Abstract
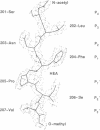
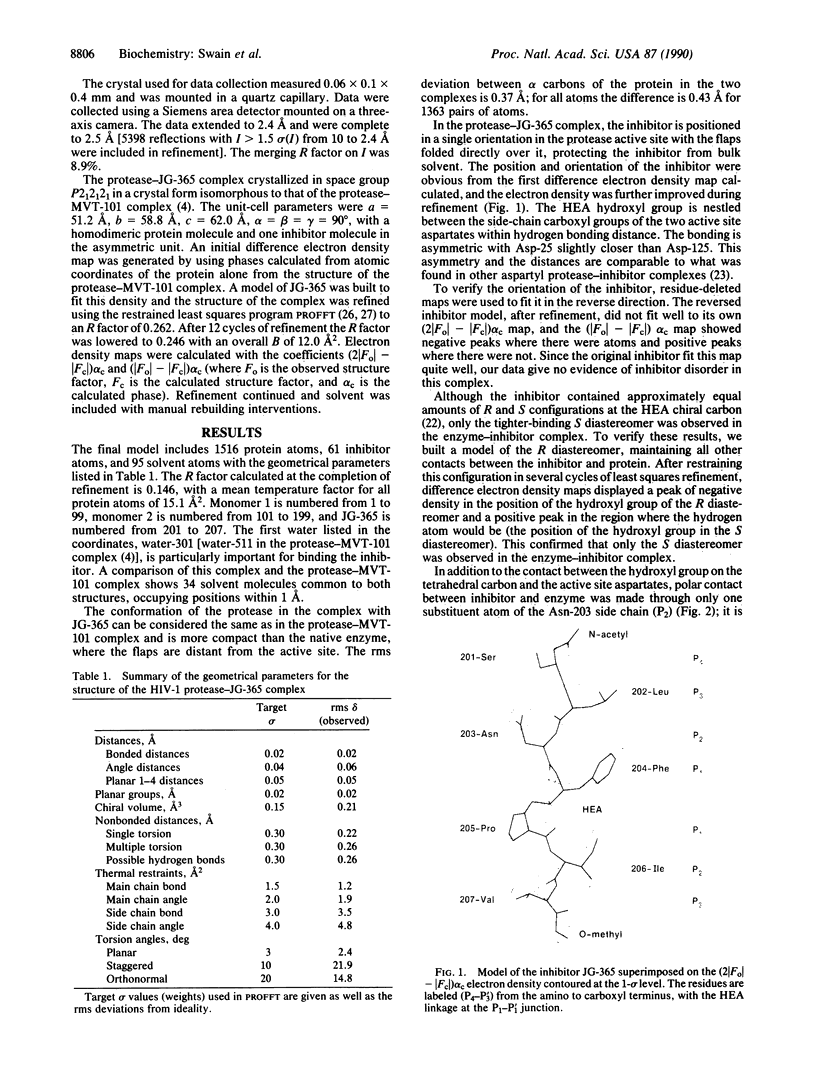
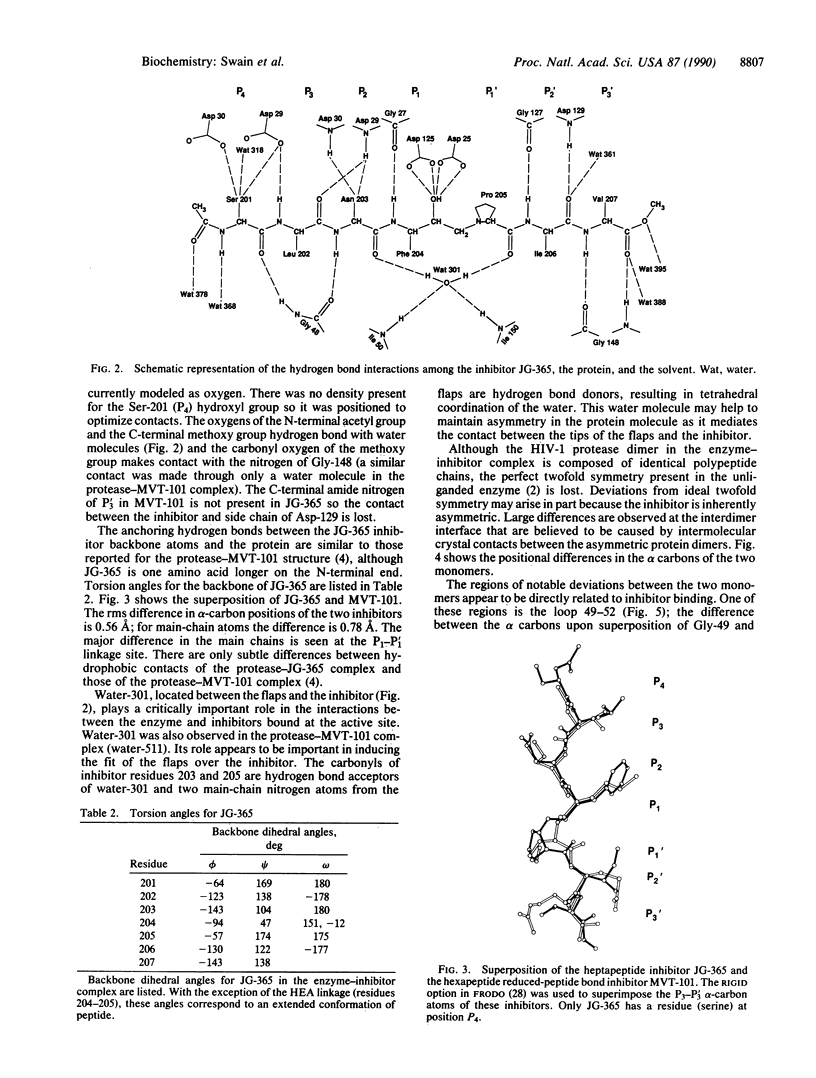
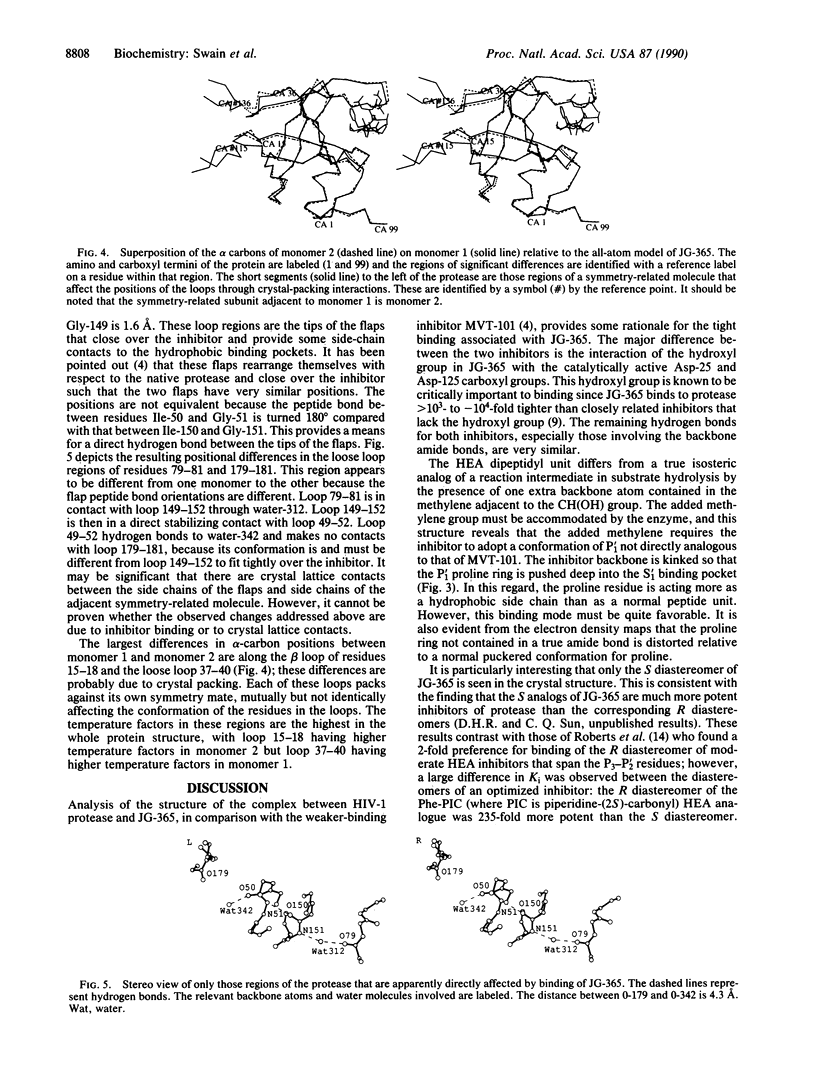
The structure of a crystal complex of the chemically synthesized protease of human immunodeficiency virus 1 with a heptapeptide-derived inhibitor bound in the active site has been determined. The sequence of the inhibitor JG-365 is Ac-Ser-Leu-Asn-Phe-psi[CH(OH)CH2N]-Pro-Ile-Val-OMe; the Ki is 0.24 nM. The hydroxyethylamine moiety, in place of the normal scissile bond of the substrate, is believed to mimic a tetrahedral reaction intermediate. The structure of the complex has been refined to an R factor of 0.146 at 2.4-A resolution by using restrained least squares with rms deviations in bond lengths of 0.02 A and bond angles of 4. The bound inhibitor diastereomer has the S configuration at the hydroxyethylamine chiral carbon, and the hydroxyl group is positioned between the active site aspartate carboxyl groups within hydrogen bonding distance. Comparison of this structure with a reduced peptide bond inhibitor-protease complex indicates that these contacts confer the exceptional binding strength of JG-365.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Billich S., Knoop M. T., Hansen J., Strop P., Sedlacek J., Mertz R., Moelling K. Synthetic peptides as substrates and inhibitors of human immune deficiency virus-1 protease. J Biol Chem. 1988 Dec 5;263(34):17905–17908. [PubMed] [Google Scholar]
- Blundell T. L., Cooper J., Foundling S. I., Jones D. M., Atrash B., Szelke M. On the rational design of renin inhibitors: X-ray studies of aspartic proteinases complexed with transition-state analogues. Biochemistry. 1987 Sep 8;26(18):5585–5590. doi: 10.1021/bi00392a001. [DOI] [PubMed] [Google Scholar]
- Dann J. G., Stammers D. K., Harris C. J., Arrowsmith R. J., Davies D. E., Hardy G. W., Morton J. A. Human renin: a new class of inhibitors. Biochem Biophys Res Commun. 1986 Jan 14;134(1):71–77. doi: 10.1016/0006-291x(86)90528-0. [DOI] [PubMed] [Google Scholar]
- Dreyer G. B., Metcalf B. W., Tomaszek T. A., Jr, Carr T. J., Chandler A. C., 3rd, Hyland L., Fakhoury S. A., Magaard V. W., Moore M. L., Strickler J. E. Inhibition of human immunodeficiency virus 1 protease in vitro: rational design of substrate analogue inhibitors. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9752–9756. doi: 10.1073/pnas.86.24.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee W. J. Renin inhibitors. Pharm Res. 1987 Oct;4(5):364–374. doi: 10.1023/a:1016422009662. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Kent S. B. Chemical synthesis of peptides and proteins. Annu Rev Biochem. 1988;57:957–989. doi: 10.1146/annurev.bi.57.070188.004521. [DOI] [PubMed] [Google Scholar]
- Lapatto R., Blundell T., Hemmings A., Overington J., Wilderspin A., Wood S., Merson J. R., Whittle P. J., Danley D. E., Geoghegan K. F. X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes. Nature. 1989 Nov 16;342(6247):299–302. doi: 10.1038/342299a0. [DOI] [PubMed] [Google Scholar]
- McQuade T. J., Tomasselli A. G., Liu L., Karacostas V., Moss B., Sawyer T. K., Heinrikson R. L., Tarpley W. G. A synthetic HIV-1 protease inhibitor with antiviral activity arrests HIV-like particle maturation. Science. 1990 Jan 26;247(4941):454–456. doi: 10.1126/science.2405486. [DOI] [PubMed] [Google Scholar]
- Meek T. D., Lambert D. M., Dreyer G. B., Carr T. J., Tomaszek T. A., Jr, Moore M. L., Strickler J. E., Debouck C., Hyland L. J., Matthews T. J. Inhibition of HIV-1 protease in infected T-lymphocytes by synthetic peptide analogues. Nature. 1990 Jan 4;343(6253):90–92. doi: 10.1038/343090a0. [DOI] [PubMed] [Google Scholar]
- Miller M., Jaskólski M., Rao J. K., Leis J., Wlodawer A. Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature. 1989 Feb 9;337(6207):576–579. doi: 10.1038/337576a0. [DOI] [PubMed] [Google Scholar]
- Miller M., Schneider J., Sathyanarayana B. K., Toth M. V., Marshall G. R., Clawson L., Selk L., Kent S. B., Wlodawer A. Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution. Science. 1989 Dec 1;246(4934):1149–1152. doi: 10.1126/science.2686029. [DOI] [PubMed] [Google Scholar]
- Moore M. L., Bryan W. M., Fakhoury S. A., Magaard V. W., Huffman W. F., Dayton B. D., Meek T. D., Hyland L., Dreyer G. B., Metcalf B. W. Peptide substrates and inhibitors of the HIV-1 protease. Biochem Biophys Res Commun. 1989 Mar 15;159(2):420–425. doi: 10.1016/0006-291x(89)90008-9. [DOI] [PubMed] [Google Scholar]
- Mous J., Heimer E. P., Le Grice S. F. Processing protease and reverse transcriptase from human immunodeficiency virus type I polyprotein in Escherichia coli. J Virol. 1988 Apr;62(4):1433–1436. doi: 10.1128/jvi.62.4.1433-1436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navia M. A., Fitzgerald P. M., McKeever B. M., Leu C. T., Heimbach J. C., Herber W. K., Sigal I. S., Darke P. L., Springer J. P. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989 Feb 16;337(6208):615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Taylor W. R. A structural model for the retroviral proteases. Nature. 1987 Sep 24;329(6137):351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- Rich D. H., Green J., Toth M. V., Marshall G. R., Kent S. B. Hydroxyethylamine analogues of the p17/p24 substrate cleavage site are tight-binding inhibitors of HIV protease. J Med Chem. 1990 May;33(5):1285–1288. doi: 10.1021/jm00167a003. [DOI] [PubMed] [Google Scholar]
- Roberts N. A., Martin J. A., Kinchington D., Broadhurst A. V., Craig J. C., Duncan I. B., Galpin S. A., Handa B. K., Kay J., Kröhn A. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990 Apr 20;248(4953):358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- Schneider J., Kent S. B. Enzymatic activity of a synthetic 99 residue protein corresponding to the putative HIV-1 protease. Cell. 1988 Jul 29;54(3):363–368. doi: 10.1016/0092-8674(88)90199-7. [DOI] [PubMed] [Google Scholar]
- Seelmeier S., Schmidt H., Turk V., von der Helm K. Human immunodeficiency virus has an aspartic-type protease that can be inhibited by pepstatin A. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6612–6616. doi: 10.1073/pnas.85.18.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suguna K., Padlan E. A., Smith C. W., Carlson W. D., Davies D. R. Binding of a reduced peptide inhibitor to the aspartic proteinase from Rhizopus chinensis: implications for a mechanism of action. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7009–7013. doi: 10.1073/pnas.84.20.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szelke M., Leckie B., Hallett A., Jones D. M., Sueiras J., Atrash B., Lever A. F. Potent new inhibitors of human renin. Nature. 1982 Oct 7;299(5883):555–557. doi: 10.1038/299555a0. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Miller M., Jaskólski M., Sathyanarayana B. K., Baldwin E., Weber I. T., Selk L. M., Clawson L., Schneider J., Kent S. B. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989 Aug 11;245(4918):616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]