Abstract
Multiple myeloma (MM) is an incurable malignant plasma cell neoplasm. Proteasome inhibitors including Bortezomib (Bz) are used to treat MM, and treatment failure due to drug resistance occurs. Bz sensitive and resistant MM cells have distinct immunophenotypic signatures that correlate with clinical outcome. These changes can be identified by fluorescence-based cytometry (FBC), however, FBC is rarely used in predicting Bz resistance. Mass cytometry (MC) is a recently developed variation of flow cytometry that detects heavy metal-ion tagged antibodies using time-of-flight mass spectrometry allowing for detection of up to 38 epitopes simultaneously in a single cell, without significant overlap, exceeding the dimensionality of FBC 3–4 fold. Here we compared FBC and MC in the immunophenotypic characterization of Bz sensitive and resistant human MM cell line U266. We show that Bz resistant cells are associated with loss of CD56 and CD66a adhesion molecules as well as an activation signature.
Keywords: Multiple myeloma, cytometry, bortezomib, drug resistance
Introduction
Multiple myeloma (MM) is a fatal malignant neoplasm hallmarked by a clonal expansion of plasma cells (PCs) in the bone marrow [1]. Typical clinical findings include the presence of a monoclonal gammopathy (M-spike), hypercalcemia, renal failure, anemia, and bone pain [1]. While MM remains an incurable disease with a median five-year survival of approximately 45% [1], novel therapeutic agents and stem cell transplantation have greatly improved overall survival, with some recent studies now reporting a median overall survival of 8 years [2]. The proteasome inhibitor class of drugs have had a major positive impact on extending the survival of MM patients [3], with the first generation including the drug bortezomib (Bz) followed by the development of second generation inhibitors such as carfilzomib and ixazomib [4,5]. While the mechanism of Bz efficacy of action on the MM cell is not fully elucidated, the unfolded protein response (UPR) has been implicated due to the abundant cytoplasm and endoplasmic reticulum engorged by immunoglobulin, rendering these cells exquisitely sensitive to proteasome inhibitors such as Bz that interfere with the UPR process [3].
Unfortunately, while many MM patients initially respond to Bz, most patients eventually relapse or progress with drug resistant disease. Currently, there are no clinically available diagnostic assays to evaluate an MM patient’s own PCs for Bz sensitivity. In prior studies, we have used murine and human MM cell lines to characterize the gene expression signatures and have identified candidate markers that differentiate Bz sensitive (BzS) from Bz resistant (BzR) cell lines [6–8]. Subsequently, we showed that the gene expression of these candidate markers in primary human MM samples correlates with overall survival in two clinical trials incorporating Bz [8]. These studies showed that these cells lines can be used as a model system to identify clinically relevant biomarkers that can be utilized in a diagnostic setting to measure the fraction of cells harboring the bortezomib resistance phenotype.
Multicolor flow cytometry (FBC) allows for the simultaneous detection of up to 6, 8, or 10 antigens in routine clinical labs. Typically these antigens are cell surface cluster of differentiation (CD) markers. The combination of CD19, CD38, CD45, and CD56 allows for the separation of neoplastic and non-neoplastic PCs by Boolean gating, a standard method to analyze FBC data [9]. FBC is an important assay in the diagnosis of MM, and is becoming increasingly important as a tool for minimal residual disease detection [9–11]. However, several limitations to FBC exist, and one limitation includes the restricted number of channels/fluorochromes that are available. One method to overcome this limitation is to run multiple assays/tubes on a single specimen, but this prevents the detection of multiple markers within a single cell simultaneously and poses difficulties with specimen quantity, prolongs sample run time, and increases the cost of the assay. Another solution to overcome the restricted number of channels/fluorochromes is to increase the number of epitopes per assay, however emission spectral overlap limits the utility of individual fluorochromes that bleed into each other. Emission spectral overlap to some extent can be managed using compensation, the process of subtracting emission signals from closely emitting fluorochromes, ensuring that the fluorescence detected is specific to a particular fluorochrome. While compensation can be successfully applied, this technique can lead to loss of signal, is highly subjective and can lead to operator and interpretation error. Thus, though FBC is expanded by the number of epitopes used, the emissions spectra of the antibody-conjugated fluorochromes has spectral overlap which creates challenges and reduces individual marker sensitivity when designing a clinical assay and for discovery of novel markers of clinical value [12,13].
In recent years a new technology has been created that overcomes these technological limits of FBC and has increased the detection capacity by 3–4 fold with negligible spectral overlap. Mass cytometry (MC) is an innovative variation of flow cytometry that detects heavy metal ion tagged antibodies using time-of-flight mass spectrometry [12]. This system of deep phenotyping incorporates numerous antibody combinations allowing for the detection of up to 38 epitopes simultaneously in a single cell, without significant overlap between channels (compared to fluorochromes in FBC) [12]. The addition of more channels using MC adds tremendous clinical benefits including identification of rare cells improving MRD detection and the potential for functional characterization using intracellular staining. Thus, MC increases the number of available channels without reducing sensitivity due to spectral overlap.
In the current study we compared the immunophenotypic signatures of both BzS and BzR U266 human MM cell lines using FBC and MC and demonstrate the utility of MC to characterize phenotypic correlates of drug resistance in human MM. To our knowledge, this study demonstrates for the first time the utility of mass cytometry in the characterization of drug resistance in MM cells.
Methods
Cell lines and culture
The U266 cell line was purchased from the American Type Culture Collection (Manassas, VA) and cultured as previously described [14] using RPMI media (Gibco/Life Technologies; Grand Island, NY) supplemented with 10% fetal serum, penicillin/streptomycin, and IL-6 (R&D Systems; Minneapolis, MN). Bortezomib (Millennium Pharmaceuticals; The Takeda Oncology Company; Cambridge, MA) was dissolved in serum-free RPMI-1640 and stored at −80°C. BzR cells were established by dose escalation of Bz once-weekly for a period of six months as previously described and cell viability assays were performed as described in Mitra, et al.[15] Cell viability was measured by CellTiter-Glo Luminescent cell viability assay according to manufacturer’s protocol (Promega Corporation; Madison, WI) using the Synergy 2 Microplate Reader (Biotek; Winooski, VT).
Flow cytometry
Cell concentrations were adjusted to between 5 ×107 to 10×107 cells per mL, and antibodies were added to cells suspended in phosphate buffered saline (PBS). Cells were stained with antibodies against the following cell-surface markers: CD19, CD20, CD38, CD45, CD56, CD138, and CD184 (CXCR4) (Becton Dickinson; Franklin Lakes, NJ). Conjugated fluorochromes included FITC, PE, PerCP-Cy5.5, PE-Cy7, APC-H7, V450, and V500 (Becton Dickinson). The stained cells were analyzed in 7-colors/channels with a FACSCanto II flow cytometer (Becton Dickinson). List-mode files were analyzed with Kaluza software, v. 1.2 (Beckman Coulter; Brea, CA) creating scatter plots.
Mass cytometry
Prior to incubation with metal-conjugated antibodies, cells were washed once in 1X PBS and re-suspended at a concentration of 1×107 cells per ml stained with a 5 uM concentration of cisplatin viability dye and incubated at 37°C for 5 minutes. Fc blocking was performed for 10 minutes at room temperature, and cell surface staining was performed by adding a cocktail of pre-conjugated antibodies including CD19 (clone HIB19, metal 169Tm), CD38 (clone HIT2, metal 172Yb), CD45 (clone HI30, metal 141Pr), CD56 (clone NCAM16.2, metal 176Yb), CD138 (clone DL-101, metal 145Nd), CD184 (CXCR4) (clone 12G5, metal 175Lu), pTyrosine (clone P-Tyr-100, metal 144Nd), CD28 (clone CD28.2, metal 160Gd), Ki67 (clone B56, metal 168Er), cleaved caspase 3 (clone D3E9, metal 142Nd), CD66 (clone CD66a-B1.1, metal 171Yb) and HLADR (clone L243, metal 174Yb) (all from Fluidigm; Sunnyvale, CA) for 30 minutes at room temperature. Cells were fixed in media containing 1 mL of 1.6% paraformaldehyde and incubated at room temperature for 30 minutes. Cells were permeabilized by first washing twice in cold 1X PBS, incubation of cells on ice for 10 minutes followed by incubation with 1 mL of methanol at 4°C and stored at −80°C until cytoplasmic staining/data acquisition. Prior to cytoplasmic staining, cells were brought to room temperature, washed twice in MaxPar Cell Staining Buffer (Fluidigm) and incubated with a cocktail of antibodies for 30 minutes at room temperature. After staining, cells were washed in MaxPar Cell Staining Buffer (Fluidigm), treated for one hour with 1 mL of intercalation solution (containing iridium isotypes 191Ir and 193Ir), washed (with MaxPar) and analyzed by the CyTOF2 instrument (Fluidigm).
SPADE plots [16] were generated using Cytobank (cytobank.org) by clustering on 14 cell surface markers (CD79b, CD27, CD10, CD28, CD184, CD117, CD138, CD45, CD200, CD19, CD66, CD38, HLADR, and CD56) with 30 target nodes and 10% downsampled events. viSNE plots [17] were generated using Cytobank by using all of the epitopes measured in this experiment and including an equal number of cells from each sample.
Results
To compare and contrast the performance of multicolor flow cytometry (FBC) to mass cytometry (MC) in the detection of cell surface and/or intracellular markers, we utilized BzS and BzR U266 human MM cell lines isolated as previously described [15,18]. As described in Mitra et al [15], the BzR cell line displays a 20-fold increase in IC50 (BzS=3.72 mM; BzR=74.70 nM) in response to Bz compared to the BzS line. The BzR resistant cells lack a PSMB5 mutation [15]. These cell lines serve as a model system allowing the direct comparison of immunophenotypic markers in BzS and BzR cell lines detected by both FBC and MC. Figure 1 depicts scatter plot data of parental (P) and Bz resistant (VR) populations collected by FBC (panels A and C) or by MC (panels B and D). Initial gating strategies differed between FBC and MC. In FBC, viable cells were included by use of Boolean gating based on increased forward scatter (FSC) and low side scatter (SSC), as cells entering apoptosis and dead cells have decreased forward scatter and increased side scatter [13,19] (Figure 1A, left panel). Doublet discrimination was sequentially performed by including singlets based on FSC and SSC height (H) versus width (W) plots (Figure 1A, right panels). In contrast, the MC technique (using the second generation of cytometers which lacks a flow cell) did not offer the feature of forward and side scatter detection provided by FBC; thus no information regarding cell size and complexity by light scatter was available in this study [20]. Singlet discrimination in MC was achieved using DNA content (Ir193). Viable cell inclusion relies on cisplatin (Pt195) exclusion (Figure 1B). Using iridium to assess DNA content and cisplatin to exclude dead cells, allows for live, single cell gating by MC analogous to the function of a flow cell in FBC. Though a similar total number of cells or events were collected for comparative studies between parental and Bz resistant populations as well as between FBC and MC, there were considerable differences in cytometer acquisition rates. Whereas a BD FACS Canto II FBC method has a maximum theoretical sample acquisition rate of 10,000 events per second (per technical specifications document, Becton Dickinson), a MC has a maximum theoretical sample acquisition rate of 1,000 events per second [20]. However, when acquiring multiple parameters, the optimal acquisition rate is reported to be 2,000,000 events per hour, which is approximately 55 events per second [20]. Indeed, in our practice, collection times on the MC were considerably longer than with the FBC instrument; the low collection speed of a MC would prove impractical for most clinical labs. For a typical standard sensitivity myeloma minimal residual disease bone marrow sample (where one would collect 500,000 events) could be collected on a FC instrument in under a minute but would require greater than 2 hours on the MC (greater than 180-fold difference).
Figure 1. Scatter plots of multicolor flow cytometry (FBC) and mass cytometry (MC).
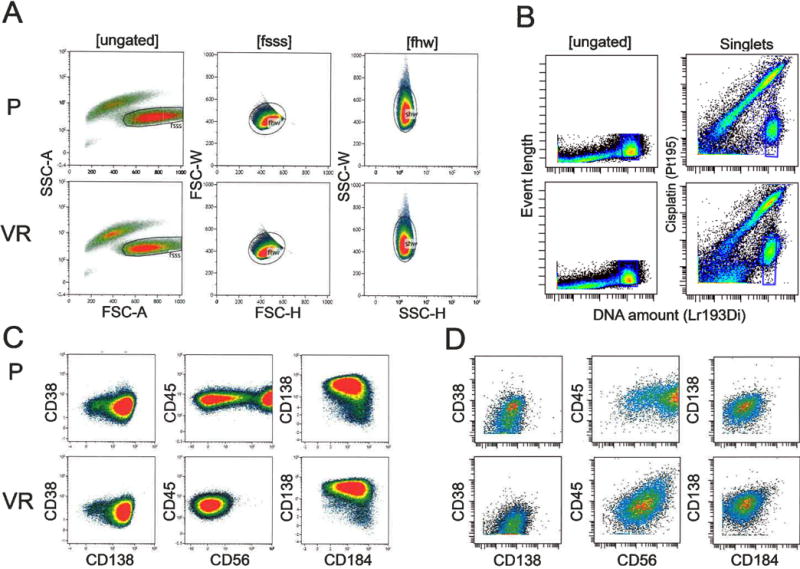
Panels A and C depict FBC, and panes B and D depict MC. In alternating rows we compared parental (P) with bortezomib/Velcade resistant (VR) cell lines. A) By FBC, both the P and VR cell lines have similar scatter characteristics. Viable cells are included by Boolean gating based on increased forward scatter (FSC) and low side scatter (SSC). Doublet discrimination is sequentially performed by including singlets based on FSC and SSC height (H) versus width (W) plots. B) In MC, live singlets are identified by DNA content (based on iridium intercalation, Ir193/195), and live cells are identified by cisplatin (PT195) exclusion. C–D) Cell surface staining characteristics of multiple myeloma markers by C) FBC and D) MC.
Next, we included 5 immunophenotypic markers (CD38, CD138, CD45, CD56, and CD184 (Cxcr4)) to directly compare the sensitivity of FBC to MC. Four of these markers (CD38, CD138, CD45, CD56) are commonly used in the characterization of plasma cell neoplasms. CD38 and CD138 have been characterized extensively and are typically brightly expressed on neoplastic and non-neoplastic plasma cells [9]. CD45 and CD56 are useful to distinguish neoplastic from non-neoplastic plasma cells. CD45 is typically dim to absent in neoplastic plasma cells, and CD56 is aberrantly expressed in a majority of neoplastic plasma cells [9]. We have previously shown in model systems that Bz resistant cell lines lose CD184 expression [8], and others have shown that a majority of plasma cells from patients with plasma cell myeloma express CD184 with variable intensity [21].
Parental specimens analyzed by each technique revealed the majority of the population as CD38+, CD138+, CD45+ and CD184+. Two distinct populations characterized as CD56 bright and CD56 dim were identified by both FBC and MC (Figure 1C, D). However, the two differentially expressing CD56 populations were more clearly distinguished using FBC in comparison to MC; this may be due in part to the PE-Cy7 fluorochrome conjugated to the anti-CD56 antibody, as PE and its conjugates tend to have bright emission intensity. Qualitative comparison of immunophenotypic staining patterns revealed a significant reduction in the amount of CD56 bright cells in the Bz resistant population compared to the parental population identified by both FBC and MC (Figure 1C, D).
A significant advantage to MC in comparison to FBC is the detection of up to 38 epitopes simultaneously in a single cell allowing the identification of rare cells and the added potential for signaling and functional characterization by intracellular staining of cytokine, transcription factor and phosphoprotein expression. We next tested the ability of MC to provide functional information using a combination of intracellular and cell surface antibodies and to further quantify immunophenotypic differences between parental and Bz resistant populations, as the multimarker intracellular staining and the overall increased number of channels provided has the potential to provided expanded information. The fold change in the expression levels between parental and Bz resistant populations of the tested antibodies was calculated from the signal intensity and plotted as a heatmap (Figure 2A) and histogram (Figure 2C). The fold change in the expression levels of CD184, pTyr (anti-phosphotyrosine, a surrogate marker for signal transduction and cell cycle progression) and CD28 was not appreciably different between parental and Bz resistant U266 cell populations, while expression levels of Ki67 (a cellular marker for proliferation), CD45, CD38, CD66a and were reduced in Bz resistant cells, most significantly in their expression of CD66a and CD56. Expression of cleaved caspase 3 (a cellular marker for apoptosis), CD138 and HLADR displayed increased expression in Bz resistant cells. Because expression of CD56 showed the most significant differences between parental and Bz resistant cells as indicated by the heatmap and dot plots (Figure 2A–B), we gated on both the CD56High and CD56Intermediate (Figure 2C) populations in the parental and Bz resistant cells and asked whether we could identify markers that associate with CD56 surface expression. The most significant differences were observed for CD38 and CD66a; reduced CD56 correlated with reduced expression of both CD38 and CD66a. Thus, CD56Intermediate parental and Bz resistant cells display reduced expression of both CD38 and CD66a. Subclonal populations with features of Bz resistant cells (CD56Intermediate, CD38Low and CD66aLow) appear to be present within the parental population.
Figure 2. MC analysis reveals multidimensional corrections of cell surface markers that differ between parental and Bz resistant cells.
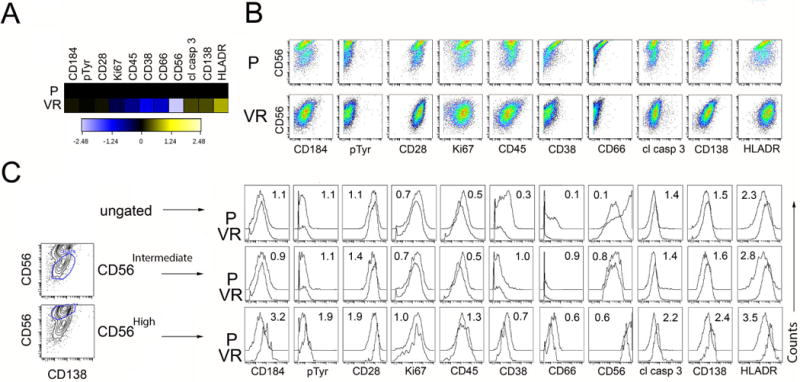
Cell lines were stained with cell surface markers, intracellular signaling intermediates, and functional markers (including phosphor-specific antibodies) and analyzed by MC. The fold change in the expression levels between parental and Bz resistant populations of the tested antibodies was calculated from the signal intensity and plotted as a heatmap (Figure 2A) and scatter plots (Figure 2B). We gated on both the CD56hi and CD56lo (Figure 2C) populations in the parental and Bz resistant cells and asked whether particular markers associate with CD56 surface expression; each marker depicted in individual histograms (Figure 2C).
FBC employs traditional methods such as Boolean gating with bivariate scatter plots for visualization of results used to create images such as those shown in Figures 1 and 2. These types of plots are standard in the clinical laboratory, but are mainly used to characterize populations in two dimensions, are user/operator dependent and introduce subjectivism. To circumvent these limitations, we next utilized spanning-tree progression analysis of density-normalized events (SPADE) tools to further define intraclonal variation among the two cell lines using data collected by MC [16].
SPADE takes multi-dimensional MC data and employs an objective and computational approach to cluster cells according to phenotypic staining characteristics to further analyze cellular heterogeneity between multiple populations. The data are then displayed in clusters (represented by circles, “nodes,” Figure 3). Each cluster represents a group of cells that display similar levels of all of the epitope that were used for clustering. Typically, cell surface markers are used for clustering such that the clusters are groups of cells with a similar immunophenotype. SPADE plots were generated by clustering on the 14 cell surface markers measured during this experiment (CD10, CD19, CD27, CD28, CD38, CD45, CD56, CD66, CD79b, CD117, CD138, CD184, CD200 and HLADR) into 30 unique nodes. The size of each node represents the number of cells within that population. Clusters that are similar to each other are arranged in proximity to one other and are connected by a line. A SPADE plot is colored according to an epitope of interest and the color represents levels of that epitope. For example, in Figure 3A, the color of the nodes represent levels of CD56 in the cells in those nodes. Figure 3B displays SPADE plots from the same experiment and the same sample, but the color represents CD38. In this way, the levels of CD38 and levels of CD56 in each node can be compared. Comparing SPADE plots across samples (as in Figure 3A left and right panels) reveals that the CD56High clusters are red in the left and orange/yellow in the right panel (as indicated by the green circles) which indicates that the Bz resistant sample expresses less CD56 in comparison to the parental population. Since the size of a node is proportional to the number of cells in a cluster, the larger parental CD56High clusters compared to the corresponding clusters in the Bz resistant sample indicates that there are fewer cells with this immunophenotype in the Bz resistant sample (Figure 3A).
Figure 3. SPADE plots reveal immunophenotypic and functional discrepancies between parental (P) and bortezomib-resistant (Bz) multiple myeloma cells.
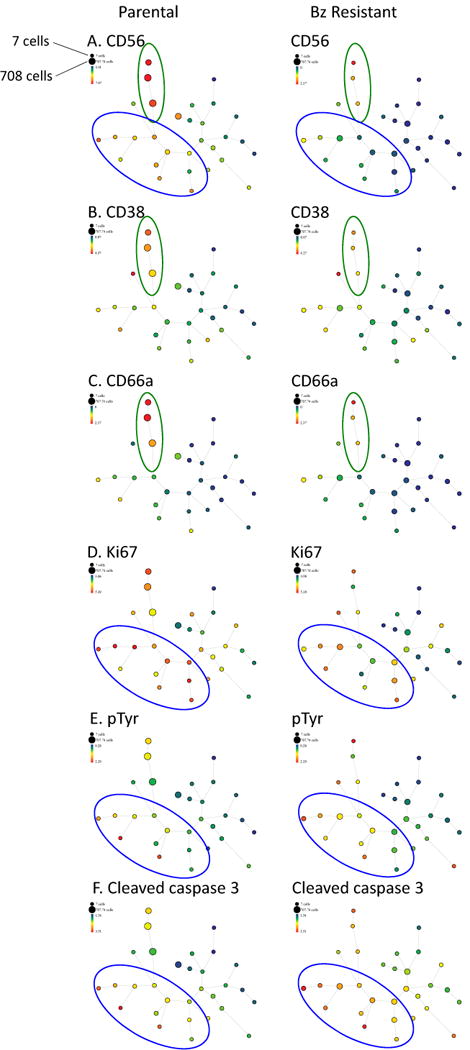
Parental and Bz-resistant multiple myeloma cells were stained with a panel of 14 cell surface antibodies and 4 functional epitopes and measured by mass cytomtery. SPADE analysis was performed on this data set by clustering on the 14 cell surface markers. Rainbow color scale reflects expression values (arcsinh difference) of each epitope. The smallest node represents 7 cells. The largest node represents 708 cells.
We used the SPADE plots to reveal the multidimensional immunophenotypic and functional differences between parental and Bz-resistant cells. The structure of the SPADE tree is maintained between comparable samples. As noted above, the size of the node represents the number of cells occupying that immunophenotypic characteristic in each sample and the color of the node in each panel represent the level of expression of the epitope being displayed. As previously noted, CD56High cells were abundant in parental cells (as indicated by the large, red clusters, Figure 3A, green oval, left panel) and rare in Bz-resistant cells (as indicated by small clusters in the corresponding branch, Figure 3A, green oval, right panel). In confirmation of our histogram analysis (Figure 2), SPADE analysis also reveals that CD38 and CD66a levels correspond with CD56 levels: they are highest in the CD56High clusters (Figure 3B–C). Relative to the parental cells, the Bz-resistant cells displayed an apparent expansion of CD56Intermediate/CD38/CD66aLow populations as indicated by the increase in size and number of blue and green nodes in the Bz resistant CD56 SPADE plot (Figure 3A, right panel, blue oval). These CD56Intermediate/CD38/CD66aLow populations can also be found in the parental cells leading us to speculate that these may represent a subset of Bz-resistant cells pre-existing within the parental cells population.
We also used SPADE to analyze the functional characteristics of immunophenotypically-defined cellular subsets. We found that the CD56Intermediate/CD38/CD66aLow populations (which are more abundant in the Bz-resistant cells) displayed the highest level of Ki67 (Figure 3D, blue oval represents CD56Intermediate/CD38/CD66aLow clusters), indicating that these are the most rapidly proliferating cellular subsets. This population also displayed relatively high levels of pTyr (Figure 3E) and cleaved caspase 3 (Figure F) suggesting that this population may be the most active cellular subset.
Another approach to visualizing high parameter data, such as our MC data, maps each individual cell into a scatter plot called [17]. This scatter plot retains all of the information of the high parameter data set and orients each individual cell in proximity to the next cell according to similarities across all of the parameters that were measured, analogous to a principal component analysis. Cells that are close together in a viSNE plot are similar to each other across all parameters measured. Like SPADE plots, viSNE plots are colored to represent levels of the epitope of interest. Since viSNE plots represent each cell individually, the heterogeneity within the system can be more readily discerned. For example, the expression level of CD56 in each cell is seen in Figure 4A. Like SPADE analysis, visNE reveals that the CD56High population is only found in the parental sample, a large portion of CD56High cells are CD38High and CD66aHigh.
Figure 4. viSNE analysis identifies a subpopulation of Bz resistant cells with correlates to a subpopulation in parental cells.
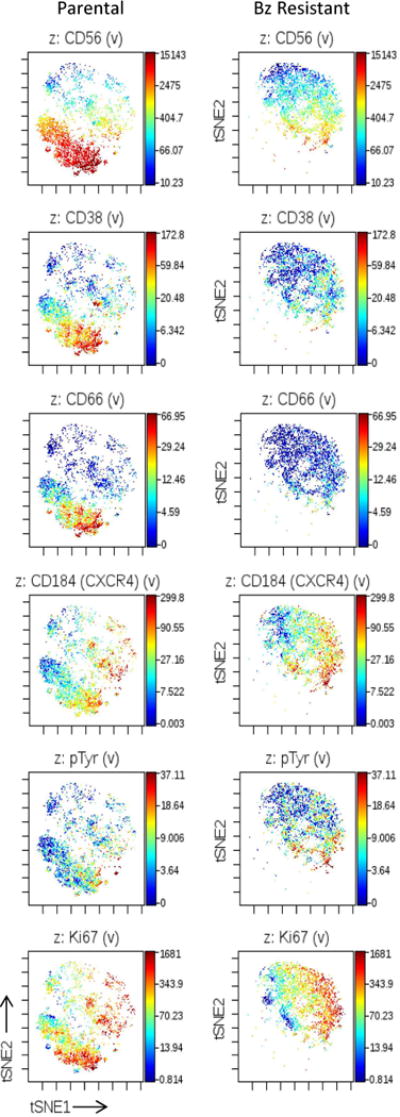
Parental and Bz-resistant multiple myeloma cells were stained with a panel of 14 cell surface antibodies and 4 functional epitopes and measured by mass cytometry. viSNE analysis was performed on these data by using all epitopes that were measured. The same number of cells from each group were sampled for creation of the viSNE plot. Rainbow color scale reflects expression values (arcsinh difference) of each epitope. Each dot represents a single measured cell. The labels at the top of each plot indicate the epitope that is displayed.
We used the viSNE plots to interrogate markers of multiple myeloma cells function and behavior. We found that CXCR4, an important mediator of bone marrow homing, can be found in parental cells, seemingly irrespective of CD56 status. In contrast, Only CD56Intermediate Bz resistant cells express CXCR4 suggesting that these CD56Intermediate cells may be able to thrive in the bone marrow microenvironment. These CXCR4High Bz resistant cells are the ones that displayed the highest levels of phosphorylated tyrosine (pTyr) and Ki67, suggesting these may harbor the most active signal transduction pathway activation and are most proliferative. Notably, viSNE analysis highlights that the CD56Intermediate/Low cells are seen in both the parental and Bz resistant samples. The distribution of their functional markers (CXCR4, pTyr, and Ki67) appears largely similar between the two samples, suggesting that the functional characteristics of the CD56Intermediate/Low cells may be similar in the two samples.
Discussion
In this study we compared the immunophenotypic signatures of populations of cells of a human multiple myeloma cell lines with sensitivity or resistance to the proteasome inhibitor bortezomib. By FBC, the biggest changes between the parental and BzR cell lines included a reduction in CD38 (slight), CD45 and CD56 surface expression. MC also confirmed decreases in surface CD38, CD45, and CD56, but the increase in dimensionality allowed MC to detect additional changes such as decreases in CD66a and increased cleaved caspase 3. MC also allowed us to find CD56 positive and negative subpopulations, and we observed that the CD56Intermediate parental and Bz cell line populations also have decreased expression of CD38 and CD66a, suggesting that a population similar to the BzR cells is present as a subclone among BzS cells in this cell line.
A key marker that distinguished the two populations by both methods was CD56. CD56 is a marker that is commonly included on clinical flow cytometry panels, as CD56 is absent on non-neoplastic plasma cells and is aberrantly expressed on approximately 70–80% of neoplastic plasma cells [22,23]. A recent study identified CD56 as one of the “top three” flow cytometric markers to demonstrate intra-clonal heterogeneity [24]. In addition, clonal heterogeneity of CD56 expression has been rarely reported using immunohistochemistry on trephine core biopsies [25].
CD56 is an isoform of the neural cell adhesion molecular (NCAM) and in normal development mediates both homotypic and heterotypic adhesion [26]. Prior studies have looked at the prognostic significance of CD56 in myeloma, and found that CD56 negative myeloma patients were more often associated with aggressive disease and a higher frequency of extramedullary involvement [26]. It should be noted, however, that these studies were conducted at a time that preceded widespread use of Bz therapy.
Although CXCR4 (CD184) did not vary appreciably between the sensitive and resistant cell line pairs in overall levels, the distribution of this marker varied among the subpopulations. In the parental cells, CXCR4 was seen in both CD56High and CD56Low population. In the Bz resistant cells, CXCR4 levels appeared to be highest in the CD56Intermediate population. CXCR4 is thought to provide important bone marrow microenvironment signals that allow tumor formation in that niche. These data may suggest that the CD56Intermediate population of Bz resistant cells are adept at tumor formation.
CD66a was shown by MC to have significantly decreased expression among resistant cells. CD66 is a member of the carcinoembryonic (CEA) and immunoglobulin superfamily [27]. The CD66 proteins are expressed as a number of isoforms, which are also called CEACAMs (for carcinoembryonic antigen cellular adhesion molecules). Previous studies have demonstrated that CD66a was present on two myeloma cell lines, including U266, as well as 5 patient samples tested, with a range of intensities [27]. While the literature discusses the therapeutic potential of CD66 as a radioimmunotherapy target, the role of CD66 in association with prognosis or chemotherapeutic sensitivity has not been described.
Importantly, we found a population of CD56IntermediateCXCR4High cells that displayed the highest levels of phosphor-tyrosine and Ki67 among Bz resistant cells. This is likely an active, proliferative population. This population, with similar staining characteristics was also found (in low abundance) in the parental cells. This could imply that the active, proliferative Bz resistant cells are pre-existing in the parental sample and that Bz treatment selects for the outgrowth of these cells. This would be an important and interesting question to pursue in further studies.
Though the precise mechanism of Bz resistance is not elucidated in the comparisons made in this study, downregulation of cell surface adhesion markers (including CD56 and CD66a) appears to be associated with Bz resistance. Plasma cells intimately interact with the bone marrow stroma and microenvironment, depending on it for its cytokine milieu. Most myeloma patients have disease restricted to the bone marrow/bone, with a few presenting as a primary plasma cell leukemia or relapsing as a secondary plasma cell leukemia [1]. Plasma cell leukemias are far less likely to express CD56 [1]. Thus, though loss of adhesion molecules is certainly associated with extramedullary disease, it may also be associated with chemotherapeutic resistance (including Bz and other proteasome inhibitors); this is further supported by our prior CD184/CXCR4 studies [8].
A key component of our study was also to compare conventional FBC to MC. MC has clear advantages over FBC as a tool for immunophenotypic discovery. The capacity to analyze numerous parameters is increased, with no need for cumbersome color compensation, etc. These experiments allowed us to describe the immunophenotypic heterogeneity of parental and Bz resistant cells. The additional dimensionality provided by MC allowed us to also infer functional differences and commonalities between these subpopulations. However, first and second generation MC instruments may not yet completely replace a clinical flow cytometry lab due to the lack of a flow cell on first/second generation instruments and the relative slowness of sample preparation and data acquisition.
One limitation of our study was the utilization of cell lines for sample comparison. Human myeloma samples contain varying percentages of plasma cells and contain heterogeneous populations of cells. Moreover, though clinical responses to drugs can be estimated based on decreasing “M-spike,” there are few standard assays to estimate in vitro drug sensitivity in purified primary human myeloma cells. Thus, for this proof of principle comparison we used established cell lines with quantifiable Bz sensitivities.
An additional limitation included our approach to FBC data analysis. While principal component analysis (PCA) is a powerful tool to analyze FBC data, not all laboratorians have the expertise to perform PCA or the software necessary to analyze the data. However, we acknowledge its utility in analyzing multiple parameters, especially when collected via multiple FBC tubes as has been done in multiple myeloma [24].
In summary, we used two different cytometric approaches to compare immunophenotypic markers of Bz sensitivity and resistance in the human myeloma cell line U266. Analytically, each method has its advantages, though the robustness of MC technology makes it useful in cytometric discovery of drug sensitivity in myeloma cell lines. Finally, our data suggest that in the U266 MM cell line, Bz resistant cells may be subclonally present among parental cells and are associated with loss of the CD56 and CD66a adhesion molecules.
Acknowledgments
Research reported in this publication was funded in part by the University of Minnesota’s Institute of Human Genetics Seed Grant and in part by the National Center for Advancing Translational Sciences of the National Institutes of Health Award Number UL1TR000114. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank and acknowledge the Mass Cytometry Shared Resource at the University of Minnesota which is supported by the Office of the Vice President for Research (U of MN).
Footnotes
Conflict of interest disclosures: Individual forms submitted for each author as per journal standards.
References
- 1.McKenna R. In: WHO Classification of Tumours of Haematopoietic and Lymphoid Tissue. Swerdlow SH, Campo E, Harris L, et al., editors. France: IARC; Oct, 2008. [Google Scholar]
- 2.Mikhael JR, Dingli D, Roy V, et al. Management of newly diagnosed symptomatic multiple myeloma: updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines 2013. Mayo Clin Proc. 2013;88:360–376. doi: 10.1016/j.mayocp.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 3.Fall DJ, Stessman H, Patel SS, et al. Utilization of translational bioinformatics to identify novel biomarkers of bortezomib resistance in multiple myeloma. J Cancer. 2014;5:720–727. doi: 10.7150/jca.9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan ML, Stewart AK. Carfilzomib: a novel second-generation proteasome inhibitor. Future Oncol. 2011;7:607–612. doi: 10.2217/fon.11.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Gomez A, Quwaider D, Canavese M, et al. Preclinical activity of the oral proteasome inhibitor MLN9708 in Myeloma bone disease. Clin Cancer Res. 2014;20:1542–1554. doi: 10.1158/1078-0432.CCR-13-1657. [DOI] [PubMed] [Google Scholar]
- 6.Stessman HA, Mansoor A, Zhan F, Linden MA, Van Ness B, Baughn LB. Bortezomib Resistance Can Be Reversed by Induced Expression of Plasma Cell Maturation Markers in a Mouse In Vitro Model of Multiple Myeloma. PloS one. 2013;8:e77608. doi: 10.1371/journal.pone.0077608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stessman HA, Baughn LB, Sarver A, et al. Profiling bortezomib resistance identifies secondary therapies in a mouse myeloma model. Molecular cancer therapeutics. 2013;12:1140–1150. doi: 10.1158/1535-7163.MCT-12-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stessman H, Mansoor A, Zhan F, et al. Reduced CXCR4 expression is associated with extramedullary disease in a mouse model of myeloma and predicts poor survival in multiple myeloma patients treated with bortezomib. Leukemia. 2013 doi: 10.1038/leu.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh C, Yohe S, Baughn LB, Linden MA. Utility of Flow Cytometry to Classify Abnormal Plasma Cell Populations in Marrow Samples Collected from Patients with Putative Plasma Cell Neoplasms. Open Journal of Blood Diseases. 2012;2:39–45. [Google Scholar]
- 10.Paiva B, Vidriales MB, Cerveró J, et al. Multiparameter flow cytometric remission is the most relevant prognostic factor for multiple myeloma patients who undergo autologous stem cell transplantation. Blood. 2008;112:4017–4023. doi: 10.1182/blood-2008-05-159624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paiva B, van Dongen JJ, Orfao A. New criteria for response assessment: role of minimal residual disease in multiple myeloma. Blood. 2015;125:3059–3068. doi: 10.1182/blood-2014-11-568907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nolan GP. Flow cytometry in the post fluorescence era. Best Pract Res Clin Haematol. 2011;24:505–508. doi: 10.1016/j.beha.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Cherian S, Wood B. Flow Cytometry in evaluation of hematopoietic neoplasms. Northfield, IL, USA: CAP Press; 2012. p. 165. [Google Scholar]
- 14.Croonquist PA, Linden MA, Zhao F, Van Ness BG. Gene profiling of a myeloma cell line reveals similarities and unique signatures among IL-6 response, N-ras-activating mutations, and coculture with bone marrow stromal cells. Blood. 2003;102:2581–2592. doi: 10.1182/blood-2003-04-1227. [DOI] [PubMed] [Google Scholar]
- 15.Mitra AK, Mukherjee UK, Harding T, et al. Single-cell analysis of targeted transcriptome predicts drug sensitivity of single cells within human myeloma tumors. Leukemia. 2016;30:1094–1102. doi: 10.1038/leu.2015.361. [DOI] [PubMed] [Google Scholar]
- 16.Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amir E-A, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsson K, Bennich H, Johansson SG, Pontén J. Established immunoglobulin producing myeloma (IgE) and lymphoblastoid (IgG) cell lines from an IgE myeloma patient. Clin Exp Immunol. 1970;7:477–489. [PMC free article] [PubMed] [Google Scholar]
- 19.Henry JB, McPherson RA, Pincus MR, Abraham NZ. Henry’s clinical diagnosis and management by laboratory methods. 22nd. Philadelphia, PA: Elsevier/Saunders; 2011. p. 1. online resource (xxi, 1543 pages) [Google Scholar]
- 20.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng W, Liu D, Fan X, et al. Potential therapeutic biomarkers in plasma cell myeloma: a flow cytometry study. Cytometry B Clin Cytom. 2013;84:222–228. doi: 10.1002/cyto.b.21083. [DOI] [PubMed] [Google Scholar]
- 22.Lin P, Owens R, Tricot G, Wilson CS. Flow cytometric immunophenotypic analysis of 306 cases of multiple myeloma. Am J Clin Pathol. 2004;121:482–488. doi: 10.1309/74R4-TB90-BUWH-27JX. [DOI] [PubMed] [Google Scholar]
- 23.Rawstron AC, Orfao A, Beksac M, et al. Report of the European Myeloma Network on multiparametric flow cytometry in multiple myeloma and related disorders. Haematologica. 2008;93:431–438. doi: 10.3324/haematol.11080. [DOI] [PubMed] [Google Scholar]
- 24.Paíno T, Paiva B, Sayagués JM, et al. Phenotypic identification of subclones in multiple myeloma with different chemoresistant, cytogenetic and clonogenic potential. Leukemia. 2015;29:1186–1194. doi: 10.1038/leu.2014.321. [DOI] [PubMed] [Google Scholar]
- 25.Quinn JG, Sadek I. Clonal heterogeneity in plasma cell myeloma. Lancet. 2015 doi: 10.1016/S0140-6736(15)00384-0. [DOI] [PubMed] [Google Scholar]
- 26.Sahara N, Takeshita A, Shigeno K, et al. Clinicopathological and prognostic characteristics of CD56-negative multiple myeloma. Br J Haematol. 2002;117:882–885. doi: 10.1046/j.1365-2141.2002.03513.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee C, Guinn BA, Brooks SE, Richardson D, Orchard K. CD66a (CEACAM1) is the only CD66 variant expressed on the surface of plasma cells in multiple myeloma: a refined target for radiotherapy trials? Br J Haematol. 2010;149:795–796. doi: 10.1111/j.1365-2141.2010.08100.x. [DOI] [PubMed] [Google Scholar]


