Abstract
Myeloid cells are important cell types that carry human cytomegalovirus. Latent viral DNA is present in CD34+ progenitor cells and their derived monocytes. However, differentiation of latently infected monocytes to mature macrophages or dendritic cells causes reactivation of latent viruses. During hematopoietic development, pluripotent genes are repressed, and lineage specific genes are activated in a step-wise manner. This process is governed by cell-type specific chromatin states. Enhancers in the hematopoietic system are highly dynamic and established by pioneer (first tier) transcription factors (TFs), which set the stage for second and third tier TF binding. In this review, we examine the epigenetic mechanisms that regulate myeloid cell development, cell identity, and activation with a special focus on factors that regulate viral gene expression and the status of viral infection in myeloid cells.
Keywords: Chromatin, enhancers, hierarchy of transcription factors, myeloid cells, cytomegalovirus
Introduction
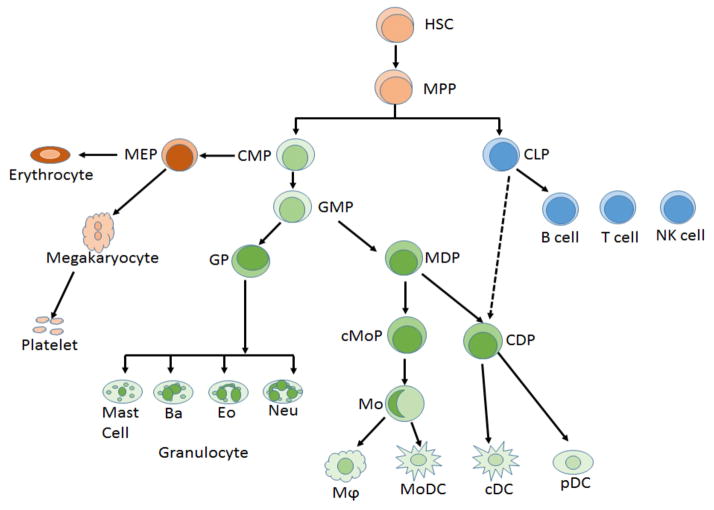
Human cytomegalovirus (HCMV) is a member of beta herpesvirus family, which infects 50–90 percent of the population. As with other herpesviruses, HCMV can establish life-long latent infection in its hosts, and reactivate from latency. Although primary infection is typically asymptomatic in immunocompetent individuals, in immunocompromised individuals, such as organ transplant recipients and AIDS patients, primary infection and reactivation impose morbidity and mortality [1, 2]. Congenital infection and neonatal infection can cause neurological defect, such as deafness and mental retardation [3]. It is well-established that myeloid lineage cells in bone marrow and peripheral blood are critical cell types carrying viral DNA in seropositive individuals [4, 5]. In undifferentiated myeloid progenitor cells and monocytes, the virus stays in latent infection, and only few viral genes are expressed. Once the myeloid cells differentiate into mature macrophages and dendritic cells (DCs), the cellular environment alters and becomes conducive to lytic gene expression, and supports lytic infection [6, 7]. Therefore, the differentiation state of myeloid cells determines the status of viral infection, i.e. latency versus reactivation. The hematopoietic system has a well-defined developmental tree that can serve as an ideal framework to study epigenetic regulation of gene expression during cell-fate decision (figure 1). In this review, we will discuss the epigenetic regulation of gene expression during the mammalian development of myeloid compartment, with a particular focus on macrophages and DCs, and the relevance of this regulation to latency and reactivation of CMV in hematopoietic system.
Figure 1.
Hierarchical model of hematopoiesis with the emphasis of myeloid cell development (shown in green color). HSC, hematopoietic stem; MPP, multipotent progenitor; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; MEP, megakaryocyte-erythrocyte progenitor; GMP, granulocyte-monocyte progenitor; GP, granulocyte progenitor; MDP, monocyte-dendritic cell progenitor; cMoP, common monocyte progenitor; CDP, common dendritic cell progenitor; Ba, basophil; Eo, eosinophil; Mϕ, macrophages; Mo, monocyte; Neu, neutrophil; MoDC, monocyte derived dendritic cells; cDC, classical dendritic cells; pDC, plasmacytoid dendritic cell.
Principle of epigenetics
In mammalian cells, nuclear DNA is organized into nucleosomes, the basic unit of chromatin, which consists of histone octamers. Each octamer contains two copies of histones H2A, H2B, H3, and H4, with approximately 147 DNA base pairs wrapping around the histone octamer [8]. In some nucleosomes, histone variants such as H2A.Z and H3.3 substitute for H2A and H3 respectively [9, 10]. The N- and C-terminal tails of histones dangle out of nucleosome cores, and are subject to a variety of post translational modifications (PTMs), such as acetylation, phosphorylation, methylation, ubiquitination, sumoylation, and crotonylation [11, 12]. Combinations of histone modifications form histone codes, which can serve as docking sites for both transcription factors (TFs) and ATP-dependent nucleosome modifying complexes, as well as for chromatin modifying enzymes, affecting nucleosome positioning and regulation of gene transcription. Acetylation of lysines in H3 and H4, trimethylation of H3 lysine 4 (H3K4me3), of H3 lysine 79 (H3K79me3), and of histone H3 lysine 36 (H3K36me3), as well as ubiquitination of H2B lysine 120 (H2B K120ub1) are generally associated with transcriptional activation. In contrast, methylation of histone H3 lysine 9 (H3K9me1/2/3), trimethylation of histone H3 lysine 27 (H3K27me3), and ubiquitination of histone H2A lysine 119 (H2A K119ub1) are markers of transcriptionally repressed genes [13].
In addition, the CpG dinucleotides in the DNA sequence are susceptible to methylation. When a methyl group is added to 5′ position of the pyrimidine ring of a cytosine, a 5-methylcytosine (5meCs) is formed, which can interfere directly with the binding of sequence-specific TFs to their target DNA and prevent transcriptional activation. 5meCs can also recruit transcriptional repressors containing methyl-CpG binding domain (MDB), which in turn, cause histone modifications characteristic of transcriptional repression, resulting in the formation of compacted chromatin [14, 15]. Thus, regulation of gene expression is determined by the chromatin state including nucleosome positioning and chromatin remodeling, which includes histone modifications and DNA methylation.
Chromatin dynamics, enhancers, and lineage-determining transcription factors
Enhancers are distal cis-regulatory regions characterized by H3K4me1 high/H3K4me3 low, and harboring binding sites of heterotypic TFs [16]. Enhancer regions are highly dynamic during hematopoietic development, and enhancer landscape strongly correlates with cell identity [17]. TFs important for myeloid differentiation include PU.1, CCAAT/enhancer binding protein (C/EBP) family (C/EBPα, C/EBP β), interferon-regulatory factor 8 (IRF8), GATA-binding protein 1 (GATA1), growth factor independence 1 (Gfi1), and CCAAT displacement protein (CDP).
PU.1 protein, encoded by Spi1 gene, is required for generation of common myeloid progenitors (CMPs) from hematopoietic stem cells (HSCs) and formation of the myeloid transcription network [18, 19]. As a member of the Ets family of TFs, PU.1 shows a specific pattern of expression in different lineages of hematopoietic cells [18, 20]. Its expression is detectable in HSCs, CMPs, common lymphoid progenitors (CLPs), and B cells, but is significantly increased in mature myeloid cells, such DCs, and macrophages [20]. A majority of myeloid lineage specific genes contain PU.1 binding sites within their enhancers and/or promoters [20]. High levels of PU.1 expression cause a differentiation bias that favors myeloid lineage production [16]. Spi1 knockout mice are devoid of mature myeloid cells, and B cells [21–24]. Hence PU.1 serves as a master regulator for myeloid lineage commitment and determination of cell identities. Upon binding to its cognate binding sites, PU.1 synergizes with other factors, such as C/EBPβ, to initiates nucleosome remodeling, leading to monomethylation of H3K4. Striking DNase I hypersensitivity spikes appear around PU.1 binding sites, indicating the chromatin in those areas is accessible to other TFs and enzymes [25]. This observation is consistent with the mechanism proposed by Miller and Windom that concurrent binding of PU.1 and C/EBPβ competes with nucleosomes to establish cell type–specific binding patterns and enhancers, which correlate with the cell-type specific profiles of gene expression [25, 26]. Moreover, PU.1 also plays a crucial role in maintaining H3K4me1 in both macrophages and DCs [16, 27].
C/EBPα is required for transition from CMPs to granulocyte-monocyte progenitors (GMPs), and from multipotent progenitors (MPPs) to common dendritic cell progenitors (CDPs). C/EBPα is a member of the basic leucine zipper transcription factor family, and is expressed in HSCs, CMPs, CDPs and GMPs [20, 28, 29]. Mice that are deficient in C/EBPα can give rise to a normal number of CMPs, but have a deficiency in GMPs and all downstream granulocytic cells [30, 31]. Similarly, C/EBPα is needed for production of CDPs from MPPs and CMPs in steady-state DC differentiation but is not required for later stages of DC maturation [29]. ChIP-seq data shows that C/EBPα binds to regulatory regions of several genes that are critical for the transition from MPP to CDP, whose expression dramatically decreases in the absence of C/EBPα [29]. Acting in concert with PU.1, C/EBPα can activate ten-eleven-translocation protein 2 (TET2), which in turn, demethylates 5meCs in the latent enhancers and promoters, and thereby activates myeloid gene transcription [28]. C/EBPα can also bind to brahma related gene 1 (BRG1), a member of SWI/SNF family of nucleosome modifying complexes to alter the positioning of nucleosomes [32, 33]. Both C/EBPα and PU.1 can bind to TATA box binding protein (TBP), and thus enable recruitment of the transcription machinery [20].
C/EBPβ is also expressed in myeloid lineage committed progenitor cells, and particularly in classical DCs, and may play an important role in DC identity determination [27, 28, 34]. Together with PU.1, C/EBPβ is present in a majority of regulatory regions, mostly enhancer regions that are bound by remainder TFs [27].
Interferon regulatory factor 8 (IRF8), a member of IRF family, is a DNA-binding TF. During differentiation of HSCs along the hematopoietic developmental tree, IRF8 is not expressed until the GMP stage in myeloid development. IRF8 is then drastically increased in monocyte-dendritic cell progenitors (MDPs), CDPs, and common monocyte progenitors (cMoPs). All types and differential stages of DCs but CD8 negative cDCs maintain high level IRF8 expression. Monocytes and tissue resident macrophages (except osteoclasts) express steady-state intermediate levels of IRF8 [35]. IRF8 plays an integral role in promoting development of monocytes, DCs, basophils (Ba) and eosinophils (Eo), but negatively regulates development of neutrophils (Neu) [35]. In cooperation with PU.1, IRF8 binds to ETS-IRF composite element (EICE) of DNA in compacted chromatin, with the help of nucleosome-remodeling complexes, destabilizes the nucleosomes and deposits H4K4me1 and H3K27ac (a marker of active enhancer), and thereby establishes and activates the enhancers to induce corresponding gene expression [35, 36]. In mononuclear phagocyte progenitors, IRF8 can interact with C/EBPα and hamper the ability of C/EBPα to bind to chromatin. In this way, IRF8 wards off C/EPBα-mediated anomalous neutrophil differentiation [37]. IRF8 plays an essential role in preserving gene expression profiles in plasmacytoid dendritic cells (pDCs), maintaining their identity, and also participate cell-specific responses to pathogen stimuli [34, 35]. IRF8 occupies more than 30,000 regulatory regions, and is significantly enriched in pDC-specific enhancers [34].
GATA1 is a member of zinc finger containing DNA binding transcription factor family that have two zinc fingers. Two zinc fingers bind to distinct target sites and have different functions. The C-zinc finger binds to the GATA consensus DNA sequence, on the top of which, the N-zinc finger of GATA1 interacts with specific DNA sequence and associates with cofactors, such as Friend of GATA1 (FOG1), to reinforce the binding and facilitate transcriptional activation [38]. GATA1 is expressed in HSCs, CMPs, CLPs, megakaryocyte-erythrocyte progenitors (MEPs), erythroid cells, megakaryocytes, and mast cells and eosinophils as well. Expression of GATA1 is essential for the development and maturation of erythroid cells and megakaryocytes [38]. On the one hand, GATA1 boosts megakaryocyte or erythroid commitment by promoting megakaryocyte-specific gene expression through recruiting BRG1, a ATPase of mammalian SWI/SNF chromatin remodeling complexes, to the promoters [17, 32]. On the other hand, GATA1 precludes the development of GMP and lymphoid cells through down regulating cofactors required by lineage-determining transcription factors of those developmental branches, such as PU.1, PAX5, and IL-7[38]. Mutations of GATA1 are associated with both acute megakaryoblastic leukemia (AMKL) and with transient abnormal myelopoiesis (TAM) in Down syndrome (DS) children [38]. Eosinophils and mast cell development is also impeded in GATA1 deficient mice, suggesting a critical role of GATA1 in development of these two types of cells [39]. Thus, GATA1 is required for differentiation of four types of hematopoietic cells.
Growth factor independence 1 (Gfi1) is a zinc-finger domain containing transcriptional repressors, is expressed in HSCs, B cells, T cells, and neutrophils [40] [41–43]. Gfi1 knockout mice exhibit defect in HSCs, lymphoid progenitors, absence of neutrophilic granulocytes, and accumulation of neutrophil precursors [41, 44], but these mice have intact CMPs and GMPs [41, 44]. These findings suggest that Gfi1 is required for neutrophil differentiation and maturation, development of CLPs, and maintenance of HSCs. Gene expression data show that the accumulated neutrophil precursors in Gfi1−/− mice express both neutrophilic markers and monocyte-specific genes, suggesting a failure in suppression of monocyte-determining gene expression [41]. Gfi1 has been shown to interact with PU.1, repressing the trans-activation effect of PU.1 to its target promoters such as Egr2 promoter, steering the development away from monocyte development and towards granulopoiesis [45]. Serial transplantation studies demonstrate that Gfi1−/− HSCs have lower self-renewal capacity than wild type HSCs, suggesting that Gfi1 plays a role in regulating HSC survival and proliferation [46].
CCAAT-displacement proteins (CDPs), also called CUX1 or Cut protein, are a family of transcription factors that contain Cut homeodomain and Cut repeats [47]. CDPs can act as transcriptional repressors or activator depending on the isoforms and promoter context. CDP function is regulated through posttranslational modifications, such as phosphorylation and acetylation [47, 48]. In hematopoietic cells, full length CDP negatively regulate the expression of cytochrome b heavy chain (gp91-phox), C/EBPε, and lactoferrin (LF) [49–52]. The DNA binding activity of CDP is diminished upon differentiation and maturation of granulocytes, macrophages, and erythrocytes [45, 53]. Evidence shows that CDP exerts its repressive function through competing with transcriptional activators and recruiting histone deacetylase (HDACs) and/or histone lysine methyltransferase to promoter and enhancer region [45, 52, 54, 55].
It is generally believed that during differentiation, chromatin is dynamically modified through sequential events, leading to a chromatin landscape specific to the cell’s identity. In this process, the pioneer TFs play an initiating role, binding to the closed chromatin, and recruiting chromatin modulating complexes/enzymes to the regulatory regions to establish cell type-specific enhancers [17, 34].
Hierarchy of transcription factor activities during immune cell responses to stimuli
Immune cells, such as macrophages and DCs, NK cells, B cells, and T cells, exist in different states, including resting state and activated state. When immune cells meet external stimuli, such as antigens or pathogen components, they launch a robust, specific and reproducible response, activating thousands of genes. Using the response of DCs or macrophages to lipopolysaccharide (LPS) stimulation as a model system, high-throughput ChIP and transcriptome studies have been performed [16, 27, 34]. These studies demonstrate that there is considerable variation among TFs in terms of binding dynamics, location, and interaction with other TFs. Combined with temporal gene expression and chromatin modification markers, these data classify the TFs into three tiers, forming a “layered architecture” [16, 27]. The first tier TFs includes PU.1 and C/EBPβ, which bind to the lineage-specific chromatin regulatory regions and loosen up the compacted chromatin during hematopoietic development. These pioneer TFs are present in the vast majority of regions bound by other TFs, but are also present in regions the other TFs do not occupy, are static in binding locations, and exhibit no alteration upon stimulation. The second tier TFs, which includes Junb, Irf4, Atf3 in DCs, and AP-1 in macrophages, occupy LPS induced genes prior to stimulation, and associate with more dynamic and specific TFs after stimulation. The third tier TFs bind, in a stimulus-dependent fashion, to a specific set of genes that share biological functions, such as inflammation or antiviral responses [16, 27].
Thus, it appears that immune cells, such as DCs and macrophages, respond to pathogenic stimuli through the three-tiered TF network [17]. The pioneer TFs (first tier) bind to compacted chromatin, recruit chromatin modifying complexes, and convert the chromatin into an accessible state so that other TFs can bind. The pioneer TFs commit the cells into specific lineages. The second tier of TFs, called “primers”, or “settlers” bind to thousands of genes in an opened state to serve as “beacons” that guide other TF binding upon stimulation. These “primers” bind to lineage-specific genes, maintain the binding sites, and act as anchoring points for the dynamic partners to dock. The third layer TFs are signal-dependent, transiently bind to specific sets of the genes that have common biological functions, such as NF-kB for inflammation, and STAT1 for defense against viruses [16, 17, 27].
Cytomegalovirus infection of hematopoietic cells
In latently infected human hosts, CD34+ progenitor cells in bone marrow and its derived CD14+ monocytes in peripheral blood are sites where latent viral genome resides [4, 56]. The latent viral genome exist as episomal DNA molecules in the nuclear of latently infected cells [57]. When latently infected CD14+ monocytes differentiate into mature dendritic cells (DCs) and/or macrophages, the latent viral genomes are reactivated, which is characterized by lytic gene expression and production of infectious viral particles [58–61]. Interestingly, in vitro infection experiments showed that HCMV can productively infect monocyte-derived macrophages (MDMs) and monocyte derived dendritic cells (MoDs) [6, 7]. These observations suggest that the differentiation process of myeloid cells is responsible for supporting viral lytic infection and reactivation as well. Furthermore, this differentiation-dependent difference in viral infection state stems from regulation of viral gene expression, which is controlled by the chromatin structure. It has been demonstrated that in latently infected CD34+ progenitor cells and CD14+ monocytes, HCMV chromatin carries repressive markers, such as H3K9Me3, H3K27me3, and is associated with transcriptional repressors such as heterochromatin protein 1 (HP1) and KAP1 [60, 62, 63]. In contrast, in mature dendritic cells, the reactivated viral chromatin carries transcriptional active markers, such as acetylated histones (AcH) and phosphorylated histone H3 [60, 64].
Cellular transcription factors expressed in myeloid lineage cells may contribute to maintenance of latency and reactivation of HCMV
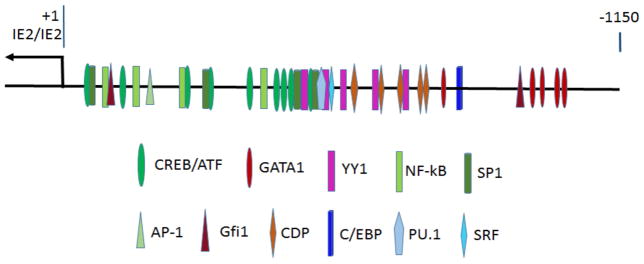
HCMV major immediate early genes (MIE), driven by major immediate early enhancer and promoter (MIEP), are the first genes expressed in both lytic infection and reactivation from latency. The MIEP is repressed in CD34+ progenitor cells and CD14+ monocytes, but is reactivated in mature DCs, suggesting that it is regulated by differentiation associated transcription factors. The promoter analysis using MatInspector of Genomatrix, a web-based bioinformatics program (http://www.Genomatrix.de), with a matrix similarity cutoff of 0.9, identified numerous putative binding site of TFs in HCMV MIEP region (Figure 2). It is striking to find that the most aforementioned transcription factors associated with myeloid differentiation and activation, including PU.1, C/EBP, Gfi1, GATA1, CDP, CREB/ATF, AP-1, and NF-kB, have binding sites in the HCMV MIEP region. Previous studies have shown that CREB/ATF, AP-1 and NF-kB can activate MIEP, whereas Gfi1 and CDP can repress the activity of MIEP, akin to roles they exhibit in myeloid cell differentiation and activation [40, 65–67]. These findings raise the possibility that regulation of viral gene expression during differentiation associated reactivation of HCMV synchronize with the cellular genes of myeloid lineage commitment and activation, using same set of TFs. Throughout the course of myeloid differentiation and activation, transcriptional repressors (CDP and Gfi1) are down regulated, which is accompanied by the gradual up regulation of transcriptional activators (PU.1, C/EBP, CREB/ATF, AP-1, and NF-kB). Therefore, it is plausible to postulate that along with myeloid lineage commitment, the latent CMV genome in hematopoietic progenitor cells are gradually reactivated in step-wise manner regulated by chromatin dynamics, which in turn, is controlled by the hierarchy of multi-layered TFs.
Figure 2.
Putative binding sites of transcription factors in the human cytomegalovirus major immediate early enhancer and promoter region. The arrow shows the IE1/IE2 gene transcription start site. The putative binding sites of transcription factors identified by MatInspector program have of matrix similarity value equal or bigger than 0.9. CREB, cAMP-responsive element binding proteins; ATF, activating transcription factor; GATA1, GATA-binding transcription factor 1; YY1, Yin and Yang1; NF-kB, Nuclear factor kappaB; SP1, Specificity Protein 1; AP-1, activating protein 1; Gfi1, growth factor independence 1; CDP, CCAAT displacement protein; C/EBP, CCAAT/enhancer binding protein; SRF, serum response element binding factor.
Perspective
The unique feature of hematopoietic system provides a pragmatic model to study epigenetic regulation of gene expression in cell fate decision. During the differentiation, the pluripotent genes are turned off, and lineage commitment genes are gradually activated in step-wise manner. This process is controlled by the multi-layered TF network. Interestingly, the infection status of CMV in myeloid cells depend on the differentiation state of the host cells. In undifferentiated progenitor cells and partially differentiated monocytes, and virus genome is kept in latent state. Whereas, in terminally differentiated DCs and macrophages, virus can launch a productive infection, and latent virus can be reactivated upon stimuli. It seems that from undifferentiated cells to fully differentiated myeloid cells, such as macrophages and DCs, the chromatin state of viral genome gradually shifts from completely closed to open for transcription. Furthermore, most differentiation-associated transcription factors have binding site not only in cellular genes but also in viral MIE gene regulatory region. Thus, we infer that the viral MIE gene is also regulated by the multi-layered TF network, and is de-repressed, and then activated in a step-wise manner. The delineation of CMV chromatin landscape in different differentiation stages of myeloid lineage cells will be required to test this hypothesis.
Acknowledgments
The research in our laboratory is funded by NIH R01 AI112911-01 and P01 AI112522-01, and the Northwestern University Comprehensive Transplant Center.
Footnotes
CONFLICT of INTEREST
The authors declare that they have no conflict of interest
References
- 1.Drew WL. Diagnosis of cytomegalovirus infection. Rev Infect Dis. 1988;10(Suppl 3):S468–76. doi: 10.1093/clinids/10.supplement_3.s468. [DOI] [PubMed] [Google Scholar]
- 2.Rubin RH. Impact of cytomegalovirus infection on organ transplant recipients. Rev Infect Dis. 1990;12(Suppl 7):S754–66. doi: 10.1093/clinids/12.supplement_7.s754. [DOI] [PubMed] [Google Scholar]
- 3.Gaytant MA, et al. Congenital cytomegalovirus infection: review of the epidemiology and outcome. Obstet Gynecol Surv. 2002;57(4):245–56. doi: 10.1097/00006254-200204000-00024. [DOI] [PubMed] [Google Scholar]
- 4.Taylor-Wiedeman J, et al. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72(Pt 9):2059–64. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 5.Larsson S, et al. Cytomegalovirus DNA can be detected in peripheral blood mononuclear cells from all seropositive and most seronegative healthy blood donors over time. Transfusion. 1998;38(3):271–8. doi: 10.1046/j.1537-2995.1998.38398222871.x. [DOI] [PubMed] [Google Scholar]
- 6.Ibanez CE, et al. Human cytomegalovirus productively infects primary differentiated macrophages. J Virol. 1991;65(12):6581–8. doi: 10.1128/jvi.65.12.6581-6588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riegler S, et al. Monocyte-derived dendritic cells are permissive to the complete replicative cycle of human cytomegalovirus. J Gen Virol. 2000;81(Pt 2):393–9. doi: 10.1099/0022-1317-81-2-393. [DOI] [PubMed] [Google Scholar]
- 8.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423(6936):145–50. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 9.Turinetto V, Giachino C. Histone variants as emerging regulators of embryonic stem cell identity. Epigenetics. 2015;10(7):563–73. doi: 10.1080/15592294.2015.1053682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santoro SW, Dulac C. Histone variants and cellular plasticity. Trends Genet. 2015;31(9):516–27. doi: 10.1016/j.tig.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnaudo AM, Garcia BA. Proteomic characterization of novel histone post-translational modifications. Epigenetics Chromatin. 2013;6(1):24. doi: 10.1186/1756-8935-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Garcia BA. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb Perspect Biol. 2015;7(9):a025064. doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO Rep. 2015;16(11):1467–81. doi: 10.15252/embr.201540945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blattler A, Farnham PJ. Crosstalk between site-specific transcription factors and DNA methylation states. J Biol Chem. 2013;288(48):34287–94. doi: 10.1074/jbc.R113.512517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18(11):6538–47. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32(3):317–28. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Winter DR, Amit I. The role of chromatin dynamics in immune cell development. Immunol Rev. 2014;261(1):9–22. doi: 10.1111/imr.12200. [DOI] [PubMed] [Google Scholar]
- 18.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7(2):105–17. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 19.Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12(15):2403–12. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenen DG, et al. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90(2):489–519. [PubMed] [Google Scholar]
- 21.Dakic A, et al. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201(9):1487–502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klemsz MJ, et al. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61(1):113–24. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 23.Scott EW, et al. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–7. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 24.McKercher SR, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. Embo j. 1996;15(20):5647–58. [PMC free article] [PubMed] [Google Scholar]
- 25.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller JA, Widom J. Collaborative competition mechanism for gene activation in vivo. Mol Cell Biol. 2003;23(5):1623–32. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garber M, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47(5):810–22. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldberg GW, Marraffini LA. Resistance and tolerance to foreign elements by prokaryotic immune systems - curating the genome. Nat Rev Immunol. 2015;15(11):717–24. doi: 10.1038/nri3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welner RS, et al. C/EBPalpha is required for development of dendritic cell progenitors. Blood. 2013;121(20):4073–81. doi: 10.1182/blood-2012-10-463448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang DE, et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997;94(2):569–74. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21(6):853–63. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Ryme J, et al. Variations in the composition of mammalian SWI/SNF chromatin remodelling complexes. J Cell Biochem. 2009;108(3):565–76. doi: 10.1002/jcb.22288. [DOI] [PubMed] [Google Scholar]
- 33.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11(2):377–89. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 34.Bornstein C, et al. A negative feedback loop of transcription factors specifies alternative dendritic cell chromatin States. Mol Cell. 2014;56(6):749–62. doi: 10.1016/j.molcel.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurotaki D, Tamura T. Transcriptional and Epigenetic Regulation of Innate Immune Cell Development by the Transcription Factor, Interferon Regulatory Factor-8. J Interferon Cytokine Res. 2016;36(7):433–41. doi: 10.1089/jir.2015.0138. [DOI] [PubMed] [Google Scholar]
- 36.Kurotaki D, et al. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. 2013;121(10):1839–49. doi: 10.1182/blood-2012-06-437863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurotaki D, et al. IRF8 inhibits C/EBPalpha activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat Commun. 2014;5:4978. doi: 10.1038/ncomms5978. [DOI] [PubMed] [Google Scholar]
- 38.Gao J, Chen YH, Peterson LC. GATA family transcriptional factors: emerging suspects in hematologic disorders. Exp Hematol Oncol. 2015;4:28. doi: 10.1186/s40164-015-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crispino JD. GATA1 in normal and malignant hematopoiesis. Semin Cell Dev Biol. 2005;16(1):137–47. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Zweidler-Mckay PA, et al. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol. 1996;16(8):4024–34. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hock H, et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18(1):109–20. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- 42.Karsunky H, et al. High levels of the onco-protein Gfi-1 accelerate T-cell proliferation and inhibit activation induced T-cell death in Jurkat T-cells. Oncogene. 2002;21(10):1571–9. doi: 10.1038/sj.onc.1205216. [DOI] [PubMed] [Google Scholar]
- 43.Hock H, Orkin SH. Zinc-finger transcription factor Gfi-1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr Opin Hematol. 2006;13(1):1–6. doi: 10.1097/01.moh.0000190111.85284.8f. [DOI] [PubMed] [Google Scholar]
- 44.Karsunky H, et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet. 2002;30(3):295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- 45.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26(47):6816–28. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 46.Hock H, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431(7011):1002–7. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 47.Sansregret, Nepveu A. The multiple roles of CUX1: Insights from mouse models and cell-based assays. Gene. 2008:412. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Li S, et al. Regulation of the homeodomain CCAAT displacement/cut protein function by histone acetyltransferases p300/CREB-binding protein (CBP)-associated factor and CBP. Proc Natl Acad Sci U S A. 2000;97(13):7166–71. doi: 10.1073/pnas.130028697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khanna-Gupta A, et al. Chromatin immunoprecipitation (ChIP) studies indicate a role for CCAAT enhancer binding proteins alpha and epsilon (C/EBP alpha and C/EBP epsilon) and CDP/cut in myeloid maturation-induced lactoferrin gene expression. Blood. 2003;101(9):3460–8. doi: 10.1182/blood-2002-09-2767. [DOI] [PubMed] [Google Scholar]
- 50.Khanna-Gupta A, et al. C/EBP epsilon mediates myeloid differentiation and is regulated by the CCAAT displacement protein (CDP/cut) Proc Natl Acad Sci U S A. 2001;98(14):8000–5. doi: 10.1073/pnas.141229598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marziali G, et al. The activity of the CCAAT-box binding factor NF-Y is modulated through the regulated expression of its A subunit during monocyte to macrophage differentiation: regulation of tissue-specific genes through a ubiquitous transcription factor. Blood. 1999;93(2):519–26. [PubMed] [Google Scholar]
- 52.Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci U S A. 2004;101(31):11257–62. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin-Soudant N, et al. CDP/Cut DNA binding activity is down-modulated in granulocytes, macrophages and erythrocytes but remains elevated in differentiating megakaryocytes. Leukemia. 2000;14(5):863–73. doi: 10.1038/sj.leu.2401764. [DOI] [PubMed] [Google Scholar]
- 54.Li S, et al. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol Chem. 1999;274(12):7803–15. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 55.Mailly F, et al. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol Cell Biol. 1996;16(10):5346–57. doi: 10.1128/mcb.16.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mendelson M, et al. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol. 1996;77(Pt 12):3099–102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 57.Bolovan-Fritts CA, Mocarski ES, Wiedeman JA. Peripheral blood CD14(+) cells from healthy subjects carry a circular conformation of latent cytomegalovirus genome. Blood. 1999;93(1):394–8. [PubMed] [Google Scholar]
- 58.Hargett D, Shenk TE. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc Natl Acad Sci U S A. 2010;107(46):20039–44. doi: 10.1073/pnas.1014509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reeves MB, Compton T. Inhibition of inflammatory interleukin-6 activity via extracellular signal-regulated kinase-mitogen-activated protein kinase signaling antagonizes human cytomegalovirus reactivation from dendritic cells. J Virol. 2011;85(23):12750–8. doi: 10.1128/JVI.05878-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reeves MB, et al. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J Gen Virol. 2005;86(Pt 11):2949–54. doi: 10.1099/vir.0.81161-0. [DOI] [PubMed] [Google Scholar]
- 61.Soderberg-Naucler C, et al. Reactivation of latent human cytomegalovirus in CD14(+) monocytes is differentiation dependent. J Virol. 2001;75(16):7543–54. doi: 10.1128/JVI.75.16.7543-7554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rauwel B, et al. Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. Elife. 2015:4. doi: 10.7554/eLife.06068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rossetto CC, Tarrant-Elorza M, Pari GS. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013;9(5):e1003366. doi: 10.1371/journal.ppat.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kew VG, et al. Mitogen and stress activated kinases act co-operatively with CREB during the induction of human cytomegalovirus immediate-early gene expression from latency. PLoS Pathog. 2014;10(6):e1004195. doi: 10.1371/journal.ppat.1004195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meier JL, Stinski MF. Regulation of human cytomegalovirus immediate-early gene expression. Intervirology. 1996;39(5–6):331–42. doi: 10.1159/000150504. [DOI] [PubMed] [Google Scholar]
- 66.Sinclair J, Sissons P. Latency and reactivation of human cytomegalovirus. J Gen Virol. 2006;87(Pt 7):1763–79. doi: 10.1099/vir.0.81891-0. [DOI] [PubMed] [Google Scholar]
- 67.Stern JL, et al. Repression of human cytomegalovirus major immediate early gene expression by the cellular transcription factor CCAAT displacement protein. Virology. 2008;378(2):214–25. doi: 10.1016/j.virol.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]