq) technologies, which allow exploration of gene structure and regulation in unprecedented detail (Fig. 1). Data generated from these advanced techniques have transformed our view of the central dogma that DNA is transcribed into RNA, which is translated into protein. Non-coding RNAs (ncRNAs) are emerging as key transcriptional and post-translational regulators, representing a large and diverse class of regulatory molecules (Alexander et al., 2010). ncRNAs make up a sizeable portion of the transcriptional landscape of the cell (Carninci & Hayashizaki, 2007; Carninci et al., 2005), but the precise functions of many non-coding elements remain largely unknown. Defining the biological roles carried out by multiple classes of ncRNAs is an expanding area of transcriptomics that will likely rival the large number and diversity represented by the proteome. Currently, ncRNAs are grouped into three general subclasses based on nucleotide number (small, 18–31 nt; medium, 31–200 nt; and long, >200 nt), with each class having regulatory potential and specific subcellular localization (Alexander et al., 2010; Costa, 2005; Dozmorov, Giles, Koelsch, & Wren, 2013). These ncRNAs include transfer and ribosomal RNA (tRNA and rRNA, respectively), small nucleolar RNA (snoRNA), microRNA (miRNA), small interfering RNA (siRNA), small nuclear (snRNA), extracellular RNA (exRNA), piRNAs, and small Cajal body-specific RNA (scaRNA), as well as different classes of long ncRNA (lncRNA), including intergenic (lincRNA) and intronic RNA. Identifying roles for ncRNAs will provide a better understanding of cellular function, as well as new insight into gene regulation of disease.
Fig. 1.
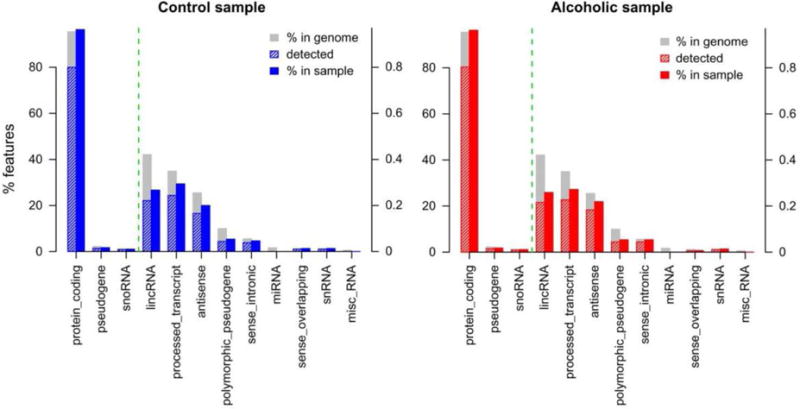
RNA features detected by RNA-seq in the prefrontal cortex of alcoholic and matched control samples. Bar plots depict the percentage of features detected in representative control (blue) and alcoholic (red) samples. The left axis shows the percentage of features for the top three biotypes and the right axis shows the percentage of remaining biotypes (separated by dotted green vertical line). Protein-coding transcripts were the predominant feature detected in both groups.
Genomic studies based on DNA and RNA sequencing have identified thousands of ncRNAs in diverse animal genomes. Until recently, the conservation of ncRNAs between human and nonhuman primates was limited compared to human-mouse conservation, due to limitations in the quality of nonhuman primate genome annotations. However, sequencing technology has resulted in more complete and correct assemblies, thus greatly improving genome annotation quality. Systematic curation efforts have enabled the development of several cross-species ncRNA databases (Ulitsky, 2016). Identification of cross-species conservation is a key question when evaluating the functional impact of specific ncRNAs. For example, if a ncRNA is associated with a human condition, it is important to know the extent to which it can be studied in model organisms. Conversely, if a ncRNA is discovered in a model organism, evidence of conservation will be critical for establishing relevance to a human condition.
Distinct ncRNA mechanisms in brain are thought to influence the development of psychiatric diseases (Kocerha, Dwivedi, & Brennand, 2015; Sartor, St Laurent, & Wahlestedt, 2012), including alcoholism (Farris & Mayfield, 2014). Rapid advances in this field have largely come from next-generation sequencing technologies such as RNA-seq that have been critical for genome-wide identification of novel transcripts (Fig. 1). Previous review articles have focused on ncRNAs (primarily miRNAs) that are altered in response to alcohol administration (Balaraman, Tingling, Tsai, & Miranda, 2013; Farris & Mayfield, 2014; Most, Workman, & Harris, 2014; Nunez & Mayfield, 2012; Pietrzykowski, 2010). This review focuses on the effects of alcohol on ncRNAs from studies published since 2012. It should be noted that there are few studies focusing on the association of genetic risk to ncRNAs in animal models, representing an area of future research.
III. MicroRNA
The expanding fields of genetics and genomics over the previous 15 years have highlighted the growing number of genes that can potentially influence alcohol-drinking behavior in humans and animal models. In particular, the discovery of miRNAs (Lee, Feinbaum, & Ambros, 1993) and their mechanisms of action are revolutionizing our understanding of gene regulation in physiology and disease (Mattick & Makunin, 2006; Morris & Mattick, 2014). These short (~17–24 nt) ncRNAs act as post-transcriptional modulators of gene expression by binding to miRNA-recognition elements in their numerous target genes. miRNA-mediated gene suppression occurs through multiple mechanisms, including interruption of translational initiation, 5′ decapping, alternative splicing, 3′ deadenylation, and exonuclease degradation (Fabian & Sonenberg, 2012; Krol, Loedige, & Filipowicz, 2010). miRNA biogenesis includes gene transcription by RNA polymerase II that typically binds to promoters near DNA sequences encoding precursor miRNAs (pre-miRNA). The resulting transcript is capped, polyadenylated, and spliced (Hammond, 2015). Many pre-miRNAs are derived from intronic transcriptional regions; however, novel mechanisms have been identified demonstrating that traditional RNA splicing events can negatively regulate the processing of pre-miRNAs that overlap exon-intron junctions (Melamed et al., 2013). These distinctly different RNA processing mechanisms (miRNA processing and RNA splicing) underscore the regulatory potential of these short ncRNAs.
miRNAs play important roles in neuronal differentiation, developmental timing, synapse function, and neurogenesis (Fiorenza & Barco, 2016). miRNAs are thought to act as ‘master regulators’ of gene expression, and a recent RNA-seq study of 13 different types of human tissue identified over 3,700 mature miRNAs (Londin et al., 2015), compared to the >2,700 listed in release 20 of miRBase (Kozomara & Griffiths-Jones, 2014). In addition, the sequence conservation across human and nonhuman primate-specific lineage is quite high (>94% of the newly discovered miRNAs). Given the vast number of miRNAs identified to date and the expression silencing of large collections of target genes, there is considerable regulatory potential of these molecules.
Alcohol-responsive miRNAs in human postmortem brain
Because of their regulatory functions, it is reasonable to expect that miRNAs are also critical mediators of alcohol’s effects. Early studies demonstrated that alcohol alters miRNA levels and miRNA-regulated systems that are associated with tolerance, gut leakiness, and neural stem cell proliferation and differentiation (Miranda et al., 2010; Pietrzykowski et al., 2008). Similarly, expression-profiling studies in postmortem brains of human alcoholics have shown that the transcriptional reprogramming that takes place is brain region-specific and may reflect both pre-existing differences in gene expression and alterations in response to alcohol consumption (Nunez & Mayfield, 2012; Nunez, Truitt, Gorini, Ponomareva, Blednov, et al., 2013). In addition, epigenetic reprogramming primarily mediated by direct methylation of DNA and acetylation, methylation, and phosphorylation of histone proteins, appears to contribute to the altered gene expression observed in alcoholics and animal models of excessive alcohol consumption (Krishnan, Sakharkar, Teppen, Berkel, & Pandey, 2014; Miranda, 2014). The first transcriptome-wide study of alcohol-responsive miRNAs in human alcoholics identified ~35 upregulated human miRNAs in the prefrontal cortex (PFC) (Lewohl et al., 2011). This study included both miRNA and mRNA whole genome microarrays and used integrative statistical analyses to demonstrate that the predicted mRNAs targeted by upregulated miRNAs were significantly over-represented among the downregulated mRNAs, with no over-representation detected among the set of significantly upregulated mRNAs. This supports a role for miRNA-dependent inhibition of gene expression in the PFC of human alcoholics. The magnitude of changes in miRNA levels was relatively small (20–30%), with few changes exceeding 40%. It is likely that the small changes detected in miRNAs, as well as mRNA expression, in PFC of human alcoholics is due to an increased expression that is localized to a specific cellular compartment, such as the neuronal synapse. Such compartmentalized, enhanced differential expression has been demonstrated in an animal model where miRNAs were isolated from synaptoneurosomes when compared to total, unfractionated tissue (Most, Leiter, Blednov, Harris, & Mayfield, 2016).
Another whole genome study examined miRNA expression from frontal cortex (Broadman area 9) of human alcoholics to further characterize the effects of alcohol consumption on predicted target mRNA expression (Manzardo, Gunewardena, & Butler, 2013). Similar to Lewohl et al. (2011), the majority of differentially expressed miRNAs were upregulated in alcohol-dependent subjects compared with controls, including a cluster of four miRNAs (miR-299-3p, miR-377, miR-379, and miR-493) from the maternally expressed 14q32 chromosomal region. The predicted mRNA targets of these upregulated miRNAs were involved in cellular adhesion, tissue differentiation, neuronal migration, myelination, and oligodendrocyte proliferation. These findings suggest that white matter abnormalities observed in alcoholism may be linked to upregulated miRNAs from this chromosomal region, and are consistent with the earlier finding that upregulation of miRNAs in the PFC of alcoholics targets mRNAs important for lipid biosynthesis and myelination (Lewohl et al., 2011).
The whole transcriptome studies outlined above profiled miRNA levels from total homogenate preparations, but we must consider that miRNA regulation of gene expression may specifically shape synaptic structure and function. Synaptoneurosomes (SNs) contain membrane vesicles of pre- and postsynaptic compartments of neurons and perisynaptic compartments of astrocytes and microglia (Raab-Graham, Haddick, Jan, & Jan, 2006), and provide a model for studying the synaptic transcriptome. Alcohol-responsive miRNAs have been examined in SNs prepared from amygdala of mice following chronic two-bottle choice (2BC) drinking (Most et al., 2016). This research was microarray-based and used different informatics approaches to identify key alcohol-sensitive miRNA-mRNA synaptic interactions. These miRNAs and mRNAs demonstrated overlapping patterns of expression that correlated with alcohol consumption, and a significant number of the alcohol-responsive mRNAs and miRNAs were unique to glutamate neurons. These findings point to the utility of the SN preparation in studying cell-specific signaling and the ability of chronic alcohol to perturb coordinated miRNA regulation of mRNAs as a mechanism to disrupt synaptic plasticity and thereby alter brain function. Some of the alcohol-sensitive synaptic miRNAs in mouse brain were also found in human (miR-18a, miR-203, miR-369*, miR-92a, and miR-423) (Lewohl et al., 2011) and rat brain (miR-137, miR-187, miR-18a, miR-34c*, miR-369*, miR-374, miR-382*, miR-423, miR-488, and miR-92b) (Tapocik et al., 2013), suggesting that overlapping alcohol-responsive miRNAs are conserved across different species and models of alcohol consumption.
Recently, bioinformatic approaches were used to overlay gene networks from both miRNAs and mRNAs in the nucleus accumbens (NAc) of postmortem brain (Mamdani et al., 2015). Weighted gene co-expression network analysis (WGCNA; Langfelder & Horvath, 2008) was used to define networks of co-expressed genes from genome-wide miRNA and mRNA datasets to identify multiple coding- and ncRNA gene modules that correlated significantly with DSM-IV alcohol dependence. This study included an additional layer of analyses that combined network hub gene expression with genome-wide genotypic data from an independent genetic sample from the Collaborative Studies on the Genetics of Alcoholism (COGA) to identify numerous mRNA and miRNA cis-eQTLs significantly enriched for alcohol dependence, representing an important step toward identification of alcohol-relevant eQTLs from gene expression in human postmortem brain. This study also identified cell-type specific mRNA modules that were enriched for both neuronal genes (primarily downregulated), as well as astrocyte and microglial marker genes (primarily upregulated). Among these modules, multiple functional categories of genes were enriched with MAPK and cytokine signaling pathways, consistent with early studies that identified genes involved in neuroimmune signaling and function from human alcoholics and rodent models of excessive drinking (Liu et al., 2006; Mayfield, Ferguson, & Harris, 2013; Mulligan et al., 2006; Robinson et al., 2014).
Alcohol-responsive miRNAs in rodent models
Rodent drinking models have been instrumental in defining alcohol-responsive miRNAs and their functional relevance based on responses to expression manipulation. These animal models are advantageous because select miRNAs can be upregulated or downregulated using viral vectors, mimics (molecules designed to simulate naturally occurring mature miRNAs), and antagomirs (artificial antisense oligonucleotides) to determine their effect on alcohol-related behaviors. Studies have demonstrated that alcohol-induced changes in miRNAs are associated with cellular tolerance to alcohol (Pietrzykowski et al., 2008), antianxiety effects (Teppen, Krishnan, Zhang, Sakharkar, & Pandey, 2015), cellular reward mechanisms (Li et al., 2013), regulation of alcohol consumption and preference (Bahi & Dreyer, 2013; Li et al., 2013; Most et al., 2016; Tapocik et al., 2014), episodes of binge drinking (Darcq et al., 2015; Nunez, Truitt, Gorini, Ponomarev, Harris, et al., 2013; Tian et al., 2016), dependence/withdrawal (Gorini, Nunez, & Mayfield, 2013; Tapocik et al., 2013, 2014), and alcohol-induced conditioned-place preference (Bahi & Dreyer, 2013). Recent research articles published since 2012 are listed in Table 1.
Table 1.
MicroRNAs implicated in alcohol studies (2012 to 2016).
| Species/Model | Region/System | miRNA(s) | Citation(s) |
|---|---|---|---|
| Human | * PFC | miR-130a ↓ | Wang, Gelernter, & Zhang, 2013 |
| * FC (Broadmann 9) | multiple ↑ | Manzardo et al., 2013 | |
| * NAc | multiple ↑↓ | Mamdani et al., 2015 | |
| Rat | * DLS | miR-124a ↓ | Bahi & Dreyer, 2013 |
| * NAc | miR-382 ↓ | Li et al., 2013 | |
| * mPFC | multiple ↑↓ | Tapocik et al., 2013 | |
| Hippocampus | miR-10a-5p, miR-26a, miR-103, miR-495 | Prins, Przybycien-Szymanska, Rao, & Pak, 2014 | |
| * mPFC | miR-206 ↑ manipulationa | Tapocik et al., 2014 | |
| Amygdala | miR-494 ↓ | Teppen et al., 2015 | |
| * PFC | miR-125a-5p ↓ | Tian et al., 2016 | |
| Mouse | * FC and midbrain | multiple ↑↓ | Gorini, Nunez, & Mayfield, 2013 |
| * FC | multiple ↑↓ | Nunez, Truitt, Gorini, Ponomareva, Harris, et al., 2013 | |
| * mPFC | miR-30a-5p ↑ manipulationa | Darcq et al., 2015 | |
| Amygdala | multiple ↑↓ | Most et al., 2016 | |
| Cell Culture | Primary mouse cortical neurons | miR-155, miR-186, miR-24, and miR-375 ↑ | Bekdash & Harrison, 2015 |
| Primary mouse microglial cells | miR-339-5p ↑ | Zhang, Wei, Di, & Zhao, 2014 | |
| Primary rat cortical neurons | miR-125a-5p ↓ manipulationa | Tian et al., 2016 |
Frontal cortex (FC); Prefrontal cortex (PFC); Medial prefrontal cortex (mPFC); nucleus accumbens (NAc); dorsal striatum (DLS)
↑upregulated; ↓downregulated; ↑↓both up- and down-regulation of multiple miRNAs
manipulation; specific manipulation with viral vectors, miRNA mimics, or antagomirs
A comprehensive and integrative analysis of alcohol-responsive miRNAs (Exiqon miRNA arrays) and proteins (2-dimensional differential in-gel electrophoresis followed by MALDI tandem mass spectrometry) from mouse cortex and midbrain was reported using a mouse model of voluntary alcohol consumption and dependence (Gorini, Nunez, et al., 2013; Gorini, Roberts, & Mayfield, 2013). Bioinformatic analyses identified modules of co-expressed miRNAs highly correlated with predicted target genes encoding differentially expressed proteins (Gorini, Roberts, et al., 2013). Overall, miRNA profiles from alcohol-dependent animals segregated into clear clusters of predominantly upregulated miRNAs that correlated with predominantly downregulated proteins. Key regulatory molecules were associated with the escalation of alcohol consumption to dependence (e.g., miR-532-3p and miR-339-5p on Pea15 in the midbrain), and included genes encoding co-expressed proteins that are targeted by the same miRNA (e.g., miR-494-3p on both Dpysl2 and Dpysl3 and miR-140-3p on co-expressed Flot1 and Dnm1 in the cortex). This analysis of global miRNA and protein expression levels from different brain regions of alcohol-dependent mice suggests a synergistic regulation of miRNAs and proteins in the behavioral transition from alcohol consumption to dependence.
A separate study utilizing a rat model of alcohol dependence examined the relationships among miRNAs and mRNAs that are correlated with genes involved in disruption of synaptic processes and neuroplasticity (Tapocik et al., 2013). Multiple miRNAs, a number of which are also differentially expressed in human alcoholics (Lewohl et al., 2011), and mRNAs were differentially expressed in the medial PFC (mPFC) in response to chronic alcohol exposure in rats. This study identified miR-206, a primarily upregulated miRNA target which has also been implicated to have functional relevance with brain-derived neurotrophic factor (BDNF) expression (Duman & Monteggia, 2006; Hansson, Rimondini, Heilig, Mathé, & Sommer, 2011). BDNF, an activity-regulated neurotrophin with known 3′-UTR binding sites for miR-206, was a confirmed downregulated mRNA. Building upon these findings, subsequent research confirmed that miR-206 is upregulated in a brain region-specific manner in response to dependence-induced drinking. Upregulation was observed only in mPFC and not in the ventral tegmental area, amygdala, or NAc (Tapocik et al., 2014). Viral-mediated overexpression of miR-206 decreased BDNF protein expression and induced escalation of alcohol self-administration in nondependent rats, further supporting a role for miR-206 in alcohol dependence. This study provides strong evidence for a direct role of miR-206 in escalation of alcohol self-administration.
Rodent studies designed to model binge-like drinking in humans have identified numerous alcohol-responsive miRNAs. For example, whole-genome miRNA studies found many differentially regulated miRNAs in binge-alcohol models in adolescent rats (Prins, Przybycien-Szymanska, Rao, & Pak, 2014; Tian et al., 2016) and adult mice (Gorini, Nunez, et al., 2013; Most et al., 2016; Nunez, Truitt, Gorini, Ponomarev, Harris, et al., 2013). Using a 2BC-drinking in the dark (DID) paradigm in mice bred for high alcohol consumption (C57BL/6J × FVB/NJ hybrid), Nunez and colleagues (Nunez, Truitt, Gorini, Ponomarev, Harris, et al., 2013) found that miRNA expression was predominantly upregulated in the PFC of alcohol-drinking mice (52 miRNA families), a finding consistent with previous results from human alcoholic PFC (Lewohl et al., 2011). In addition, approximately 40–80% of the common differentially expressed genes were changed in opposite directions in human and mouse models, with the majority of genes being upregulated in the alcohol-treated mouse brain but downregulated in human alcoholic brain. These results demonstrate 1) a high degree of conservation of alcohol-responsive genes and pathways from mouse to human and 2) unique differences in the direction of gene expression changes between the two models. These distinctions may be a consequence of the duration of alcohol exposure, which lasted for most of the adult life in the case of the human alcoholics (long-term adaptations), but only 20 days during early adulthood in the binge-drinking mice (early stages of adaptation). Co-expression network analysis identified modules of genes affected by alcohol and miRNAs with biological functions that include synaptic vesicle-mediated transport, endocytic recycling, and neuroimmune signaling mediated by chemokine and Toll-like receptor signaling. The pathways that were both upregulated by alcohol and over-targeted by upregulated miRNAs were enriched in downregulated modules, suggesting that these may be involved in the early stages of chronic alcohol consumption and trigger a miRNA adaptive response aimed at counteracting such activation. miR-34c-5p and let-7g-5p, in particular, appear to be central regulators of hub genes in the alcohol-responsive gene modules. Differentially expressed and PCR-validated miRNA families that were identified in both mouse and human included let-7, miR-7, miR-15, miR-101, miR-140, and miR-152, and the validated miRNAs specific to the mouse model included miR-195 (member of the miR-15 family) and miR-541 family members. There was also significant overlap among differentially expressed mouse miRNA families and those reported in PFC of ethanol-treated rats (Tapocik et al., 2013). The miRNA families with altered expression in both mice and rats included miR-7, miR-9, miR-10, miR-15, miR-17, miR-26, miR-29, miR-30, miR-101, miR130, miR-181, miR-204, miR-339, miR-340, miR-368, miR-434, and miR-467. Overall, these results underscore the relevance of gene regulation by miRNAs in response to alcohol consumption and suggest conservation of alcohol-responsive miRNA regulatory pathways from rodent to human.
In contrast to the effects of alcohol on BDNF signaling pathways outlined above (Tapocik et al., 2013, 2014), BDNF mRNA increased in the dorsal striatum of rodents following moderate levels of alcohol intake in a chronic continuous-access test (Bahi & Dreyer, 2013) and an operant model of self-administration (Jeanblanc et al., 2009), and the increase was attenuated by prolonged daily exposure to alcohol (Jeanblanc et al., 2009). Prolonged access also resulted in decreased BDNF levels in the cortex, suggesting that binge alcohol consumption dysregulates BDNF expression in corticostriatal brain pathways (Jeanblanc et al., 2009). A subsequent study by the same group found that BDNF mRNA levels were also decreased in mPFC using a model that mimics binge alcohol drinking in humans (Darcq et al., 2015), and these changes were accompanied by increased expression levels of miR-3-a-5p, miR-195 and miR-1. A similar increase in miR-1 was seen in the PFC of human alcoholics (Lewohl et al., 2011). Detailed follow-up studies indicated that overexpression of miR-30a-5p (using viral vectors) in the mPFC decreased BDNF expression and increased alcohol intake and preference, whereas knockdown (LNA-miRNA; antagomers) decreased excessive alcohol drinking (Darcq et al., 2015). These findings, as well as those from adolescent binge models (Prins et al., 2014), suggest that there is a dynamic miRNA-dependent regulation of the BDNF pathway that is associated with the transition from moderate to uncontrolled alcohol intake.
IV. Long Non-coding RNA
Long non-coding RNAs (lncRNAs; >200 nt) are the most abundant class of RNAs. Compared to the approximately 20,000 protein-coding genes in humans (Carninci & Hayashizaki, 2007; Jia et al., 2010; Ravasi, 2006), current annotations indicate that there are >16,000 lncRNAs, the majority of which show tissue- and temporal-specific expression in the CNS (Derrien et al., 2012). lncRNAs are critical to normal cellular function, including the regulation of gene expression (Wang & Chang, 2011), cell proliferation and differentiation (Guttman et al., 2011), as well as the pathophysiology of disease (Wapinski & Chang, 2011). The expression level of lncRNAs is highest in brain (Derrien et al., 2012), and similar to protein-coding genes; they are regulated by neuronal activity (Barry et al., 2014) and show cell type- and brain region-specific expression patterns (Belgard et al., 2011; Mercer, Dinger, Sunkin, Mehler, & Mattick, 2008), suggesting that their expression is important in discrete CNS functions. Although the physiological roles of lncRNAs are still emerging, they appear to be involved in cis-regulation of neighboring genes (Goff et al., 2015; Zuo et al., 2016) and the regulation of gene expression involved in adaptive behavior (Bekdash & Harrison, 2015; Spadaro et al., 2015). The extent to which specific lncRNAs contribute to the development and maintenance of alcohol- or other substance-abuse disorders is currently an under-investigated area, with the potential to uncover novel regulatory gene networks involved in the addictive process.
According to the positional relationship between lncRNAs and their associated protein-coding genes, lncRNAs are classified as intergenic (lincRNA; located between protein-coding genes and at least 1 kb away from the nearest protein-coding genes), intronic (located within the intron of annotated protein-coding genes), anti-sense (RNA molecules that are transcribed from the opposite strand of many protein-coding genes), and sense overlapping (considered transcript isoforms of protein-coding mRNAs) (Zuo et al., 2016).
lncRNA and psychiatric disease
Data from psychiatric and substance-abuse disorders (Sartor et al., 2012) clearly demonstrate regulatory roles for these molecules; however, there are few studies that establish a direct causal link between lncRNAs and regulation of drinking in animal models or humans. As data rapidly accumulate, it will remain a challenge to determine which lncRNAs are associated with specific disease states and to identify the mechanisms and regulatory pathways that underlie these conditions. Current approaches such as sequence classification, association studies, differential expression, and informatic analyses have been useful in predicting the regulatory potential of lncRNAs in psychiatric conditions, including Alzheimer’s disease (Faghihi et al., 2008; Magistri, Velmeshev, Makhmutova, & Faghihi, 2015), schizophrenia (Barry et al., 2014; Perkins, Jeffries, & Sullivan, 2005), autism spectrum disorders (Ziats & Rennert, 2013), as well as substance dependence (Sartor et al., 2012; Sullivan, Daly, & O’Donovan, 2012; Zuo et al., 2016) discussed in the following section.
lncRNA and alcohol dependence
To date, there have been few studies of how lncRNAs are regulated by alcohol exposure, but alterations in lncRNAs have been reported following nicotine, heroin, and cocaine exposure (Sartor et al., 2012; Zuo et al., 2016). The majority of alcohol research has focused on miRNAs; however, there is accumulating evidence that lncRNAs are also involved in synaptic plasticity changes associated with drug abuse and dependence. These transcripts are known to mediate epigenetic factors that affect gene expression (Khalil et al., 2009; Lee, 2012; Wang et al., 2011), act as endogenous competitors (Cesana et al., 2011), regulate alternative splicing events (Barry et al., 2014; Massone et al., 2011; Tripathi et al., 2010), control neuronal development (Pollard et al., 2006), and guide synaptic plasticity (Bond et al., 2009). These diverse roles suggest that lncRNA expression may prove to have significant regulatory control in alcohol dependence and other psychiatric diseases.
Transcriptome studies
There are few studies of lncRNA dysregulation in alcohol dependence, and none that identify mechanisms underlying functional changes in response to chronic alcohol use. Interestingly, there may be overlap in drug-responsive lncRNAs altered by alcohol dependence and other substance-abuse disorders. For example, nuclear-enriched abundant transcript 2 (NEAT2), an lncRNA important in synaptogenesis (Bernard et al., 2010), was upregulated in human alcoholic brain (Kryger, Fan, Wilce, & Jaquet, 2012). NEAT2, and other related lncRNAs, were also upregulated in the NAc of heroin (e.g., NEAT1, MIAT, and MEG3) and cocaine abusers (e.g., MIAT, MEG3, and EMX2OS) (Albertson et al., 2004; Michelhaugh et al., 2011). Thus, generalized substance abuse/dependence may produce overlapping changes in related lncRNAs.
Transcriptional network analyses using RNA-seq have also been used to identify networks of co-expressed genes that include lincRNAs such as LINC00092, LINC00174, and LINC00284 (Farris, Arasappan, Hunicke-Smith, Harris, & Mayfield, 2014). The function of these ncRNAs is currently unknown, but may involve gene regulation in human disease (Mercer et al., 2009). The co-expression patterns of these lincRNAs suggest a cooperative role in cellular plasticity that shapes patterns of human alcohol consumption. RNA-seq analysis of postmortem alcoholic brain tissue (Farris & Mayfield, unpublished) identified over 130 differentially expressed intergenic lncRNAs (p < 0.05) across four brain regions. The RNA-seq analysis was based on conservative estimates of gene expression according to the RefSeq database, which focuses on curated genome sequences.
Epigenetic studies (methylation)
Epigenetic studies have established a link between alcohol use and hypomethylation of genomic regions critical for embryonic development (Ouko et al., 2009). There was a pattern of increased demethylation at H19, a lncRNA known to regulate cell proliferation and imprinting, that correlated with alcohol consumption (Gabory et al., 2009), demonstrating an association between chronic alcohol use and demethylation of normally hypermethylated regions of DNA.
Genome-wide association studies
A survey of genome-wide association studies (GWAS) of alcohol dependence identified potential biological functions for the reported risk variants (Zuo et al., 2014). For example, variants located within an alcohol dehydrogenase (ADH) cluster included a large antisense-overlapping lncRNA, LOC100507053 (Xu et al., 2015), covering multiple loci for ADH genes that form a risk-genomic region for alcohol dependence identified by GWAS and candidate gene studies (Gelernter et al., 2009; Li, Zhao, & Gelernter, 2011; Xu et al., 2015). This class of antisense-overlapping lncRNAs frequently uses diverse transcriptional and post-transcriptional regulatory mechanisms to fulfill a wide variety of biological roles (Sartor et al., 2012; St Laurent, Wahlestedt, & Kapranov, 2015), and thus may significantly regulate cellular responses to alcohol.
V. Conclusions
Recent technological advances in genetics and genomics have dramatically changed our view of cellular regulation. RNA-seq and other technologies have identified several classes of ncRNAs and expanded a new area of transcriptomics that is not only revolutionizing our understanding of the diversity of the proteome and cellular regulatory mechanisms, but also our insight into the genetic regulation of disease. Because ncRNAs have proven to be important for normal development and synaptic physiology, it follows that they are also key regulators in pathological conditions such as alcoholism (Fig. 2). Studies have identified alcohol-sensitive miRNAs and lincRNAs that are involved in the transition from excessive alcohol consumption to dependence, potentially silencing large collections of target genes. The regulation of gene expression by ncRNAs plays an important role in adaptive behavior, but the extent to which these molecules contribute to the development and maintenance of alcohol dependence or other substance-abuse disorders remains largely unknown. A better understanding of mechanisms involved in alcohol-mediated ncRNA changes could advance therapeutic strategies for this disease. This review has outlined numerous ncRNAs that respond to various challenges by alcohol, and examples are outlined above demonstrating that ncRNA manipulation can alter drinking behavior. Clearly, additional work is required to move this type of basic research to a clinical setting. There has been a tremendous effort to develop synthetic or expressed ncRNA mimics to treat other disease conditions, particularly cancer (Matsui & Corey, 2016; Roberts & Wood, 2013), and a number of biotechnology companies have focused on developing oligonucleotide-based drug chemistries for use in clinical trials (Matsui & Corey, 2016). This novel use of synthetic oligonucleotides to influence ncRNA and ncRNA activity is an exciting prospect for future AUD therapies.
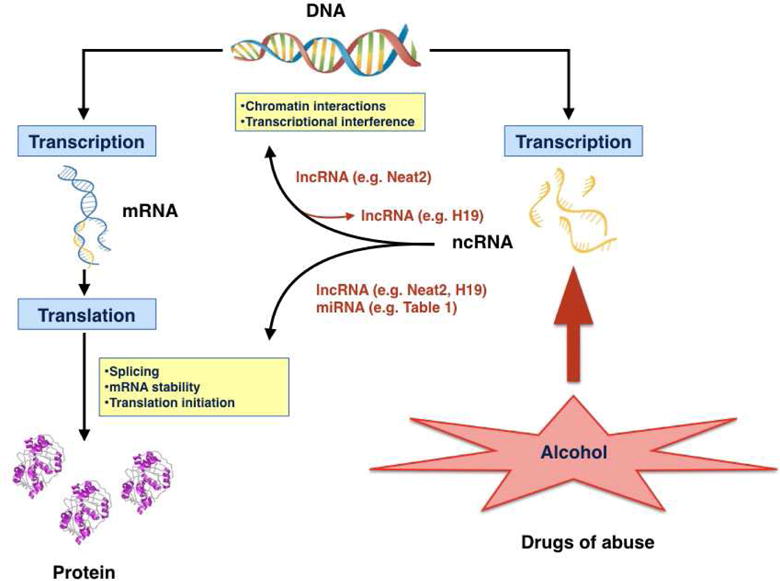
Fig. 2.
Cartoon summary of alcohol-mediated regulation of ncRNAs. The blue and yellow boxes represent the “central dogma” concept of gene expression at the transcriptional or translational levels, and the potential mechanisms for ncRNA regulation are illustrated in red. Examples of lncRNA-mediated regulation of epigenetic factors (Neat2) and alcohol-induced changes in lncRNA methylation (H19) are shown. Alcohol may regulate both lncRNAs and miRNAs at the protein translational level.
Highlights.
Technological advances in the previous decade have increasingly highlighted the role of non-protein-coding RNAs (ncRNAs) in the regulation of gene expression and function.
These recently characterized molecules were discovered to mediate diverse processes in the central nervous system, from normal development and physiology to regulation of disease, including alcoholism and other psychiatric disorders.
Acknowledgments
This work was supported by National Institute on Alcohol Abuse and Alcoholism Grants R01 AA012404, U01 AA020926, and P01 AA020683.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albertson DN, Pruetz B, Schmidt CJ, Kuhn DM, Kapatos G, Bannon MJ. Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. Journal of Neurochemistry. 2004;88(5):1211–1219. doi: 10.1046/j.1471-4159.2003.02247.x. http://doi.org/10.1046/j.1471-4159.2003.02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nature Reviews Genetics. 2010;11(8):559–571. doi: 10.1038/nrg2814. http://doi.org/10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- Bahi A, Dreyer JL. Striatal modulation of BDNF expression using microRNA124a-expressing lentiviral vectors impairs ethanol-induced conditioned-place preference and voluntary alcohol consumption. The European Journal of Neuroscience. 2013;38(2):2328–2337. doi: 10.1111/ejn.12228. http://doi.org/10.1111/ejn.12228. [DOI] [PubMed] [Google Scholar]
- Balaraman S, Tingling JD, Tsai PC, Miranda RC. Dysregulation of microRNA expression and function contributes to the etiology of fetal alcohol spectrum disorders. Alcohol Research: Current Reviews. 2013;35(1):18–24. doi: 10.35946/arcr.v35.1.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Molecular Psychiatry. 2014;19(4):486–494. doi: 10.1038/mp.2013.45. http://doi.org/10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- Bekdash RA, Harrison NL. Downregulation of Gabra4 expression during alcohol withdrawal is mediated by specific microRNAs in cultured mouse cortical neurons. Brain and Behavior. 2015;5(8):e00355–n/a. doi: 10.1002/brb3.355. http://doi.org/10.1002/brb3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, et al. A Transcriptomic Atlas of Mouse Neocortical Layers. Neuron. 2011;71(4):605–616. doi: 10.1016/j.neuron.2011.06.039. http://doi.org/10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. The EMBO Journal. 2010;29(18):3082–3093. doi: 10.1038/emboj.2010.199. http://doi.org/10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nature Neuroscience. 2009;12(8):1020–1027. doi: 10.1038/nn.2371. http://doi.org/10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Current Opinion in Genetics & Development. 2007;17(2):139–144. doi: 10.1016/j.gde.2007.02.008. http://doi.org/10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science (New York, NY) 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. http://doi.org/10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. http://doi.org/10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. http://doi.org/10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357(2):83–94. doi: 10.1016/j.gene.2005.06.019. http://doi.org/10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Darcq E, Warnault V, Phamluong K, Besserer GM, Liu F, Ron D. MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Molecular Psychiatry. 2015;20(10):1219–1231. doi: 10.1038/mp.2014.120. http://doi.org/10.1038/mp.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Research. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. http://doi.org/10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov MG, Giles CB, Koelsch KA, Wren JD. Systematic classification of non-coding RNAs by epigenomic similarity. BMC Bioinformatics. 2013;14(Suppl 14)(14):S2. doi: 10.1186/1471-2105-14-S14-S2. http://doi.org/10.1186/1471-2105-14-S14-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biological Psychiatry. 2006;59(12):1116–1127. doi: 10.1016/j.biopsych.2006.02.013. http://doi.org/10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature Structural & Molecular Biology. 2012;19(6):586–593. doi: 10.1038/nsmb.2296. http://doi.org/10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nature Medicine. 2008;14(7):723–730. doi: 10.1038/nm1784. http://doi.org/10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD. Transcriptome organization for chronic alcohol abuse in human brain. Molecular Psychiatry. 2014;20(11):1438–1447. doi: 10.1038/mp.2014.159. http://doi.org/10.1038/mp.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, Mayfield RD. RNA-Seq Reveals Novel Transcriptional Reorganization in Human Alcoholic Brain. International Review of Neurobiology. 2014;116:275–300. doi: 10.1016/B978-0-12-801105-8.00011-4. http://doi.org/10.1016/B978-0-12-801105-8.00011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorenza A, Barco A. Role of Dicer and the miRNA system in neuronal plasticity and brain function. Neurobiology of Learning and Memory. 2016;135:3–12. doi: 10.1016/j.nlm.2016.05.001. http://doi.org/10.1016/j.nlm.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forné T, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development (Cambridge, England) 2009;136(20):3413–3421. doi: 10.1242/dev.036061. http://doi.org/10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, Poling J, et al. Dense genomewide linkage scan for alcohol dependence in African Americans: significant linkage on chromosome 10. Biological Psychiatry. 2009;65(2):111–115. doi: 10.1016/j.biopsych.2008.08.036. http://doi.org/10.1016/j.biopsych.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Groff AF, Sauvageau M, Trayes-Gibson Z, Sanchez-Gomez DB, Morse M, et al. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proceedings of the National Academy of Sciences. 2015;112(22):6855–6862. doi: 10.1073/pnas.1411263112. http://doi.org/10.1073/pnas.1411263112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini G, Nunez YO, Mayfield RD. Integration of miRNA and protein profiling reveals coordinated neuroadaptations in the alcohol-dependent mouse brain. PloS One. 2013;8(12):e82565. doi: 10.1371/journal.pone.0082565. http://doi.org/10.1371/journal.pone.0082565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini G, Roberts AJ, Mayfield RD. Neurobiological signatures of alcohol dependence revealed by protein profiling. PloS One. 2013;8(12):e82656. doi: 10.1371/journal.pone.0082656. http://doi.org/10.1371/journal.pone.0082656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. http://doi.org/10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. An overview of microRNAs. Advanced Drug Delivery Reviews. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. http://doi.org/10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson AC, Rimondini R, Heilig M, Mathé AA, Sommer WH. Dissociation of antidepressant-like activity of escitalopram and nortriptyline on behaviour and hippocampal BDNF expression in female rats. Journal of Psychopharmacology (Oxford, England) 2011;25(10):1378–1387. doi: 10.1177/0269881110393049. http://doi.org/10.1177/0269881110393049. [DOI] [PubMed] [Google Scholar]
- Jeanblanc J, He DY, Carnicella S, Kharazia V, Janak PH, Ron D. Endogenous BDNF in the dorsolateral striatum gates alcohol drinking. Journal of Neuroscience. 2009;29(43):13494–13502. doi: 10.1523/JNEUROSCI.2243-09.2009. http://doi.org/10.1523/JNEUROSCI.2243-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA (New York, NY) 2010;16(8):1478–1487. doi: 10.1261/rna.1951310. http://doi.org/10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. http://doi.org/10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocerha J, Dwivedi Y, Brennand KJ. Noncoding RNAs and neurobehavioral mechanisms in psychiatric disease. Molecular Psychiatry. 2015;20(6):677–684. doi: 10.1038/mp.2015.30. http://doi.org/10.1038/mp.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42(Database issue):D68–73. doi: 10.1093/nar/gkt1181. http://doi.org/10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HR, Sakharkar AJ, Teppen TL, Berkel TDM, Pandey SC. The epigenetic landscape of alcoholism. International Review of Neurobiology. 2014;115:75–116. doi: 10.1016/B978-0-12-801311-3.00003-2. http://doi.org/10.1016/B978-0-12-801311-3.00003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics. 2010;11(9):597–610. doi: 10.1038/nrg2843. http://doi.org/10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kryger R, Fan L, Wilce PA, Jaquet V. MALAT-1, a non protein-coding RNA is upregulated in the cerebellum, hippocampus and brain stem of human alcoholics. Alcohol (Fayetteville, NY) 2012;46(7):629–634. doi: 10.1016/j.alcohol.2012.04.002. http://doi.org/10.1016/j.alcohol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(1):559. doi: 10.1186/1471-2105-9-559. http://doi.org/10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science (New York, NY) 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. http://doi.org/10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD. Up-Regulation of MicroRNAs in Brain of Human Alcoholics. Alcoholism: Clinical and Experimental Research. 2011 doi: 10.1111/j.1530-0277.2011.01544.x. http://doi.org/10.1111/j.1530-0277.2011.01544.x. [DOI] [PMC free article] [PubMed]
- Li D, Zhao H, Gelernter J. Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biological Psychiatry. 2011;70(6):504–512. doi: 10.1016/j.biopsych.2011.02.024. http://doi.org/10.1016/j.biopsych.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li J, Liu X, Qin S, Guan Y, Liu Y, et al. MicroRNA expression profile and functional analysis reveal that miR-382 is a critical novel gene of alcohol addiction. EMBO Molecular Medicine. 2013;5(9):1402–1414. doi: 10.1002/emmm.201201900. http://doi.org/10.1002/emmm.201201900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology. 2006;31(7):1574–1582. doi: 10.1038/sj.npp.1300947. http://doi.org/10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Londin E, Loher P, Telonis AG, Quann K, Clark P, Jing Y, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proceedings of the National Academy of Sciences. 2015;112(10):E1106–15. doi: 10.1073/pnas.1420955112. http://doi.org/10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistri M, Velmeshev D, Makhmutova M, Faghihi MA. Transcriptomics Profiling of Alzheimer’s Disease Reveal Neurovascular Defects, Altered Amyloid-β Homeostasis, and Deregulated Expression of Long Noncoding RNAs. Journal of Alzheimer’s Disease: JAD. 2015;48(3):647–665. doi: 10.3233/JAD-150398. http://doi.org/10.3233/JAD-150398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamdani M, Williamson V, McMichael GO, Blevins T, Aliev F, Adkins A, et al. Integrating mRNA and miRNA Weighted Gene Co-Expression Networks with eQTLs in the Nucleus Accumbens of Subjects with Alcohol Dependence. PloS One. 2015;10(9):e0137671. doi: 10.1371/journal.pone.0137671. http://doi.org/10.1371/journal.pone.0137671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzardo AM, Gunewardena S, Butler MG. Over-expression of the miRNA cluster at chromosome 14q32 in the alcoholic brain correlates with suppression of predicted target mRNA required for oligodendrocyte proliferation. Gene. 2013;526(2):356–363. doi: 10.1016/j.gene.2013.05.052. http://doi.org/10.1016/j.gene.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massone S, Vassallo I, Fiorino G, Castelnuovo M, Barbieri F, Borghi R, et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiology of Disease. 2011;41(2):308–317. doi: 10.1016/j.nbd.2010.09.019. http://doi.org/10.1016/j.nbd.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Matsui M, Corey DR. Non-coding RNAs as drug targets. Nature Reviews Drug Discovery. 2016 doi: 10.1038/nrd.2016.117. http://doi.org/10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed]
- Mattick JS, Makunin IV. Non-coding RNA. Human Molecular Genetics. 2006;15(90001):R17–29. doi: 10.1093/hmg/ddl046. Spec No 1. http://doi.org/10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Mayfield J, Ferguson L, Harris RA. Neuroimmune signaling: a key component of alcohol abuse. Current Opinion in Neurobiology. 2013;23(4):513–520. doi: 10.1016/j.conb.2013.01.024. http://doi.org/10.1016/j.conb.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed Z, Levy A, Ashwal-Fluss R, Lev-Maor G, Mekahel K, Atias N, et al. Alternative splicing regulates biogenesis of miRNAs located across exon-intron junctions. Molecular Cell. 2013;50(6):869–881. doi: 10.1016/j.molcel.2013.05.007. http://doi.org/10.1016/j.molcel.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Reviews Genetics. 2009;10(3):155–159. doi: 10.1038/nrg2521. http://doi.org/10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proceedings of the National Academy of Sciences. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. http://doi.org/10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. Journal of Neurochemistry. 2011;116(3):459–466. doi: 10.1111/j.1471-4159.2010.07126.x. http://doi.org/10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC. MicroRNAs and ethanol toxicity. International Review of Neurobiology. 2014;115:245–284. doi: 10.1016/B978-0-12-801311-3.00007-X. http://doi.org/10.1016/B978-0-12-801311-3.00007-X. [DOI] [PubMed] [Google Scholar]
- Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, Keshavarzian A, et al. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcoholism: Clinical and Experimental Research. 2010;34(4):575–587. doi: 10.1111/j.1530-0277.2009.01126.x. http://doi.org/10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nature Reviews Genetics. 2014;15(6):423–437. doi: 10.1038/nrg3722. http://doi.org/10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Most D, Leiter C, Blednov YA, Harris RA, Mayfield RD. Synaptic microRNAs Coordinately Regulate Synaptic mRNAs: Perturbation by Chronic Alcohol Consumption. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology. 2016;41(2):538–548. doi: 10.1038/npp.2015.179. http://doi.org/10.1038/npp.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Most D, Workman E, Harris RA. Synaptic adaptations by alcohol and drugs of abuse: changes in microRNA expression and mRNA regulation. Frontiers in Molecular Neuroscience. 2014;7:85. doi: 10.3389/fnmol.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(16):6368–6373. doi: 10.1073/pnas.0510188103. http://doi.org/10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez YO, Mayfield RD. Understanding Alcoholism Through microRNA Signatures in Brains of Human Alcoholics. Frontiers in Genetics. 2012;3:43. doi: 10.3389/fgene.2012.00043. http://doi.org/10.3389/fgene.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA, et al. Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC Genomics. 2013;14(1):725. doi: 10.1186/1471-2164-14-725. http://doi.org/10.1186/1471-2164-14-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: implications for fetal alcohol spectrum disorders. Alcoholism: Clinical and Experimental Research. 2009;33(9):1615–1627. doi: 10.1111/j.1530-0277.2009.00993.x. http://doi.org/10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- Perkins DO, Jeffries C, Sullivan P. Expanding the “central dogma”: the regulatory role of nonprotein coding genes and implications for the genetic liability to schizophrenia. Molecular Psychiatry. 2005;10(1):69–78. doi: 10.1038/sj.mp.4001577. http://doi.org/10.1038/sj.mp.4001577. [DOI] [PubMed] [Google Scholar]
- Pietrzykowski AZ. The role of microRNAs in drug addiction: a big lesson from tiny molecules. International Review of Neurobiology. 2010;91:1–24. doi: 10.1016/S0074-7742(10)91001-5. http://doi.org/10.1016/S0074-7742(10)91001-5. [DOI] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, et al. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59(2):274–287. doi: 10.1016/j.neuron.2008.05.032. http://doi.org/10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins SA, Przybycien-Szymanska MM, Rao YS, Pak TR. Long-term effects of peripubertal binge EtOH exposure on hippocampal microRNA expression in the rat. PloS One. 2014;9(1):e83166. doi: 10.1371/journal.pone.0083166. http://doi.org/10.1371/journal.pone.0083166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443(7108):167–172. doi: 10.1038/nature05113. http://doi.org/10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PCG, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science (New York, NY) 2006;314(5796):144–148. doi: 10.1126/science.1131693. http://doi.org/10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- Ravasi T. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Research. 2006;16(1):11–19. doi: 10.1101/gr.4200206. http://doi.org/10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TC, Wood MJA. Therapeutic targeting of non-coding RNAs. Essays in Biochemistry. 2013;54:127–145. doi: 10.1042/bse0540127. http://doi.org/10.1042/bse0540127. [DOI] [PubMed] [Google Scholar]
- Robinson G, Most D, Ferguson LB, Mayfield J, Harris RA, Blednov YA. Neuroimmune pathways in alcohol consumption: evidence from behavioral and genetic studies in rodents and humans. International Review of Neurobiology. 2014;118:13–39. doi: 10.1016/B978-0-12-801284-0.00002-6. http://doi.org/10.1016/B978-0-12-801284-0.00002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor GC, St Laurent G, Wahlestedt C. The Emerging Role of Non-Coding RNAs in Drug Addiction. Frontiers in Genetics. 2012;3:106. doi: 10.3389/fgene.2012.00106. http://doi.org/10.3389/fgene.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaro PA, Flavell CR, Widagdo J, Ratnu VS, Troup M, Ragan C, et al. Long Noncoding RNA-Directed Epigenetic Regulation of Gene Expression Is Associated With Anxiety-like Behavior in Mice. Biological Psychiatry. 2015;78(12):848–859. doi: 10.1016/j.biopsych.2015.02.004. http://doi.org/10.1016/j.biopsych.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends in Genetics: TIG. 2015;31(5):239–251. doi: 10.1016/j.tig.2015.03.007. http://doi.org/10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nature Reviews Genetics. 2012;13(8):537–551. doi: 10.1038/nrg3240. http://doi.org/10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapocik JD, Barbier E, Flanigan M, Solomon M, Pincus A, Pilling A, et al. microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. Journal of Neuroscience. 2014;34(13):4581–4588. doi: 10.1523/JNEUROSCI.0445-14.2014. http://doi.org/10.1523/JNEUROSCI.0445-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapocik JD, Tapocik JD, Solomon M, Solomon M, Flanigan M, Flanigan M, et al. Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. The Pharmacogenomics Journal. 2013;13(3):286–296. doi: 10.1038/tpj.2012.17. http://doi.org/10.1038/tpj.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teppen TL, Krishnan HR, Zhang H, Sakharkar AJ, Pandey SC. The Potential Role of Amygdaloid MicroRNA-494 in Alcohol-Induced Anxiolysis. Biological Psychiatry. 2015;80:711–719. doi: 10.1016/j.biopsych.2015.10.028. http://doi.org/10.1016/j.biopsych.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Ye X, Hou X, Yang X, Yang J, Wu C. SVCT2, a potential therapeutic target, protects against oxidative stress during ethanol-induced neurotoxicity via JNK/p38 MAPKs, NF-κB and miRNA125a-5p. Free Radical Biology & Medicine. 2016;96:362–373. doi: 10.1016/j.freeradbiomed.2016.03.039. http://doi.org/10.1016/j.freeradbiomed.2016.03.039. [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Molecular Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. http://doi.org/10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nature Reviews Genetics. 2016;17(10):601–614. doi: 10.1038/nrg.2016.85. http://doi.org/10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- Wang F, Gelernter J, Zhang H. Differential Expression of miR-130a in Postmortem Prefrontal Cortex of Subjects with Alcohol Use Disorders. Journal of Addiction Research & Therapy. 2013;4(155) doi: 10.4172/2155-6105.1000155. http://doi.org/10.4172/2155-6105.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. http://doi.org/10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. http://doi.org/10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends in Cell Biology. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. http://doi.org/10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, et al. Genomewide Association Study for Maximum Number of Alcoholic Drinks in European Americans and African Americans. Alcoholism: Clinical and Experimental Research. 2015;39(7):1137–1147. doi: 10.1111/acer.12751. http://doi.org/10.1111/acer.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wei G, Di Z, Zhao Q. miR-339-5p inhibits alcohol-induced brain inflammation through regulating NF-κB pathway. Biochemical and Biophysical Research Communications. 2014;452(3):450–456. doi: 10.1016/j.bbrc.2014.08.092. http://doi.org/10.1016/j.bbrc.2014.08.092. [DOI] [PubMed] [Google Scholar]
- Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. Journal of Molecular Neuroscience: MN. 2013;49(3):589–593. doi: 10.1007/s12031-012-9880-8. http://doi.org/10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Lu L, Tan Y, Pan X, Cai Y, Wang X, et al. Genome-wide association discoveries of alcohol dependence. The American Journal on Addictions/American Academy of Psychiatrists in Alcoholism and Addictions. 2014;23(6):526–539. doi: 10.1111/j.1521-0391.2014.12147.x. http://doi.org/10.1111/j.1521-0391.2014.12147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Tan Y, Wang Z, Wang KS, Zhang X, Chen X, et al. Long noncoding RNAs in psychiatric disorders. Psychiatric Genetics. 2016;26(3):109–116. doi: 10.1097/YPG.0000000000000129. http://doi.org/10.1097/YPG.0000000000000129. [DOI] [PMC free article] [PubMed] [Google Scholar]


