Summary
The maintenance of tissue homeostasis is critically dependent on the function of tissue-resident immune cells and the differentiation capacity of tissue-resident stem cells (SCs). How immune cells influence the function of SCs is largely unknown. Regulatory T cells (Tregs) in skin preferentially localize to hair follicles (HFs), which house a major subset of skin SCs (HFSCs). Here, we mechanistically dissect the role of Tregs in HF and HFSC biology. Lineage-specific cell depletion revealed that Tregs promote HF regeneration by augmenting HFSC proliferation and differentiation. Transcriptional and phenotypic profiling of Tregs and HFSCs revealed that skin-resident Tregs preferentially express high levels of the Notch ligand family member, Jagged 1 (Jag1). Expression of Jag1 on Tregs facilitated HFSC function and efficient HF regeneration. Taken together, our work demonstrates that Tregs in skin play a major role in HF biology by promoting the function of HFSCs.
Introduction
Forkhead box P3 (FOXP3)-expressing regulatory T (Treg) cells are a specialized subset of CD4+ T cells that play a major role in establishing and maintaining immune tolerance. In the steady-state, the majority of these cells reside in primary and secondary lymphoid organs. However, subsets of Tregs stably reside in specific peripheral tissues, and an emerging body of literature suggests that tissue-resident Tregs have specialized functions that are unique to the tissues in which they reside. In visceral adipose tissue, Tregs express the peroxisome proliferator-activated receptor- γ, enabling them to function in lipid and glucose metabolism (Cipolletta et al., 2012). In addition, Tregs in muscle and lung express high levels of the epidermal growth factor ligand, amphiregulin, conferring the ability to mediate tissue protection and maintain barrier integrity (Arpaia et al., 2015; Burzyn et al., 2013; Villalta et al., 2014).
Both murine and human skin contain a large number of tissue-resident Tregs (Clark et al., 2006; Sanchez Rodriguez et al., 2014; Scharschmidt et al., 2015). However, the function of these cells in this tissue are only beginning to be elucidated. Early in neonatal life, a wave of highly activated Tregs accumulates in skin and play a major role in establishing immune tolerance to skin commensal microbes (Scharschmidt et al., 2015). In adult life, a subset of Tregs in skin facilitates wound healing through increased expression of the epidermal growth factor receptor (EGFR) (Nosbaum et al., 2016). Tregs that reside in skin predominantly localize around hair follicles (HFs) (Chow et al., 2013; Gratz et al., 2013; Sanchez Rodriguez et al., 2014). Hair follicles are highly specialized organelles that are in a perpetual state of growth and regeneration. In addition to Tregs, a major epithelial stem cell (SC) population localizes to HFs (HFSCs) (Blanpain and Fuchs, 2009). These cells play an indispensable role in HF regeneration and repair of the epidermal barrier after injury (Ito et al., 2005). Interestingly, several studies link Tregs with HF biology. Genome wide association studies in alopecia areata (AA), a disorder of HF regeneration, have revealed single nucleotide polymorphisms in genes involved in the differentiation and function of Tregs, including IL-2, the high-affinity IL-2 receptor alpha (CD25), CTLA-4, Eos (IKZF4) and Foxp3 (Petukhova et al., 2010). In addition, pharmacologic augmentation of Tregs in humans with low-dose IL-2 was highly efficacious in treating AA (Castela et al., 2014). Despite these associations, a functional link between Tregs and HFs has yet to be established.
Given that Tregs in skin localize to HFs, polymorphisms in genes intimately associated with Treg function are increased in patients with AA and Treg enhancement promotes hair regeneration in this disease, we sought to determine whether Tregs play a functional role in HF biology. Immunophenotypic profiling revealed that Treg numbers and activation in skin tightly correlate with specific phases of the HF cycle. Lineage-specific depletion of these cells resulted in a marked attenuation of HF regeneration. Mechanistically, we found that Tregs promote HF cycling by enhancing the activation and differentiation of HFSCs. An unbiased discovery approach revealed that Tregs in skin express high levels of the Notch ligand, Jagged 1 (Jag1), and that signaling through the Notch pathway is a major mechanism by which Tregs promote HFSC function. Our studies reveal a mechanistic link between skin-resident Tregs and HFSCs that is critical for tissue function.
Results
Highly activated Tregs accumulate in Telogen skin
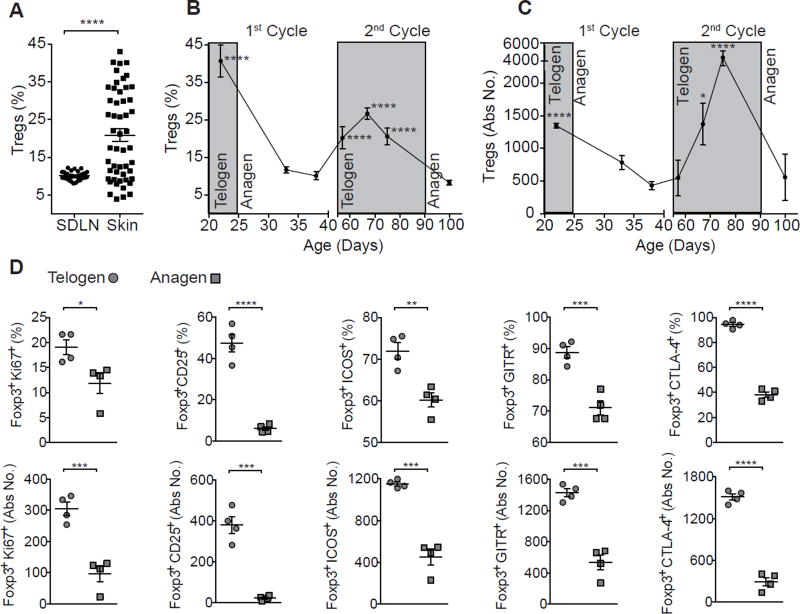
Hair follicles in mammalian skin undergo bouts of regeneration, cycling between highly synchronized phases of quiescence (telogen) and growth (anagen), ultimately resulting in the generation of a newly formed hair shaft (Müller-Röver et al., 2001). To test the hypothesis that Tregs play a functional role in HF biology, we began by performing comprehensive immune profiling of Tregs in murine skin at specific stages of the synchronous HF cycle (Müller-Röver et al., 2001). The proportion of CD4+Foxp3+ Tregs in skin draining lymph nodes (SDLNs) of adult C57BL/6 mice showed little variability (Figure 1A). In contrast, the accumulation of Tregs in dorsal skin was highly variable; however, this variance tightly correlated with HF cycling (Figures 1A-1C and S1A). Tregs were significantly more abundant in the telogen phase of the HF cycle when compared to anagen (Figures 1B and S1A). To ensure that these results were not an artifact of the tissue digestion required for flow cytometric analysis and to confirm that Tregs were accumulating around HFs, we quantified Tregs during telogen and anagen phases of the HF cycle by immunofluorescent microscopy. These studies revealed an ~3-fold higher accumulation around HFs in telogen skin relative to anagen skin, confirming our flow cytometric results (Figures S1B–S1C). While Tregs are uniquely more abundant in telogen skin relative to anagen skin, Foxp3 negative T effector cells (Teff) and CD8+ T cells show an inverse relationship to Tregs, and dermal gamma-delta T cells show little fluctuation with the HF cycle (Figures S1D). In addition, the proliferative index and activation state of skin Tregs (as evidenced by expression of Ki67, CD25, ICOS, GITR, and CTLA-4) correlated with stage of the HF cycle, as Tregs displayed a highly activated phenotype in telogen skin relative to anagen skin (Figures 1D and S1E). Taken together, these results suggest that Treg abundance and activation in skin tightly correlate with the HF cycle, with highly activated Tregs preferentially accumulating around HFs during late telogen.
Figure 1. Treg accumulation and activation in skin correlates with the HF Cycle.
(A) Treg cell abundance in skin draining lymph nodes (SDLNs) and skin of adult 4–14 week old WT mice as measured by flow cytometry. Pre-gated on live CD45+CD3+CD4+ cells. Flow cytometric profiling of Treg (B) frequency and (C) absolute numbers from dorsal skin of C57BL/6 mice at specific stages of the synchronous HF cycle (n = 5–12 mice per time point). Shaded areas represent telogen phase and unshaded areas represent anagen phase. Treg associated activation markers were assessed: CD25, ICOS, Ki67, CTLA-4, and GITR. (D) Summary of percent and absolute cell number (Abs No.) quantification of Treg activation marker expression in telogen and anagen skin. One representative experiment of three is shown. Data are mean ± s.e.m. *P<0.05, **P<0.01 ***P<0.001, ****P<0.0001; One-way ANOVA 1st telogen vs 1st anagen and 2nd telogen vs 2nd anagen (B, C), Student’s unpaired t-test (D). See also Figure S1.
Regulatory T cells are required for hair regeneration
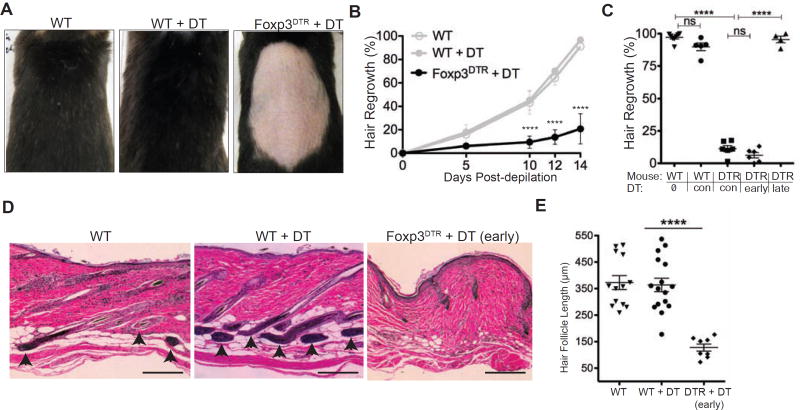
Given that both Treg abundance and activation correlated with HF stage, we sought to determine whether these cells play a functional role in HF cycling. To do so, we employed a well-established model of depilation-induced HF regeneration (Müller-Röver et al., 2001). In this model, mice with dorsal skin HFs in telogen are depilated to remove hair shafts. This treatment rapidly initiates the telogen-to-anagen transition homogenously across the entire depilated dorsum (i.e., anagen induction), and thus begins the process of hair regeneration. To determine if Tregs play a role in this process, we utilized mice transgenic for the diphtheria toxin receptor under the control of the Foxp3 promoter (Foxp3DTR) (Kim et al., 2007). These mice allow for robust depletion of Tregs following administration of Diphtheria toxin (DT). Importantly, Tregs begin to repopulate lymph nodes and peripheral tissues soon after the last DT treatment, permitting the study of transient Treg loss at specific times during HF regeneration (Kim et al., 2007). We monitored these mice for dorsal skin pigmentation, which is the earliest clinical sign of anagen entry (Müller-Röver et al., 2001), as well as clinical hair regrowth. Ablation of Tregs markedly reduced anagen induction and subsequent hair regrowth when compared to control wild type (WT) mice treated with or without DT (Figures 2A-2B and S2A–S2B). Whereas control mice had efficient anagen induction and completed hair regrowth by 14 days post-depilation, mice depleted of Tregs showed a marked diminution of anagen induction and had less than 20% hair regrowth at 14 days (Figures 2A-2B and S2A–S2D).
Figure 2. Tregs are required for hair regeneration.
Foxp3DTR mice or control WT mice were treated with DT on days −2, −1, and depilated on day 0 to induce anagen. DT administration was continued from Day 1 and then every other day until the termination of the experiment at day 14. (A) Representative photos and (B) kinetics of hair regrowth in WT and Treg depleted (Foxp3DTR) mice (n = 3–4 mice per group). Tregs were depleted either up to day 4 (early), from day 7 onwards (late) or constitutively (con) throughout the experimental period. (C) Hair regrowth at day 14 in WT and Foxp3DTR mice with DT treatments as indicated. (D), Representative H&E staining of skin from WT and Foxp3DTR mice on day 14. Arrows indicate anagen HF extension into dermal adipose. Scale Bars, 100 µ m. (E), Quantification of HF length on day 14. Data are mean ± s.e.m. ****P<0.0001, ns = not significant; One-way ANOVA (B, C, E). Foxp3DTR + DT vs WT + DT (B). See also Figure S2.
In these initial experiments, Tregs were continually depleted (i.e., DT administered every 2 days for 14 days). Because a major function of Tregs is to suppress inflammation, mice deficient in these cells for prolonged periods of time develop multiorgan autoimmunity (Kim et al., 2007). Thus, we set out to determine whether attenuation of HF cycling in the absence of Tregs was a result of prolonged systemic inflammation. In addition, we wanted to precisely define a potential ‘window’ of time for Treg requirement in HF cycling. To do so, we treated WT or Foxp3DTR mice with DT ‘early’ after depilation (up to 3 days post-depilation), ‘late’ after depilation (starting at 7 days post-depilation), or throughout the entire 14-day period of hair regeneration. Transient depletion of Tregs early after depilation fully recapitulated the attenuation of HF regeneration observed with constant Treg depletion. In contrast, late Treg depletion had no effect on hair regrowth (Figures 2C and S2C–S2D). Histologic examination of skin showed a marked diminution of anagen HFs in ‘early’ Treg depleted mice. Whereas HFs in control mice displayed an elongated phenotype extending deep into the dermal adipose (indicative of anagen induction) (Müller-Röver et al., 2001), HFs from Treg-depleted mice were significantly shorter in length and failed to extend beyond the superficial dermis (Figures 2D-2E and S2E–S2F). Consistent with these findings, mice that lack all T cells (Rag2−/−) have a significant delay in anagen induction capacity in response to depilation-induced hair regeneration, as well as during the natural synchronous HF cycle (Figures S2G–S2J). Taken together, these results indicate that Tregs play a major role in facilitating HF regeneration by promoting the telogen-to-anagen transition.
Regulatory T cells in skin preferentially localize to the HFSC niche
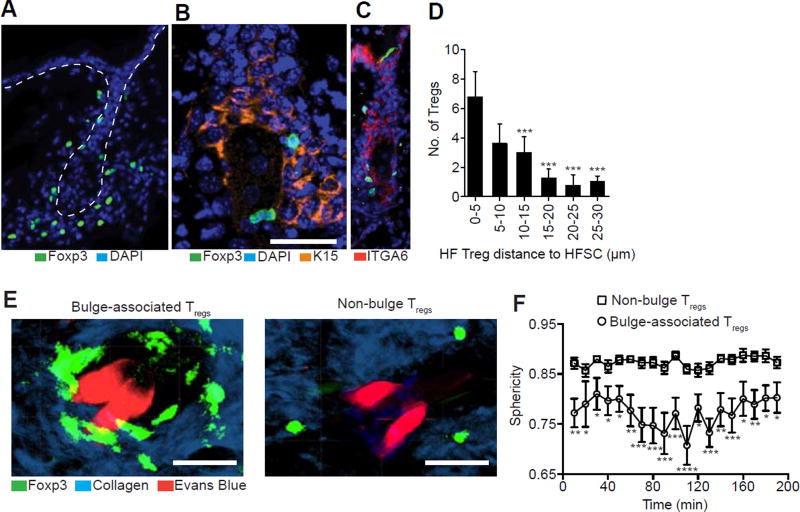
Hair follicle stem cells play a major role in HF regeneration (Blanpain and Fuchs, 2009). The transition of telogen HFs to anagen HFs is mediated by the activation, proliferation and differentiation of HFSCs (Blanpain et al., 2006; Morris et al., 2004; Trempus et al., 2003). Because Tregs in skin localize to HFs (Chow et al., 2013; Gratz et al., 2013; Sanchez Rodriguez et al., 2014) and facilitate the telogen-to-anagen transition, we set out to determine if Tregs in skin co-localize with HFSCs. Immunofluorescence microscopy of dorsal skin of Foxp3GFP reporter mice (Lin et al., 2007) revealed that Tregs predominantly reside proximal to the lower portions of telogen HFs, clustering around the bulge region (Figures 3A), a well-established niche for HFSCs (Morris et al., 2004; Trempus et al., 2003). Co-staining of cytosolic expressed GFP with markers of bulge resident SCs (Morris et al., 2004), Keratin-15, and ITGA6, revealed a sub-population of bulge-associated Tregs in close association with HFSCs (Figures 3B-3C and S3). Quantification of Foxp3-GFP+ Tregs within 30µm of follicular epithelium demonstrated that the majority of follicular Tregs reside within 0–5 µm of bulge HFSCs (Figure 3D).
Figure 3. Tregs in skin preferentially reside in close approximation to HFSCs.
(A) Representative immunofluorescent image of Foxp3+ Tregs in telogen skin of Foxp3GFP reporter mice. Dashed line indicates outline of HF. ‘B’ indicates bulge region. Treg cell co-staining with (B) Keratin-15 (K15) and (C) integrin-alpha 6 (ITGA6). (D) Quantification of follicular Treg distance to bulge HFSCs. Data are combined counts from >10 sections. (E) Maximal intensity projection profiles of bulge-associated and non-bulge associated Tregs from intravital imaging of Foxp3GFP mice compiled from images recorded every 10 minutes during a 200 min time-lapse. HF Vasculature is labelled with Evans Blue dye (red). (F) Quantification of cell sphericity between bulge-associated and non-bulge associated Tregs during a 200 min time-lapse. Scale Bars, 50 µm. Data are mean ± s.e.m. *P<0.05, **P<0.01 ***P<0.001, ****P<0.001; One-way ANOVA, 0–5 µm vs. all other groups (D); Two-way ANOVA, bulge- vs. non-bulge Tregs at each respective time-point (F). One representative experiment of three with n = 3 mice total. See also Figure S3.
To determine the dynamic behavior of Tregs in skin with high hair density (i.e., dorsal skin), we established a non-invasive vacuum suction approach for live intravital 2-photon imaging of Tregs in dorsal skin of Foxp3GFP reporter mice. This technique was adapted from approaches utilized in other tissues (Boldajipour et al., 2016; Broz et al., 2014; Headley et al., 2016; Lin et al., 2007; Lindsay et al., 2015; Looney et al., 2011; Sano et al., 2016; Vinegoni et al., 2012). Evans blue dye was utilized to ensure all intravascular Tregs were excluded while all cells located in the extravascular space were included for analysis. Utilizing this system, we observed that bulge-associated Tregs differ markedly with respect to shape and behavior when compared to Tregs found >20 µm away from follicular epithelium. Bulge-associated Tregs displayed a more amoeboid cell morphology with increased protrusive activity (Figures 3E and movies S1 and S2). This differential cell shape was quantified in individual skin Tregs by applying a measure of relative sphericity (Matheu et al., 2013; Zaid et al., 2014). Cells that are in contact with other cells typically have characteristic dynamic behavior. Specifically, cell-to-cell contacts alter the shape of cells such that they are no longer spherical (Lecuit and Lenne, 2007). When compared to non-bulge Tregs, the sphericity of bulge-associated Tregs was significantly lower at all time points analyzed (Figure 3F). While this parameter is not a specific measure of Treg function, our findings indicate that Tregs in and around the HF niche are more dynamically active when compared to Tregs found outside this region. Taken together with images obtained by static immunofluorescence microscopy, these results suggest that Tregs are highly active in the bulge region of the HF and have the potential to communicate with bulge-associated HFSCs.
Regulatory T cells promote HFSC proliferation and differentiation
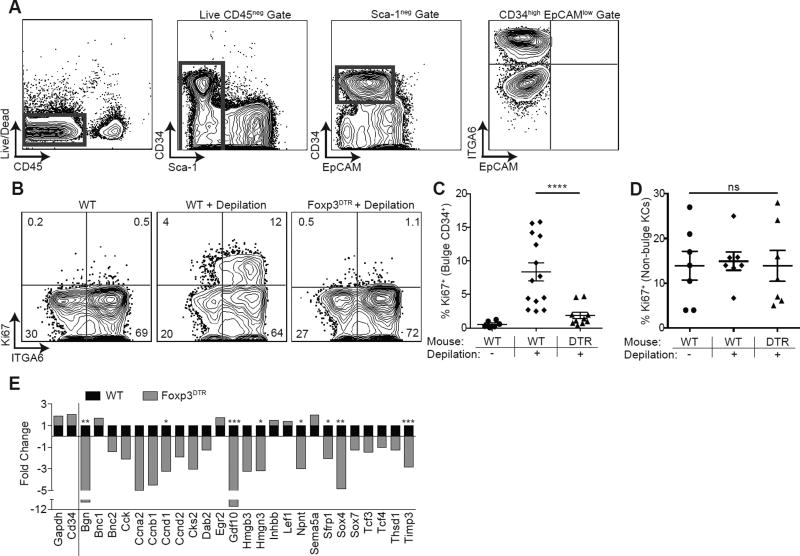
Given that HFs from Treg-depleted mice failed to regenerate, Tregs reside in the bulge region of the HF in close association with HFSCs and activation of HFSCs is necessary and required for HF regeneration, we tested whether Tregs mediate their effects by influencing the function of HFSCs. Upon anagen induction, HFSCs are activated, begin to proliferate and eventually differentiate to form all cell lineages of the newly generated HF (Blanpain et al., 2006; Morris et al., 2004; Trempus et al., 2003). Because Tregs function early to facilitate anagen induction, we set out to determine their influence on HFSC activation during the telogen-to-anagen transition. To do so, we utilized a previously established flow cytometric approach to delineate HFSCs (Nagao et al., 2012). Epidermal cell suspensions were prepared from dorsal skin and stained for CD45, Sca-1, EpCAM, CD34 and integrin α6 (ITGA6). Bulge HFSCs were defined as CD45negSca-1negEpCAMlowCD34high cells (Figure 4A). High levels of ITGA6 expression on this population distinguishes basal bulge residing HFSCs from those in the suprabasal layer (Trempus et al., 2003). Analysis of mid-telogen WT skin confirmed the presence of quiescent bulge-resident HFSCs (Cotsarelis et al., 1990), as evidenced by lack of expression of the proliferative marker Ki67 (Figures 4B-4C). Consistent with previous reports, HFSC activation closely followed depilation-induced anagen induction (Chen et al., 2012), with an increased percentage of HFSCs expressing Ki67 early after depilation (Figures 4B-4C). Induction of HFSC proliferation was significantly attenuated in mice transiently depleted of Tregs in the early window of time after depilation (Figures 4B-4C). This proliferative defect appeared to be selective for the bulge HFSC compartment, as no difference in Ki67 expression was observed in non-bulge keratinocytes between Treg-sufficient and Treg-depleted mice (Figures 4D). Absolute cell numbers of HFSCs were unchanged between all treatment groups, suggesting that HFSCs are not DT-sensitive or directly depleted in Foxp3DTR mice (Figure S4A). Absence of Ki67 in the bulge region of the HF in Treg depleted mice was confirmed by histological staining of dorsal skin sections (Figures S4B).
Figure 4. Tregs are required for HFSC proliferation and differentiation.
(A) Flow cytometric gating strategy to identify CD34+ integrin α6high (ITGA6) bulge HFSCs. Foxp3DTR mice or control mice were treated with DT on days −2, −1, depilated on day 0 to induce anagen and DT administered again on days 1 and 3 (i.e., early regimen). (B) Representative flow cytometric plots of Ki67 expression in bulge HFSCs between WT and FoxpDTR mice 4 days after depilation. (C) Flow cytometric quantification of Ki67+ bulge HFSCs and (D) non-bulge keratinocytes 4 days after depilation. RNA sequencing was performed on FACS purified bulge HFSCs at day 4 post-depilation from control (WT) or Treg depleted (Foxp3DTR) mice. (E) Fold change in HFSC differentiation genes in WT and FoxpDTR mice, expressed as fold change relative to WT (where a value of 1 = no change). Genes to the left of the solid line represent control genes. Significance values are calculated based on transcript expression level. Data are mean ± s.e.m. *P<0.05, **P<0.01 ***P<0.001, **** P<0.0001, ns = no significant difference, One-way ANOVA (C and D); RNA-Seq Differential Expression analysis, WT vs. Foxp3DTR for each respective gene (E). See also Figure S4.
To determine if Tregs play a similar role in steady-state mediated HFSC activation during the natural telogen-to-anagen transition, we transiently administered DT or PBS (vehicle control) to Foxp3DTR mice to deplete Tregs during 1st telogen from postnatal day 21 (P21) to P24. Relative to littermate controls, DT-treated Foxp3DTR mice fail to enter 1st anagen as evidenced by a lack of dorsal skin pigmentation and significantly diminished proliferative capacity of HFSCs (Figures S4C–S4E). While the absolute number of CD34+ HFSCs was equivalent, the proportion and absolute number of proliferating Ki67+ HFSCs were significantly lower in Treg depleted mice (Figures S4F–S4H). These data provide evidence that Tregs play a role in promoting the telogen-to-anagen transition during the natural HF cycle.
To investigate the in vivo frequency of proliferative HFSCs, we performed short term nucleotide labelling via injection of 5-ethynyl-2-deoxyuridine (EdU) at day 3 post-anagen entry and measured incorporation into HFSCs 24-hours later. While Treg sufficient mice efficiently incorporated EdU, the median fluorescence intensity (MFI) of HFSCs in Treg-deficient mice was significantly less (Figures S4I–S4J). These results indicate that in the absence of Tregs, HFSCs are unable to undergo multiple rounds of proliferation during this 24-hour chase period. These findings are consistent with experiments showing reduced Ki67+ HFSCs by flow cytometry (Figures 4B-4C).
To elucidate if Tregs influence HFSC differentiation, we performed whole transcriptome RNA sequencing (RNAseq) of bulge HFSCs. Lineage tracing studies in the SC niche have identified a role for ITGA6-expressing basal cell progenitors and their progeny in adopting a HF fate (Ito et al., 2005; Jaks et al., 2008; Levy et al., 2005; Morris et al., 2004; Tumbar et al., 2004). We therefore analyzed CD34+ITGA6high basal bulge HFSCs purified from Treg-sufficient and Treg-depleted mice 4 days post-depilation. Consistent with our Ki67 flow cytometric data, HFSC from Treg-depleted mice showed a significant reduction in genes associated with cell proliferation (Table S1). In addition, these HFSCs also showed a reduction in genes associated with the differentiation of HFSCs to keratinocyte lineages, including Bgn, Bnc1, Cks2, Hmgn3, Gdf10, and Sox4 (Figure 4E) (Lowry et al., 2005). Collectively, these results suggest that a major mechanism by which Tregs mediate HF cycling is by promoting the activation and differentiation of bulge HFSCs.
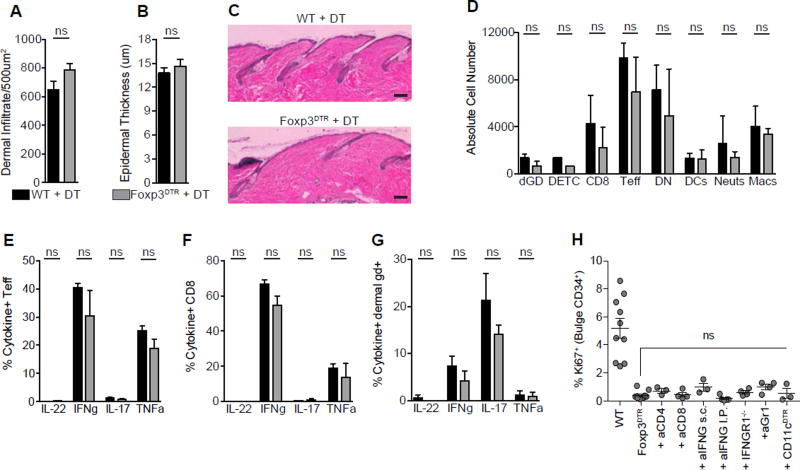
Transient Treg depletion results in minimal skin inflammation
We next set out to determine the cellular and molecular mechanisms by which Tregs influence HFSC function. Because a major mechanism by which Tregs mediate their effects is through suppression of inflammation, we first set out to define the inflammatory milieu in skin after transient Treg depletion ‘early’ after depilation. Surprisingly, classic hallmarks of skin inflammation, such as epidermal hyperplasia and immune cell infiltrate, were not significantly different between control mice and mice transiently depleted of Tregs (Figures 2D and 5A–5C). Consistent with these findings, flow cytometric quantification of the absolute numbers of dermal γδ T cells, dendritic epidermal T cells, cytotoxic CD8+ T cells, CD4+Foxp3− T effector cells, dendritic cells, neutrophils, and macrophages revealed no differences between Treg-depleted and Treg-sufficient mice (Figures 5D). In addition, skin effector T cell production of interleukin (IL)-22, interferon-gamma, IL-17 and tumor necrosis factor-alpha were similar between control and transient Treg-depleted mice (Figures 5E-5G and S5). To functionally determine if inflammatory cells play a role in Treg-mediated activation of HFSCs, we co-depleted Tregs with specific immune cell subsets to determine if HFSC function could be rescued in the absence of Tregs. HFSC proliferation was not restored following co-depletion of Tregs with CD4+ T cells, CD8+ T cells, Gr-1 expressing neutrophils, or CD11c-expressing myeloid cells (Figure 5H). In addition, both antibody neutralization and genetic deletion of the interferon-γ pathway, a major effector cytokine suppressed by Tregs (Nosbaum et al., 2016; Sojka and Fowell, 2011), did not rescue HFSC activation (Figure 5H). These findings suggest that suppression of inflammation is not a major mechanism by which Tregs promote HFSC function early in the process of hair regeneration.
Figure 5. Transient Treg loss results in minimal skin inflammation.
Control wild-type (WT) or Foxp3DTR mice were depilated and treated with DT according to the ‘early’ depletion protocol. (A) Total dermal infiltrate and (B) epidermal hyperplasia in DT treated control (WT + DT) and Tregs depleted (Foxp3DTR + DT) dorsal skin on day 4 as measured by routine histology. (C) Representative H&E staining of skin from WT and Foxp3DTR mice on day 4. (D) The absolute cell number of innate and adaptive immune cell subsets in skin as measured by flow cytometry. Single cell suspensions from day 1 skin were stimulated with PMA/ionomycin and the production of IL-22, IFNg, IL-17 and TNFalpha was assessed by flow cytometry. The proportion of cytokine producing (E) CD4+ Teff Cells, (F) CD8+ T cells, and (G) dermal γδ+ T cells in dorsal skin. (H) Flow cytometric quantification of Ki67+ bulge HFSCs at day 4 post-depilation in immune cell depleted and interferon-γ neutralized (or interferon-γ signaling deficient) mice. Scale Bars, 50 µm. Data are mean ± s.e.m. ns = no significant difference, Unpaired Student’s t-test (A, B); Two-way ANOVA (D–G); One-way ANOVA (H). Combined data from two experiments, with n = 3–4 mice per group. See also Figure S5.
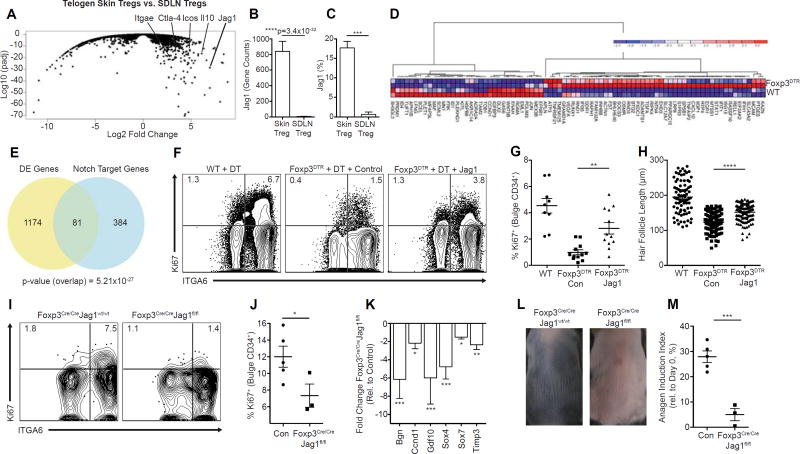
Regulatory T cells in skin preferentially express the Notch ligand Jagged 1
In an attempt to delineate potential mechanisms by which Tregs promote HFSC function, we compared the transcriptome of Tregs isolated from telogen skin with Tregs isolated from SDLNs (Table S2). We hypothesized that pathways involved in this process would be preferentially expressed in skin Tregs when compared to Tregs found in SDLNs. Consistent with heightened activation of Tregs in tissues (Burzyn et al., 2013; Feuerer et al., 2009; Sanchez Rodriguez et al., 2014), differential expression analyses revealed increased abundance of genes associated with Treg function in skin compared to SDLN Tregs, including CTLA-4, ICOS and IL-10 (Figure 6A). Interestingly, Jag1, a ligand of the Notch signaling pathway, was among the highest differentially expressed genes between skin and SDLN Tregs (Figure 6A). Tregs in skin expressed approximately 150-fold more Jag1 transcript than SDLN Tregs (p-value=3.4×10−32; Figure 6B). Assessment of Jag1 protein expression by flow cytometry confirmed greater expression in skin Tregs relative to SDLN Tregs, and increased expression on Tregs in telogen skin as compared to Tregs in anagen skin (Figure 6C, and S6A–S6D). In addition, Treg-lineage expression of Jag1 was independently validated by qPCR, relative to CD4+ Teff cells and CD8+ T cells (Figures S6E). Aside from preferential expression on skin Tregs, Jag1 was an especially interesting candidate given the established role of the Notch pathway in HFSC biology (Blanpain et al., 2006; Estrach et al., 2006; Vauclair et al., 2005). Having identified skin Treg expression of Jag1, we next sought to determine if Notch target gene transcripts (Meier-Stiegen et al., 2010) were differentially expressed in HFSCs in the presence or absence of Tregs. Genome wide RNAseq analysis of HFSCs isolated 4 days after anagen induction in the presence or absence of Tregs revealed a differential signature of Notch target genes (Figures 6D-6E). Of the 1174 differentially expressed genes between the two groups, 84 genes (~7%) were known transcriptional targets of the Notch signaling pathway (p-value of the overlap = 5.21×10−27). Collectively, these results indicate that Tregs in murine skin preferentially express the Notch ligand, Jag1, and that in the absence of Tregs, Notch signaling is significantly altered in HFSCs.
Figure 6. Treg expression of Jagged-1 (Jag1) is required for efficient HFSC activation and anagen induction.
RNA sequencing was performed on telogen skin Tregs and Tregs isolated from SDLNs. (A) Volcano plot comparing expression profile of skin versus SDLN Tregs. (B) Raw gene counts of Jag1 transcripts as quantified by RNA sequencing (n = 4 for skin Tregs and n = 3 for SDLN Tregs). (C) Flow cytometric quantification of Jag1 expression on SDLN Tregs and skin Tregs (n = 4 for both skin and SDLN Tregs). RNA sequencing was performed on FACS purified bulge HFSCs at day 4 post-depilation from control (WT) or Treg depleted (Foxp3DTR) mice. (D) Hierarchial clustering of differential expression of Notch target genes in HFSCs sequenced from control (WT) and Treg depleted (DTR) mice, depicted as a heat map (E) Venn diagram depicting the overlap between the total differentially expressed (DE) genes and known Notch target genes. P-value represents the significance of the overlap as determined by a chi-squared test. Jag1-Fc coated or control Fc coated beads were administered subcutaneously in Treg depleted mice on days −2, −1, 1 and 3. All mice were depilated on day 0. (F) Representative flow cytometric plots 4 days post-depilation gated on HFSCs from WT and Foxp3DTR mice treated with control or Jag1-Fc coated beads. (G) Quantification of Ki67+ bulge HFSCs and (H) HF length 4 days post-depilation. Control (i.e., Foxp3Cre/CreJag1wt/wt) or Foxp3Cre/CreJag1fl/fl mice were depilated to induce anagen. (I) Representative flow cytometric plots and (J) quantification of Ki67+ bulge HFSCs. (K) Fold change in HFSC differentiation genes as measured by qRT-PCR. (L) Representative photos and (M) quantification of skin pigmentation. RNA-Seq experiments were conducted using 2–4 biological samples (A, D, and E). Data are combined from three independent experiments (F–H). One representative experiment of two (I–M). Data are mean ± s.e.m. *P<0.05, **P<0.01 ***P<0.001, ****P<0.001, RNA-Seq Differential Expression analysis, Skin Tregs vs. SDLN Tregs (B); Unpaired t-test (C, J, and M) ; One-way ANOVA (G, H, and K). See also Figure S6.
Regulatory T cell expression of Jag1 promotes HFSC function
To functionally determine if Notch signaling plays a role in Treg-mediated enhancement of HFSC activation, we attempted to rescue the proliferative defect observed in HFSCs in Treg-depleted mice by the exogenous addition of Jag1 (Vas et al., 2004). Microbeads coated with Jag1-Fc or control Fc were subcutaneously administered to DT-treated Foxp3DTR mice and activation of HFSCs was quantified by flow cytometry 4 days post-depilation. In these experiments, exogenous Jag1 was able to partially rescue HFSC activation and induction of anagen in the absence of Tregs (Figure 6F-6H). To definitively test whether Jag1 expression on Tregs plays a role in HFSC differentiation and anagen induction, mice expressing a Jag1 conditional allele (Brooker et al., 2006) (Jag1fl/fl) were crossed to Foxp3-cre mice (Rubtsov et al., 2008) to specifically ablate Jag1 expression in Tregs. Foxp3Cre/CreJag1fl/fl mice in telogen phase of the HF cycle were depilated and HFSC proliferation, differentiation and kinetics of anagen induction were quantified. Early after depilation, Foxp3Cre/CreJag1fl/fl mice had significantly attenuated proliferative capacity of integrin-α6high CD34+ bulge HFSCs when compared to age- and gender- matched littermate controls (Figure 6I-6J). Absolute cell numbers of HFSCs in Foxp3CreJag1fl/fl mice and the ratio of Treg-to-HFSCs both in the steady-state and post-depilation were unchanged relative to Foxp3Cre/Jag1wt/wt mice (Figure S6F–S6H), suggesting that Jag1 loss in Tregs does not alter the overall composition of Tregs and/or HFSCs in skin.
In addition, relative to control mice, deletion of Jag1 in Tregs resulted in a significant reduction in the expression of key epidermal differentiation genes, including Bgn, Ccnd1, Gdf10, Sox4, Sox7 and Timp3 (Figure 6K). Given the early requirement for Tregs in HFSC activation, we assayed the kinetics of anagen induction by quantifying dorsal skin pigmentation (the earliest clinical marker of initiation of this process) (Müller-Röver et al., 2001). Consistent with the diminished proliferative and differentiation capacity of HFSCs, anagen entry in Foxp3Cre/CreJag1fl/fl dorsal skin was significantly attenuated relative to control dorsal skin (Figure 6L-6M), revealing a role for Jag1 expression on Tregs in facilitating anagen induction. Taken together, our results suggest that the Jag1-Notch pathway plays a role in the ability of skin Tregs to promote HFSC function.
Discussion
There is an emerging body of literature suggesting that Tregs represent a highly complex and heterogeneous lymphocyte lineage that possess specialized functions in the tissues in which they reside (Panduro et al., 2016). Here, we describe a previously unrecognized role of skin-resident Tregs in promoting epithelial SC function. We demonstrate a critical requirement for Tregs in HF regeneration through facilitation of epithelial SC proliferation and differentiation. Consistent with studies examining Tregs in muscle and lung (Arpaia et al., 2015; Burzyn et al., 2013; Villalta et al., 2014), we establish a role for Tregs in mediating a vital regenerative function of skin. The fact that skin Tregs facilitate HF regeneration by promoting HFSC activation is consistent with the observation that Tregs in bone marrow co-localize with hematopoietic SCs and support their function in this tissue (Fujisaki et al., 2011). Collectively, these findings suggest that a fundamental property of both intra- and extra- lymphoid Tregs is to influence SC biology and supports the view that a functional relationship between Tregs and SCs is a generalizable concept intrinsic to many tissues.
The finding that short-term depletion of Tregs did not alter accumulation or activation of the major immune cell subsets in skin suggests that transient Treg loss does not result in overt skin inflammation. Consistent with this observation, co-depletion of inflammatory cells (and a major inflammatory pathway) with Tregs did not restore HFSC function. Taken together, these results suggest that suppression of inflammation is not the major mechanism by which Tregs promote HFSC proliferation and differentiation. Furthermore, Treg lineage specific deletion of Jag1 largely resembled loss of the entire Treg population, with respect to HFSC proliferation, differentiation, and the capacity to induce HF entry into anagen. While our data do not definitively rule out the potential for Tregs to exert their effects on epithelial SCs by suppressing immune activation, they suggest that Tregs in skin have the ability to modulate the biology of tissue SCs independently of this function. Whether this is mediated by a subset of Tregs or the bulk skin-resident Treg population and whether expression of Jag1 is the only mechanism that Tregs utilize in this process remain to be determined.
A classic function of HFSCs is to drive HF cycling and HF regeneration (Blanpain and Fuchs, 2009; Morris et al., 2004; Trempus et al., 2003). However, an alternative function of these cells is to aid in epidermal barrier repair after injury (Ito et al., 2005). Unlike HF cycling, epidermal barrier repair is associated with marked tissue inflammation. Thus, in addition to their role in promoting HF cycling, it is interesting to speculate that skin Tregs also play a role in facilitating the function of HFSCs in the context of epidermal barrier repair. In this capacity, Tregs may regulate both inflammatory and non-inflammatory pathways to mediate their effects. It is currently unknown whether Tregs in skin influence HFSC biology in contexts other than HF cycling.
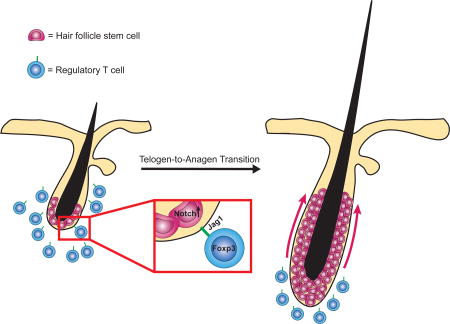
Our results highlight the Notch pathway as a mechanism employed by skin-resident Tregs to facilitate HFSC function during HF cycling. Skin Tregs in proximity to the HF are activated, by currently unknown mechanisms, as the follicle transitions from telogen to anagen. These activated Tregs produce the Notch ligand, Jag1, which stimulates the proliferation and differentiation of HFSCs and drives progression through the anagen phase. The ability of one immune cell population in skin to regulate the behavior of SCs in that tissue raises the novel possibility that immune control of stem cell function may be a general paradigm applicable to stem cells in other tissues. It will be important to determine if this principle extends to human diseases of epithelial dysfunction, and if Tregs can be exploited to develop new therapies for SC-mediated tissue regenerative disorders.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Rosenblum (Michael.Rosenblum@ucsf.edu).
Experimental Models and Subject Details
Experimental Animals
Mice were bred and/or maintained in the UCSF specific pathogen-free facility in accordance with the guidelines of the Laboratory Animal Resource Center and Institutional Animal Care and Use Committee of the University of California San Francisco (UCSF). All mice used in experiments were socially houses under a 12 hr light/dark cycle. All experiments were initiated in the telogen phase of the HF cycle in 7–10 week old mice, unless otherwise specified.
Method Details
Anagen induction
Dorsal hair was first shortened with clippers before applying depilatory cream (Nair) to the shaved region for a period of exactly 30 seconds before wiping clean. For monitoring of clinical hair regrowth, standardised pictures were taken with a ruler on the day of depilation (day 0) and then at the indicated time points until day 14. Anagen induction was quantified using intensity analysis on ImageJ software (v1.46r, NIH, USA) at each time point or as a percent of pigmented dorsal skin relative to baseline (day 0). All experiments conducted in Foxp3Cre/CreJag1fl/fl or control mice (i.e., Foxp3Cre/CreJag1wt/wt or Foxp3wt/wtJag1fl/fl) were harvested on day 10 post depilation. For monitoring of the synchronous HF cycle, dorsal hair was clipped and pictures taken at the indicated time points for quantification. For flow cytometric analysis and cell sorting for RNA-sequencing experiments, epidermal cells were prepared from mice harvested at day 4 post-depilation.
In vivo Treg cell depletion
The optimal dose of each DT Lot (Sigma) was initially determined by assessing the efficiency of skin Treg depletion by flow cytometry. Accordingly, Foxp3DTR and control mice were injected i.p. with DT at 15 ng/g or 30ng/g body weight, according to three regimens: mice were injected with DT on day −2 and day −1 prior to depilation (performed on day 0) and then every other day until day 4 (early; 4 doses total) or day 14 (constitutive; 10 doses total). For late Treg depletion studies, DT was administered first on day 7 and then every other day until day 14 (4 doses total). For Treg depletion during the natural HF cycle in Foxp3DTR mice, DT or PBS was injected at postnatal day 21 (P21), P22, and P24.
Immune cell co-depletions
All antibodies for depletion studies were purchased from BioXcell (West Lebanon, NH, USA). Anti-CD4 (GK1.5, 400 µg per injection), anti-CD8 (2.43, 400 µg per injection), anti-interferon gamma (IFNG, XMG1.2, 500 µg per injection), anti-Gr1 (RB6–8C5, 100 µg per injection) were administered i.p with DT according to the early Treg cell ablation regimen in Foxp3DTR mice. For subcutaneous anti-IFNG administration, 100 µg was injected directly into dorsal skin on the day of depilation (day 0) and then on day 1 and day 3. For IFNGR1−/− hosts, anti-CD4 was administered on days −2, −1, day 1 and day 3 to deplete CD4+ T-cells. For CD11cDTR/GFP hosts, 15ng/g DT and anti-CD4 was administered on days −2, −1, day 1 and day 3 to deplete CD11c–expressing myeloid cells and CD4+ T-cells, respectively.
Treg Conditional Jag1 Deletion
All experimental mice were generated from breeders that consisted of Foxp3-Cre homozygous/Jag1 heterozygous females (Foxp3Cre/CreJag1wt/fl) crossed to Foxp3-Cre hemizygous/Jag1 heterozygous males (Foxp3CreJag1wt/fl). Genotyping of the resulting offspring determined that all female mice were homozygous for Foxp3-Cre and all males hemizygous for Foxp3-Cre. For the floxed Jag1 allele mice were either wild-type (Jag1wt/wt), heterozygous (Jag1wt/fl), or homozygous (Jag1fl/fl). Unless otherwise stated, only age, gender, and littermate matched Foxp3-Cre homozygous (or hemizygous for males)/Jag1-wild-type mice were used as controls (Foxp3Cre/CreJag1wt/wt) and Foxp3-Cre homozygous (or hemizygous for males) Jag1 flox-homozygous mice for the experimental group (Foxp3Cre/CreJag1fl/fl).
Tissue processing
Preparation of single cell suspensions from SDLNs or full thickness skin excised at indicated time points during the synchronous HF cycle for Treg analysis or cell sorting by flow cytometry was performed as previously described(Scharschmidt et al., 2015). Briefly, Isolation of cells from axillary, brachial and inguinal lymph nodes (referred to as skin draining lymph nodes, SDLNs) for flow cytometry was performed by mashing tissue over 100 µm sterile filters For isolation of dorsal skin cells, mouse dorsal skin was harvested and lightly defatted. It was then minced finely with scissors and re-suspended in a 50ml conical with 3 ml of digestion mix (composed of 2mg/ml collagenase XI, 0.5mg/ml hyaluronidase and 0.1mg/ml DNase in RPMI with 1% Hepes, 1% penicillin-streptomycin and 10% fetal calf serum), followed by incubation in a shaking incubator at 37° C at 250 rpm for 50 minutes. An additional 20 m l of RPMI/Hepes/P-S/FCS media was then added and the 50ml conical was shaken by hand for 30–45 seconds. Another 20 ml of media was added and the suspension filtered through a sterile 100µm cell strainer followed by a 40el cell strainer into a new 50ml conical. The suspension was then pelleted and re-suspended in PBS for cell counting and staining. Epithelial cell preparations from depilated mice were prepared for flow cytometric analysis or HFSC sorting as previously described(Nagao et al., 2012). Briefly, mouse dorsal skin was harvested and lightly defatted before placing upon 2 ml of Trypsin-EDTA (0.5%, 10x, ThermoFisher), followed by incubation at 37°C for 1 hour. Epidermal cells were then scraped off into a petri dish containing RPMI/Hepes/P-S/FCS media and the cell suspension then filtered through a sterile 100µm cell strainer followed by a 40el cell strainer into a new 50ml conical. The suspension was then pelleted and re-suspended in PBS for cell counting and staining. Following isolation from the tissue, cells were labeled stained in PBS for 30 min at 4° C with surface anti bodies and a live dead marker (Ghost Dye™ Violet 510, Tonbo Biosciences). For intracellular staining, cells were fixed and permeabilized using reagents and protocol from the Foxp3 staining buffer kit (eBioscience). Fluorophore-conjugated antibodies specific for mouse cell surface antigens and intracellular transcription factors were purchased from eBioscience, BD Biosciences or Biolegend as detailed in the Key Resources Table. Samples were run on a Fortessa (BD Biosciences) in the UCSF Flow Cytometry Core. Treg cells from skin and SDLNs were sorted on a FACS Aria2 (BD Biosciences). For all experiments, voltages were standardized using SPHERO Rainbow calibration particles (BD Biosciences). An automated cell counter (NucleoCounter® NC-200™, Chemometec) was used to enumerate cell numbers from single cell suspensions and for calculations of absolute cell numbers. Flow cytometry data was analyzed using FlowJo software (FlowJo, LLC). Strict dead cell and doublet cell exclusion criteria were included for all immune cell analysis, followed by pre-gating for all hematopoietic cells as CD45+. Lymphoid cells were gated as γδ-TCR−CD3− double negative cells (DN), γδ-TCR+CD3+ dermal γδ T cells (dGD), γδ-TCRhiCD3hi dendritic epidermal T cells (DETCs), CD3+CD8+ T cells (CD8), CD3+CD4+Foxp3− T effector cells (Teff), and CD3+CD4+Foxp3+ regulatory T cells (Treg). Myeloid cells were all pre-gated as γδ-TCR−CD3− double negative and then gated as CD11c+MHC-Class II+ dendritic cells (DCs), Ly-6G+CD11b+ neutrophils (Neuts), and Ly-6G−CD11b+ClassII+F4/80+ macrophages (Macs). For nucleotide incorporation experiments, Click-iT® Plus EdU Flow Cytometry Assay Kits (Fisher) was used as per the manufacturer’s protocol.
Histology
For histopathology, skin tissue was fixed in 10% formalin and paraffin-embedded, sectioned and stained with hematoxylin and eosin by the UCSF Mouse Pathology Core. Histopathology images were acquired on a Leica microscope using a DS-Ri1 camera and NIS-Elements software (Nikon). H&E quantifications of HF length, dermal infiltrate and epidermal hyperplasia were performed using ImageJ64 software (NIH, USA). For immunofluorescent tissue staining in Figure 1A, dorsal skin from Foxp3GFP mice was first fixed in 2% PFA for 6–8 hours, washed with PBS and left in 30% sucrose overnight before embedding in OCT and freezing in a isopentane solution cooled over liquid nitrogen. 12ours, washed with PBS and left in 3SuperFrost slides (VWR), and stained with rabbit anti-GFP (Invitrogen, A11122) at 1:200 and chicken anti-Keratin-15 (Biolegend, Poly19339) at 1:400 or Rat anti-mouse/human ITGA6 (BD, 555734) at 1:300. Primary signal was amplified with Goat anti-Rabbit Alexa-488 and Goat anti-Chicken Alexa-555 or Goat anti-Rat Alexa-555 at 1:1000 (all from Invitrogen). Slides were then washed in PBS and mounted with DAPI containing medium. All images were acquired on a Zeiss Imager M2 fluorescent microscope with Apotome. Ki67 tissue staining was performed by the UCSF Dermatapathology Core using a Dako Link-48 stainer. Briefly, antigen retrieval was performed in Dako pH9 Retrieval Solution at 95° C for 1 hour and then stained with anti-Ki67 antibody (Dako, catalog # GA626) and the chromogen DAB. For detection, Dako Envision Dual Link was used. Finally, slides were counter-stained with hematoxylin.
In vivo Jag1 rescue
Foxp3DTR mice depleted of Tregs according to the early regimen were injected subcutaneously on days −2, −1, 1, and 3 into four adjacent dorsal skin sites with 1 µg control IgG1-Fc or Jag1-Fc fusion protein (both from R&D) conjugated to Affi-gel blue beads (100–200 mesh; Bio-Rad), as previously described (Chen et al., 2012). Briefly, per mouse: 1 µg of protein (in 10 µl) was soaked with 5 µl Affi-gel blue beads (corresponding to ~2000 beads) for 1 hour at 37° C. Total suspensi ons of 15 µl were then transferred into 29G 3/10 cc insulin syringes (BD) for subcutaneous injection of mice under anesthesia. HFSC Ki67 expression was assessed by flow cytometry on day 4.
RNA-Sequencing analysis and Quantitative PCR
Sorted cell populations were flash frozen in liquid nitrogen and sent overnight on dry ice to Expression Analysis, Quintiles (Morrisville, NC). RNA samples were converted into cDNA libraries using the Illumina TruSeq Stranded mRNA sample preparation kit. (Illumina). RNA was isolated by Expression Analysis using Qiagen RNeasy Spin Column and was quantified via Nanodrop ND-8000 spectrophotometer. RNA quality was checked by Agilent Bioanalyzer Pico Chip. cDNA was created from 220 pg of input RNA with the SMARTer Ultra Low input kit and sequenced to a 25M read depth with Illumina RNASeq. Reads were aligned to Ensembl mg GRCm38.p4 reference genome with TopHat software (v. 2.0.12). SAM files were generated with SAMtools from alignment results. Read counts were obtained with htseq-count (0.6.1p1) with the union option. Differential expression was determined using the R/Bioconducter package DESeq2 (Ref 2). Differentially expressed genes were analyzed by Ingenuity Pathway Analysis. Using the differentially expressed genes (p <0.05) from the HFSC dataset, a python script was written to find the overlap between the DE list and known Notch target genes(Meier-Stiegen et al., 2010). To determine significance in overlap between the two, a chi-squared test was performed. The overlapping genes between the two sets were clustered in Cluster 3.0 and visualized as a heat map using gene pattern’s HierarchicalClusteringViewer(de Hoon et al., 2004; Reich et al., 2006). For assessment of differentiation associated genes in Foxp3Cre/CreJag1fl/fl or control mice (i.e., Foxp3Cre/CreJag1wt/wt or Foxp3wt/wtJag1fl/fl), RNA was isolated from 1×106 epidermal cells on day 10 post depilation, using a column based kit (PureLink RNA Mini Kit, Thermo Fisher). For assessment of Jag1 expression on T cell subsets, Tregs, Teffs and CD8+ T cells were sorted from SDLNs of WT mice and RNA isolated as described above. RNA was then transcribed (iScript CDNA synthesis Kit, Bio-Rad) and the expression of differentiation genes or Jag1 for T cell subsets was assessed relative to Gapdh that was duplexed in every reaction. The following Taqman Gene expression assays were used (Thermo Fisher): Mm99999915_g1 (Gapdh), Mm01191753_m1 (Bgn), Mm00432359_m1 (Ccnd1), Mm01220860_m1 (Gdf10), Mm00486320_s1 (Sox4), Mm00776876_m1 (Sox7), Mm00441826_m1 (Timp3), and Mm00496902_m1 (Jag1). Data are presented as negative fold change of Delta-Delta CT or as standardized arbitrary units (AU).
Intravital 2-photon imaging of Tregs in dorsal skin
Two-photon imaging instrumentation has been previously described(Friedman et al., 2010). Dorsal hair of 7–10 week old Foxp3GFP mice was clipped and depilated to induce anagen. Mice were then placed on a custom heated microscope stage under anesthesia. The custom suction window on the microscope stage was then placed into position over the depilated dorsum, and suction was applied to gently immobilize the depilated skin(Thornton et al., 2012). The microscope objective was then lowered into position directly over the suction window with an embedded 12 mm coverslip. 10 µg Evans Blue dye was injected into the retro-orbital vein immediately before image acquisition. All images were analyzed using Imaris Software (Bitplane). HFs were visualized using second harmonic generated collagen. Bulge-associated Tregs were denoted as within a <20 µm radius of individual HFs and non-bulge associated Tregs as >20 µm from a HF. To determine in vivo changes in Treg cell shape, the sphericity of individual Tregs was calculated over the time-lapse period, as previously described(Thornton et al., 2012).
Statistical analyses
Statistical analyses were performed with Prism software package version 6.0 (GraphPad). P values were calculated using two-tailed unpaired or paired Student’s t-test. Sample size for animal experiments was determined based upon pilot experiments. Mice cohort size was designed to be sufficient to enable accurate determination of statistical significance. No animals were excluded from the statistical analysis, unless due to technical errors. Mice were randomly assigned to treatment or control groups, while ensuring inclusion criteria based on gender, age and hair cycle. Investigators were blinded for all tissue staining and clinical picture quantifications. Appropriate statistical analyses were applied, assuming a normal sample distribution. All in vivo experiments were conducted with at least two independent cohorts. RNA-Seq experiments were conducted using 2–4 biological samples (as indicated in figure legends) from indicated cohorts.
Data and Software Availability
RNA sequencing data has been deposited in NCBI GEO with the following IDs: GSE76102 and GSE76138 and are available at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76102, and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76138.
Supplementary Material
Time-lapse recording from 2-photon imaging of Foxp3GFP dorsal skin showing bulge-associated Tregs (<20 µm from individual HFs). Tregs are depicted in green and second harmonic generated collagen in blue.
Differentially expressed genes between hair follicle stem cells (HFSCs) isolated from WT and Treg depleted mice.
Differentially expressed genes between skin and skin draining lymph node (SDLN) Tregs.
Time-lapse recording from 2-photon imaging of Foxp3GFP dorsal skin showing non-bulge associated Tregs (>20 µm from individual HFs). Tregs are depicted in green and second harmonic generated collagen in blue.
Treg cell abundance and activation in dorsal skin of adult wild-type (WT) C57BL/6 mice at specific stages of the synchronous HF cycle was assessed using flow cytometry. (A) Representative flow plots of skin Tregs profiled during the synchronous HF cycle. Pre-gated on live CD45+CD3+CD4+ cells. (B) Representative images of skin Tregs from dorsal skin harvested on post-natal day 21 (telogen) and post-natal day 30 (anagen). Arrows depict Foxp3+ Treg cells. Asterisks denote autofluorescent hair shafts. Scale Bars, 100 µm. (C) Quantification of absolute cell numbers of Foxp3+ Tregs per field of view in dorsal skin. (D) T cell subsets in dorsal skin of adult WT C57BL/6 mice. Data are shown as a proportion of CD3+ T cells. Shaded areas represent telogen phase and unshaded areas represent anagen phase. (E) Representative flow plots of negative control (dendritic epidermal T cells, DETCs) and CD4+ T cell gates from telogen and anagen dorsal skin. One representative experiment of two is shown (A); n = 3–5 mice per time point combined (C-D). Unpaired Students t-test (C). ****P<0.00., Data are mean ± s.e.m.
Control (WT) or Foxp3DTR mice were treated with DT on days −2, −1, depilated on day 0 to induce anagen and DT administration continued on day 1 followed by every other day up to day 4. (A) Representative photos and (B) quantification of dorsal skin pigmentation at day 10 in WT and Foxp3DTR mice with DT treatments as indicated. Tregs were depleted either up to day 4 (early), from day 7 onwards (late) or constitutively (con) throughout the experimental period until the termination of the experiment at day 14. (C) Representative photos and (D) quantification of hair regrowth at day 14 in WT and Foxp3DTR mice with DT treatments as indicated. (E) Representative H&E staining of skin from WT and Foxp3DTR mice on day 14. Arrows indicate anagen HF extension into dermal adipose. (F) Quantification of HF length on day 14. WT and Rag2−/− mice were clinically assessed for spontaneous anagen induction during the natural HF cycle by monitoring skin pigmentation from 2nd telogen (Postnatal age of 80 days) through 2nd anagen (110 days of age). (G) Representative photos of WT and Rag2−/− mice at postnatal age of 110 days and (H) quantification of anagen induction by skin pigmentation intensity. WT and Rag2−/−mice with all dorsal HFs in 2nd telogen (Postnatal age of 80 days) were depilated to induce anagen. Anagen induction was monitored. (I) Representative photos in WT and Rag2−/− mice at 14 days post depilation and (J) quantification of anagen induction by skin pigmentation intensity analysis. One representative of three experiments shown. Scale Bars, 100 µm. n = 4–5 mice per group. One-way ANOVA (B, D, and F), Two-way ANOVA (H and J). ns = not significant, *P<0.05, **P<0.01 ***P<0.001, ****P<0.0001. Data are mean ± s.e.m.
Representative immunofluorescent image of Foxp3+ Tregs in telogen skin of Foxp3GFP reporter mice co-stained with Keratin-15 (K15). Arrows depict Foxp3+ Treg cells. Asterisks denote autofluorescent hair shafts. Scale Bar, 100 µm.
Foxp3DTR or control mice were treated with DT on days −2, −1, depilated on day 0 to induce anagen and Diphtheria toxin (DT) administered again on days 1 and 3 (i.e., early regimen). (A) Quantification of total CD34+ bulge HFSCs in WT and Foxp3DTR mice 4 days after depilation. (B) Representative histological images of Ki67 staining of WT and Foxp3DTR mice 10 days after depilation. DT or PBS (as control) was injected intraperitoneally into Foxp3DTR mice to deplete Tregs during 1st telogen from postnatal day 21 (P21) to P24, and harvested at P32 for analysis. (C) Representative photos and (D) quantification of anagen induction in PBS treated (Foxp3DTR) and DT treated (Foxp3DTR + DT) mice. (E) Representative flow plots and quantification of (F) absolute number of HFSCs. Quantification of Ki67+ HFSC (G) frequency and (H) absolute cell numbers. DT or PBS (as control) was injected intraperitoneally into Foxp3DTR mice to deplete Tregs during 1st telogen from postnatal day 21 (P21) to P24. EdU (5-ethynyl-2′-deoxyuridine) was injected intraperitoneally at P31 and mice harvested at P32 for analysis. (I) Representative histogram plot of EdU staining from PBS treated (Foxp3DTR) and DT treated (Foxp3DTR + DT) mice. (J) Summary of median fluorescence intensity (MFI) of EdU expression. One representative experiment of three (A-B) and two (C-J) is shown. n = 4 mice per group. ns = no significant difference, One-way ANOVA (A), Unpaired Students t-test (D, F-H, and J). *P<0.05, ***P<0.001, Data are mean ± s.e.m.
Control wild-type (WT) or Foxp3DTR mice were depilated and treated with DT according to the ‘early’ depletion protocol. Single cell suspensions from day 1 skin were stimulated with PMA/ionomycin and the production of IL-22, IFNg, IL-17 and TNF-alpha was assessed by flow cytometry. Representative flow cytometry plots of cytokine induction are shown. One representative experiment of two; n = 3–5 mice per group.
T cell subsets from wild-type C57BL/6 mice were assessed for Jag1 expression by flow cytometry. (A) Representative histogram plots of isotype staining and Jag1 staining from indicated T cell populations. (B) Summary of median fluorescence intensity (MFI) of Jag1 expression relative to isotype control MFI. (C) Representative histogram plots of isotype and Jag1 staining of telogen and anagen skin resident Tregs. (D) Summary of Jag1 MFI expression relative to isotype control. (E) Jag1 expression via qRT-PCR, expressed in arbitrary units (AU) for all populations tested. Quantification of (F) total bulge HFSCs and (G) HFSC:Treg ratio in control (Foxp3Cre/CreJag1wt/wt) or Treg-Jag1 deleted mice (Foxp3Cre/CreJag1fl/fl) in steady state non-depilated skin of 8 week old mice. (H) HFSC:Treg ratio assessed on day 10 post depilation. One representative experiment of two is shown. One-way ANOVA (B and E), Unpaired Student’s t-test (D, F-H). n = 3–5 mice per group. ns = no significant difference, **P<0.01, ***P<0.001, Data are mean ± s.e.m.
Acknowledgments
We thank Matthew F. Krummel and the Biological Imaging Development Center for discussion and technical assistance with 2-photon imaging. We thank C. Benetiz for assistance with animal husbandry and Lokeshchandra Kalekar for designing the graphical abstract. Flow Cytometry data was generated in the UCSF Parnassus Flow Cytometry Core which is supported by the Diabetes Research Center (DRC) grant, NIH P30 DK063720. Histology was performed with assistance from the UCSF Mouse Pathology Core which is supported by NIH 5P30CA082103-15. N.A. is supported by a Marie Curie International Outgoing Fellowship: FP7-PEOPLE-2012-IOF, Project No. 327244. This work was primarily funded by M.D.R. grants: NIH K08-AR062064, Burroughs Wellcome Fund CAMS-1010934, NIH DP2-AR068130, NIH R21-AR066821, Scleroderma Research Foundation grant, a National Psoriasis Foundation Translational Grant and a Dermatology Foundation Stiefel Scholar Award in Autoimmune & Connective Tissue Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
N.A. designed the studies, performed the experiments and analyzed the data. N.A. and M.D.R. wrote the manuscript. R.S.R. assisted with optimization of flow cytometry panels, data analysis, and performed RNAseq analysis. M.L.P. and R.R.R.G assisted with tissue staining. H.A.T. performed Treg sorting for RNAseq and assisted with mouse experiments. K.T., A.N., and B.Z. assisted with mouse experiments. R.A., K.L., and W.L. performed HFSC RNAseq analysis. K.C. assisted with 2-photon imaging. M.M.L. and M.R.T. performed HF quantification studies. T.S. designed and oversaw Treg RNA-seq studies. M.B. and R.P. assisted with HF morphometry. F.O.N, G.C. and A.K.A. were involved in study design. M.D.R. oversaw all study design and data analysis.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinforma. Oxf. Engl. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S, Treuting PM, Rudensky AY. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Pasolli HA, Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20:3022–3035. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldajipour B, Nelson A, Krummel MF. Tumor-infiltrating lymphocytes are dynamically desensitized to antigen but are maintained by homeostatic cytokine. JCI Insight. 2016;1:e89289. doi: 10.1172/jci.insight.89289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Dev. Camb. Engl. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, Barczak A, Rosenblum MD, Daud A, Barber DL, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26:638–652. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castela E, Le Duff F, Butori C, Ticchioni M, Hofman P, Bahadoran P, Lacour J-P, Passeron T. Effects of low-dose recombinant interleukin 2 to promote T-regulatory cells in alopecia areata. JAMA Dermatol. 2014;150:748–751. doi: 10.1001/jamadermatol.2014.504. [DOI] [PubMed] [Google Scholar]
- Chen T, Heller E, Beronja S, Oshimori N, Stokes N, Fuchs E. An RNA interference screen uncovers a new molecule in stem cell self-renewal and long-term regeneration. Nature. 2012;485:104–108. doi: 10.1038/nature10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow Z, Mueller SN, Deane JA, Hickey MJ. Dermal regulatory T cells display distinct migratory behavior that is modulated during adaptive and innate inflammation. J. Immunol. Baltim. Md 1950. 2013;191:3049–3056. doi: 10.4049/jimmunol.1203205. [DOI] [PubMed] [Google Scholar]
- Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RA, Chong B, Mirchandani N, Brinster NK, Yamanaka K-I, Dowgiert RK, Kupper TS. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. Baltim. Md 1950. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Dev. Camb. Engl. 2006;133:4427–4438. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RS, Beemiller P, Sorensen CM, Jacobelli J, Krummel MF. Real-time analysis of T cell receptors in naive cells in vitro and in vivo reveals flexibility in synapse and signaling dynamics. J. Exp. Med. 2010;207:2733–2749. doi: 10.1084/jem.20091201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki J, Wu J, Carlson AL, Silberstein L, Putheti P, Larocca R, Gao W, Saito TI, Lo Celso C, Tsuyuzaki H, et al. In vivo imaging of Treg cells providing immune privilege to the haematopoietic stem-cell niche. Nature. 2011;474:216–219. doi: 10.1038/nature10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz IK, Truong H-A, Yang SH-Y, Maurano MM, Lee K, Abbas AK, Rosenblum MD. Cutting Edge: memory regulatory t cells require IL-7 and not IL-2 for their maintenance in peripheral tissues. J. Immunol. Baltim. Md 1950. 2013;190:4483–4487. doi: 10.4049/jimmunol.1300212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headley MB, Bins A, Nip A, Roberts EW, Looney MR, Gerard A, Krummel MF. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature. 2016;531:513–517. doi: 10.1038/nature16985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinforma. Oxf. Engl. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgård R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Lenne P-F. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 2007;8:633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- Levy V, Lindon C, Harfe BD, Morgan BA. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev. Cell. 2005;9:855–861. doi: 10.1016/j.devcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R and 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinforma. Oxf. Engl. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Haribhai D, Relland LM, Truong N, Carlson MR, Williams CB, Chatila TA. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- Lindsay RS, Corbin K, Mahne A, Levitt BE, Gebert MJ, Wigton EJ, Bradley BJ, Haskins K, Jacobelli J, Tang Q, et al. Antigen recognition in the islets changes with progression of autoimmune islet infiltration. J. Immunol. Baltim. Md 1950. 2015;194:522–530. doi: 10.4049/jimmunol.1400626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney MR, Thornton EE, Sen D, Lamm WJ, Glenny RW, Krummel MF. Stabilized imaging of immune surveillance in the mouse lung. Nat. Methods. 2011;8:91–96. doi: 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Blanpain C, Nowak JA, Guasch G, Lewis L, Fuchs E. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19:1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu MP, Teijaro JR, Walsh KB, Greenberg ML, Marsolais D, Parker I, Rosen H, Oldstone MBA, Cahalan MD. Three phases of CD8 T cell response in the lung following H1N1 influenza infection and sphingosine 1 phosphate agonist therapy. PloS One. 2013;8:e58033. doi: 10.1371/journal.pone.0058033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Stiegen F, Schwanbeck R, Bernoth K, Martini S, Hieronymus T, Ruau D, Zenke M, Just U. Activated Notch1 target genes during embryonic cell differentiation depend on the cellular context and include lineage determinants and inhibitors. PloS One. 2010;5:e11481. doi: 10.1371/journal.pone.0011481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Müller-Röver S, Handjiski B, van der Veen C, Eichmüller S, Foitzik K, McKay IA, Stenn KS, Paus R. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- Nagao K, Kobayashi T, Moro K, Ohyama M, Adachi T, Kitashima DY, Ueha S, Horiuchi K, Tanizaki H, Kabashima K, et al. Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat. Immunol. 2012;13:744–752. doi: 10.1038/ni.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosbaum A, Prevel N, Truong H-A, Mehta P, Ettinger M, Scharschmidt TC, Ali NH, Pauli ML, Abbas AK, Rosenblum MD. Cutting Edge: Regulatory T Cells Facilitate Cutaneous Wound Healing. J. Immunol. Baltim. Md 1950. 2016;196:2010–2014. doi: 10.4049/jimmunol.1502139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panduro M, Benoist C, Mathis D. Tissue Tregs. Annu. Rev. Immunol. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, Kim H, Singh P, Lee A, Chen WV, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466:113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Sanchez Rodriguez R, Pauli ML, Neuhaus IM, Yu SS, Arron ST, Harris HW, Yang SH-Y, Anthony BA, Sverdrup FM, Krow-Lucal E, et al. Memory regulatory T cells reside in human skin. J. Clin. Invest. 2014;124:1027–1036. doi: 10.1172/JCI72932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Kobayashi T, Negoro H, Sengiku A, Hiratsuka T, Kamioka Y, Liou LS, Ogawa O, Matsuda M. Intravital imaging of mouse urothelium reveals activation of extracellular signal-regulated kinase by stretch-induced intravesical release of ATP. Physiol. Rep. 2016:4. doi: 10.14814/phy2.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt TC, Vasquez KS, Truong H-A, Gearty SV, Pauli ML, Nosbaum A, Gratz IK, Otto M, Moon JJ, Liese J, et al. A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity. 2015;43:1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka DK, Fowell DJ. Regulatory T cells inhibit acute IFN-γ synthesis without blocking T-helper cell type 1 (Th1) differentiation via a compartmentalized requirement for IL-10. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18336–18341. doi: 10.1073/pnas.1110566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton EE, Looney MR, Bose O, Sen D, Sheppard D, Locksley R, Huang X, Krummel MF. Spatiotemporally separated antigen uptake by alveolar dendritic cells and airway presentation to T cells in the lung. J. Exp. Med. 2012;209:1183–1199. doi: 10.1084/jem.20112667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinforma. Oxf. Engl. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempus CS, Morris RJ, Bortner CD, Cotsarelis G, Faircloth RS, Reece JM, Tennant RW. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J. Invest. Dermatol. 2003;120:501–511. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vas V, Szilágyi L, Pálóczi K, Uher F. Soluble Jagged-1 is able to inhibit the function of its multivalent form to induce hematopoietic stem cell self-renewal in a surrogate in vitro assay. J. Leukoc. Biol. 2004;75:714–720. doi: 10.1189/jlb.1003462. [DOI] [PubMed] [Google Scholar]
- Vauclair S, Nicolas M, Barrandon Y, Radtke F. Notch1 is essential for postnatal hair follicle development and homeostasis. Dev. Biol. 2005;284:184–193. doi: 10.1016/j.ydbio.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Villalta SA, Rosenthal W, Martinez L, Kaur A, Sparwasser T, Tidball JG, Margeta M, Spencer MJ, Bluestone JA. Regulatory T cells suppress muscle inflammation and injury in muscular dystrophy. Sci. Transl. Med. 2014;6:258ra142. doi: 10.1126/scitranslmed.3009925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinegoni C, Lee S, Gorbatov R, Weissleder R. Motion compensation using a suctioning stabilizer for intravital microscopy. Intravital. 2012;1:115–121. doi: 10.4161/intv.23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaid A, Mackay LK, Rahimpour A, Braun A, Veldhoen M, Carbone FR, Manton JH, Heath WR, Mueller SN. Persistence of skin-resident memory T cells within an epidermal niche. Proc. Natl. Acad. Sci. U. S. A. 2014;111:5307–5312. doi: 10.1073/pnas.1322292111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse recording from 2-photon imaging of Foxp3GFP dorsal skin showing bulge-associated Tregs (<20 µm from individual HFs). Tregs are depicted in green and second harmonic generated collagen in blue.
Differentially expressed genes between hair follicle stem cells (HFSCs) isolated from WT and Treg depleted mice.
Differentially expressed genes between skin and skin draining lymph node (SDLN) Tregs.
Time-lapse recording from 2-photon imaging of Foxp3GFP dorsal skin showing non-bulge associated Tregs (>20 µm from individual HFs). Tregs are depicted in green and second harmonic generated collagen in blue.
Treg cell abundance and activation in dorsal skin of adult wild-type (WT) C57BL/6 mice at specific stages of the synchronous HF cycle was assessed using flow cytometry. (A) Representative flow plots of skin Tregs profiled during the synchronous HF cycle. Pre-gated on live CD45+CD3+CD4+ cells. (B) Representative images of skin Tregs from dorsal skin harvested on post-natal day 21 (telogen) and post-natal day 30 (anagen). Arrows depict Foxp3+ Treg cells. Asterisks denote autofluorescent hair shafts. Scale Bars, 100 µm. (C) Quantification of absolute cell numbers of Foxp3+ Tregs per field of view in dorsal skin. (D) T cell subsets in dorsal skin of adult WT C57BL/6 mice. Data are shown as a proportion of CD3+ T cells. Shaded areas represent telogen phase and unshaded areas represent anagen phase. (E) Representative flow plots of negative control (dendritic epidermal T cells, DETCs) and CD4+ T cell gates from telogen and anagen dorsal skin. One representative experiment of two is shown (A); n = 3–5 mice per time point combined (C-D). Unpaired Students t-test (C). ****P<0.00., Data are mean ± s.e.m.
Control (WT) or Foxp3DTR mice were treated with DT on days −2, −1, depilated on day 0 to induce anagen and DT administration continued on day 1 followed by every other day up to day 4. (A) Representative photos and (B) quantification of dorsal skin pigmentation at day 10 in WT and Foxp3DTR mice with DT treatments as indicated. Tregs were depleted either up to day 4 (early), from day 7 onwards (late) or constitutively (con) throughout the experimental period until the termination of the experiment at day 14. (C) Representative photos and (D) quantification of hair regrowth at day 14 in WT and Foxp3DTR mice with DT treatments as indicated. (E) Representative H&E staining of skin from WT and Foxp3DTR mice on day 14. Arrows indicate anagen HF extension into dermal adipose. (F) Quantification of HF length on day 14. WT and Rag2−/− mice were clinically assessed for spontaneous anagen induction during the natural HF cycle by monitoring skin pigmentation from 2nd telogen (Postnatal age of 80 days) through 2nd anagen (110 days of age). (G) Representative photos of WT and Rag2−/− mice at postnatal age of 110 days and (H) quantification of anagen induction by skin pigmentation intensity. WT and Rag2−/−mice with all dorsal HFs in 2nd telogen (Postnatal age of 80 days) were depilated to induce anagen. Anagen induction was monitored. (I) Representative photos in WT and Rag2−/− mice at 14 days post depilation and (J) quantification of anagen induction by skin pigmentation intensity analysis. One representative of three experiments shown. Scale Bars, 100 µm. n = 4–5 mice per group. One-way ANOVA (B, D, and F), Two-way ANOVA (H and J). ns = not significant, *P<0.05, **P<0.01 ***P<0.001, ****P<0.0001. Data are mean ± s.e.m.
Representative immunofluorescent image of Foxp3+ Tregs in telogen skin of Foxp3GFP reporter mice co-stained with Keratin-15 (K15). Arrows depict Foxp3+ Treg cells. Asterisks denote autofluorescent hair shafts. Scale Bar, 100 µm.
Foxp3DTR or control mice were treated with DT on days −2, −1, depilated on day 0 to induce anagen and Diphtheria toxin (DT) administered again on days 1 and 3 (i.e., early regimen). (A) Quantification of total CD34+ bulge HFSCs in WT and Foxp3DTR mice 4 days after depilation. (B) Representative histological images of Ki67 staining of WT and Foxp3DTR mice 10 days after depilation. DT or PBS (as control) was injected intraperitoneally into Foxp3DTR mice to deplete Tregs during 1st telogen from postnatal day 21 (P21) to P24, and harvested at P32 for analysis. (C) Representative photos and (D) quantification of anagen induction in PBS treated (Foxp3DTR) and DT treated (Foxp3DTR + DT) mice. (E) Representative flow plots and quantification of (F) absolute number of HFSCs. Quantification of Ki67+ HFSC (G) frequency and (H) absolute cell numbers. DT or PBS (as control) was injected intraperitoneally into Foxp3DTR mice to deplete Tregs during 1st telogen from postnatal day 21 (P21) to P24. EdU (5-ethynyl-2′-deoxyuridine) was injected intraperitoneally at P31 and mice harvested at P32 for analysis. (I) Representative histogram plot of EdU staining from PBS treated (Foxp3DTR) and DT treated (Foxp3DTR + DT) mice. (J) Summary of median fluorescence intensity (MFI) of EdU expression. One representative experiment of three (A-B) and two (C-J) is shown. n = 4 mice per group. ns = no significant difference, One-way ANOVA (A), Unpaired Students t-test (D, F-H, and J). *P<0.05, ***P<0.001, Data are mean ± s.e.m.
Control wild-type (WT) or Foxp3DTR mice were depilated and treated with DT according to the ‘early’ depletion protocol. Single cell suspensions from day 1 skin were stimulated with PMA/ionomycin and the production of IL-22, IFNg, IL-17 and TNF-alpha was assessed by flow cytometry. Representative flow cytometry plots of cytokine induction are shown. One representative experiment of two; n = 3–5 mice per group.
T cell subsets from wild-type C57BL/6 mice were assessed for Jag1 expression by flow cytometry. (A) Representative histogram plots of isotype staining and Jag1 staining from indicated T cell populations. (B) Summary of median fluorescence intensity (MFI) of Jag1 expression relative to isotype control MFI. (C) Representative histogram plots of isotype and Jag1 staining of telogen and anagen skin resident Tregs. (D) Summary of Jag1 MFI expression relative to isotype control. (E) Jag1 expression via qRT-PCR, expressed in arbitrary units (AU) for all populations tested. Quantification of (F) total bulge HFSCs and (G) HFSC:Treg ratio in control (Foxp3Cre/CreJag1wt/wt) or Treg-Jag1 deleted mice (Foxp3Cre/CreJag1fl/fl) in steady state non-depilated skin of 8 week old mice. (H) HFSC:Treg ratio assessed on day 10 post depilation. One representative experiment of two is shown. One-way ANOVA (B and E), Unpaired Student’s t-test (D, F-H). n = 3–5 mice per group. ns = no significant difference, **P<0.01, ***P<0.001, Data are mean ± s.e.m.