Abstract
A screening for oxalate degrading abilities was initially carried on within Lactic Acid Bacteria cultures of different food origin. Seventy-nine strains were drop-inoculated onto MRS agar plates containing calcium oxalate. By comparing colonies diameters, 31 strains were used to inoculate, in parallel, MRS and MRS modified by sodium oxalate addition. Differences in the strains’ growth were assessed by colony forming unit counts. For two strains, the growth in oxalate enriched medium was significantly higher; while, for eleven strains an opposite behaviour was recorded. Two strains – probiotic Lactobacillus rhamnosus LbGG and Enterococcus faecalis 59 – were chosen. The first strain appeared to be able to metabolize oxalate more efficiently than the other tested cultures, while strain 59 appeared unable to gather advantage by oxalates and, indeed, appeared to be inhibited by the salt presence in the medium. Outcomes revealed that higher glucose concentrations may favour oxalates utilization. In MRS with oxalate, but without glucose, citrate was completely metabolized. Evaluation along time confirmed that the oxalate degradation is more significant in presence of glucose. Outcomes may represent a good start for the development of a safe and even probiotic culture able to lower the oxalates content of food.
Key words: Oxalate, Lactic acid bacteria, Probiotic culture
Competing interest statement
Conflict of interest: the authors declare no potential conflict of interest.
Introduction
Oxalate is a toxic compound ubiquitous in the plant kingdom and widely consumed in normal human diets as a component of fruits, vegetables, grains and nuts (Hodgkinson, 1977). The normal daily intake of oxalate ranges from 70 to 920 mg, but strongly increases in vegetarians (Turroni et al., 2007). Oxalic acid is a highly oxidized organic compound and is a strong chelator of cations, especially Ca2+. These properties result in limited possibilities for its catabolism and also make oxalate toxic, especially for mammalians. The accumulation of oxalic acid may lead to several pathologic conditions, including hyperoxaluria, urolithiasis, renal failure, cardiomyopathy and cardiac conductance disorders (Turroni et al., 2007).
The current therapeutic drug treatment options for primary hyperoxaluria and/or the dietetic regimen for patients with secondary hyperoxaluria do not always lead to adequate success, e.g. sufficient reduction of urinary oxalate excretion. Hence, new treatment options are clearly needed. In addition, an oxalate-free diet is difficult to achieve and would probably be deficient in essential micronutrients. The discovery of oxalate-degrading bacteria in the human gastro intestinal tract has opened the way to several investigations regarding their potential role in reducing the urinary excretion of oxalic acid (Giardina et al., 2014). Treatment with oxalate degrading bacteria could be such a new therapeutic option in patients with both form of hyperoxaluria. The most studied oxalate-degrading bacterium is Oxalobacter (O.) formigenes, a Gram-negative strictly anaerobic bacterium, found in the colon of humans and other vertebrates (Weese et al., 2004). The pathway for oxalate utilization in O. formigenes involves two enzymes: formyl-CoA transferase and oxalyl-CoA decarboxylase (Turroni et al., 2007) responsible for the conversion in CO2 and formate further metabolized and excreted via faeces. Treatments with O. formigenes have been proposed for patients with either absorptive and/or dietary, hence secondary, hyperoxaluria: colon recolonization by O. formigenes may lead to up a 50% reduction of urinary oxalate excretion (Hatch et al., 2011; Jiang et al., 2011).
Apart from O. formigenes, strains within Bifidobacterium and Lactobacillus (Lb.) genera are known to efficiently metabolize oxalate in dogs (Gnanandarajah et al., 2012). As matter of fact, several surveys were focused on the search of oxalates degradation abilities in Lactic acid Bacteria (LAB) strains (Hokama et al., 2000; Hoppe et al., 2005; Lieske et al., 2005; Turroni et al., 2007; Murphy et al., 2009; Ren et al., 2011). Anbazhagan et al. (2013) generated a recombinant Lb. plantarum strain able to efficiently degrade oxalate up to 90%, against a 15% of not modified bacteria. Since the use of genetically modified organisms is not allowed, a search for oxalate degrading activities within LAB of food origin was attempted.
Materials and methods
Seventy-nine LAB strains were dropinoculated onto MRS (Oxoid, Basingstoke, UK) agar plates containing 50 mmol/L of calcium oxalate. After incubation at 30°C for 48 hours, the presence of clear areas around to bacterial colonies was considered indication of oxalate degradation.
Selected strains were tested for sodium oxalate degradation in MRS broth containing 20 mmol/L of sodium oxalate (MRS-ox) as described by Ren et al. (2011). Isolates were cultured twice in MRS and then used to inoculate in parallel MRS and MRS-ox broth. After incubation at 37°C for 72 h, bacterial loads were assessed by drop counts on MRS agar. After centrifugation at 12,000 rpm for 15 min, supernatants were examined by high performance liquid chromatography (HPLC) according to Blaiotta et al. (2012). Analysis was performed on samples diluted in mobile phase 1:5 (vol/vol) and filtered by AcroDisc millipore (0.2 μm) by system (Gilson 307 Series HPLC system; Gilson, Middleton, WI, USA) fitted with column (MetCarb68 Hcolumn 6.5×300 mm, Varian) in an oven thermostated at 65°C. Columns were eluted at 0.4 mL/min by a solution 0.25 N H2SO4 in ultrapure water. A refractometer (RID 133, Gilson) was used as detector. Standards were used for quantification of different sugars and acids in the samples. Each experiment was performed in triplicate. For two strains, trials were repeated with MRS-ox broths with different glucose concentrations (0, 5 or 10 g/L). After 6, 24, 30, 48, 54 and 72 hours of incubation at 37°C, the bacterial populations were quantified by MRS counts and O.D. determination, while oxalate concentrations were assessed by HPLC.
Results
MRS agar containing calcium oxalate did not allow a clear discrimination of oxalate-degrading stains. All 79 LAB cultures grew on the agar plates and in no case the development of a clear halo surrounding colonies was evident (data not shown). By comparing the diameter of the colonies, 31 strains were selected (Table 1). Cultures were used to inoculate, in parallel, MRS-ox, namely MRS modified by sodium oxalate addition, and unmodified MRS. CFU counts were used to compare strains. Around 68% of the strains presented an higher growth in MRS broth base than in MRS-ox, but such difference proved to be significant (P<0.05) for 11 strains (Enterococcus spp. 11, 16, 26 and 59, Ec. durans 17, Ec. faecalis 62, Lb. johnsonii La1, St. thermophilus DSM8713 and DSM20617, Lb. reuteri Bio and Lb. casei Lbc496). Similarly, 11 strains showed a better growth in MRS-ox, but the difference was significant for just two strains: Lb. rhamnosus LbGG and St. salivarius DSM20479 (Table 1).
Table 1.
Growth in MRS and MRS-added of sodium oxalate of strains used in this study.
| Strain | Source | Taxon | CFU/mL | |
|---|---|---|---|---|
| MRS | MRS-ox | |||
| 8 | Ricciardi et al. (2014) | Ec. hirae | 7.85±0.08 | 7.54±0.16 |
| 11 | Ricciardi et al. (2014) | Enterococcus spp. | 7.56±0.17 | 6.46±0.14 |
| 15 | Ricciardi et al. (2014) | Ec. durans | 8.00±0.10 | 8.07±0.15 |
| 16 | Ricciardi et al. (2014) | Enterococcus spp. | 8.28±0.17 | 7.07±0.19 |
| 17 | Ricciardi et al. (2014) | Ec. durans | 7.78±0.18 | 6.53±0.13 |
| 24 | Ricciardi et al. (2014) | Ec. hirae | 8.65±0.12 | 8.52±0.11 |
| 26 | Ricciardi et al. (2014) | Enterococcus spp. | 8.43±0.12 | 6.54±0.17 |
| 30 | Ricciardi et al. (2014) | Enterococcus spp. | 6.46±0.08 | 6.96±0.17 |
| 31 | Ricciardi et al. (2014) | Ec. durans | 7.63±0.08 | 7.24±0.10 |
| 35 | Ricciardi et al. (2014) | Ec. hirae | 8.70±0.16 | 8.30±0.16 |
| 36 | Ricciardi et al. (2014) | Enterococcus spp. | 8.35±0.11 | 8.40±0.12 |
| 51 | Ricciardi et al. (2014) | Enterococcus spp. | 8.24±0.19 | 8.65±0.17 |
| 59 | Ricciardi et al. (2014) | Enterococcus spp. | 8.77±0.19 | 6.76±0.13 |
| 61 | Ricciardi et al. (2014) | Enterococcus spp. | 8.10±0.18 | 7.48±0.17 |
| 62 | Ricciardi et al. (2014) | Ec. faecalis | 8.10±0.13 | 7.00±0.14 |
| La1 | Nestlé, Vevey, Switzerland | Lb. johnsonii | 8.28±0.17 | 7.18±0.14 |
| YIT9029 | Yakult Honsha Co. Ltd., Tokyo, Japan | Lb. casei Shirota | 8.26±0.20 | 8.75±0.18 |
| DN-114 001 | Danone, Paris, France | Lb. casei | 8.50±0.21 | 8.86±0.12 |
| DSM20479 | Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany | St. salivarius | 7.26±0.20 | 8.57±0.16 |
| DSM8713 | Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany | St. thermophilus | 8.03±0.17 | 6.50±0.16 |
| DSM20617 | Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany | St. thermophilus | 8.45±0.19 | 7.38±0.23 |
| LbGG | Blaiotta et al. (2012) | Lb. rhamnosus | 7.77±0.21 | 8.53±0.24 |
| VT1 | Blaiotta et al. (2012) | Lb. rhamnosus | 8.69±0.18 | 8.88±0.23 |
| RBT739 | Blaiotta et al. (2012) | Lb. rhamnosus | 8.38±0.14 | 8.83±0.15 |
| RBM526 | Blaiotta et al. (2012) | Lb. rhamnosus | 7.84±0.10 | 7.83±0.14 |
| OM13 | Aponte et al. (2012) | Lb. pentosus | 7.78±0.16 | 7.77±0.20 |
| Lbc491 | Aponte et al. (2008) | Lb. casei | 7.50±0.17 | 7.60±0.19 |
| Lbc496 | Aponte et al. (2008) | Lb. casei | 8.46±0.19 | 7.58±0.13 |
| Bio | BioGaia AB, Stockholm, Sweden | Lb. reuteri | 7.22±0.14 | 5.26±0.20 |
| ATCC393 | American Type Culture Collection | Lb. casei | 8.12±0.21 | 8.03±0.15 |
| Lyofast LC4P1 Sacco, Cadorago (CO), Italy | Lb. casei | 8.68±0.23 | 8.28±0.13 | |
CFU, colony forming units; MRS-ox, MRS-added of sodium oxalate. Strains code, species identity and source are reported.
Strains LbGG, DSM 20479 and RBT739, all characterized by a better growth in presence of oxalate and Ec. faecalis 59, showing an opposite behaviour were selected for further experiments. CFU counts confirmed previous outcomes, while the oxalate degradation, as revealed by HPLC analyses proved to be more efficient in strain LbGG rather than in RBT739 (Figure 1). Strain 59 showed a reduced consumption of sugar, followed by a lower production of lactic acid. The influence of the glucose on the utilization of sodium oxalate was evaluated by repeating the same set of trials in MRS-ox with decreasing glucose concentrations, namely half content (5 g/L) and without (0 g/L) glucose (Figure 2). The presence of glucose seemed to exert a substantial effect on the oxalate consumption, since in broth without glucose, oxalate consumption was similar for both strains (Figure 2).
Figure 1.
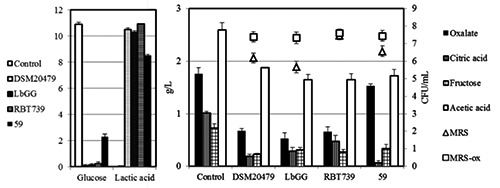
Oxalate degrading ability of Lb. rhamnosus strains LbGG and RBT739, St. salivarius DSM 20479, and Ec. faecalis 59 after 24 h of incubation. Evolution of reducing sugars, organic acids by high performance liquid chromatography and of microbial loads in MRS and MRS-added of sodium oxalate are reported. Un-inoculated broth served as control.
Figure 2.
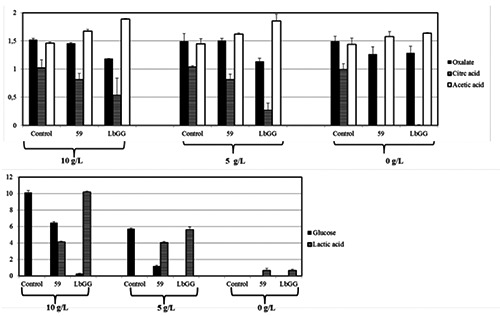
Oxalate degrading ability of Lb. rhamnosus LbGG and E. faecalis 59 in MRSadded of sodium oxalate with decreasing glucose concentrations (10, 5, 0 g/L). Content of glucose, lactic, acetic and citric acids by high performance liquid chromatography are reported. Un-inoculated broth served as control.
The analysis in MRS-ox with different glucose concentration was then repeated, by monitoring strains along incubation up to 72 hours (Figure 3). Cultures turbidity showed the same trend in the broth culture with 5 and 10 g/L of glucose, with a higher level of turbidity in broth cultures with the highest glucose concentration (data not shown). LbGG showed an humble growth in broth without glucose as revealed by CFU counts and pH monitoring (Figure 3). The cells number increased after the first 24 hours of incubation, afterward lactobacilli loads started to drop. Actually, populations decline in MRS without glucose started at 24 hour, while in the media with sugar, the drop begins after 48 hours. The increase of LAB populations was of almost one log for trials glucose-added. pH trends confirmed LAB counts evolution. HPLC analysis revealed an higher lactic acid production as long as the concentration of glucose increases. On the other hand, the acid acetic production registered a fluctuating trend along time.
Figure 3.
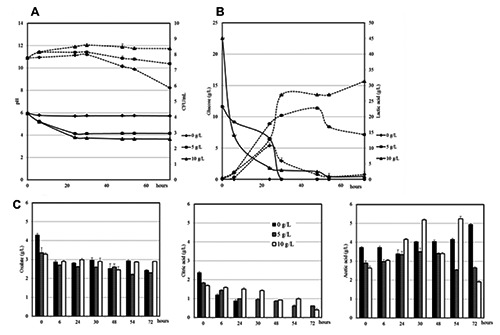
Evolution of pH (continuous lines) and MRS counts (dashed lines) (A); glucose (continuous lines) and lactic acid (dashed lines) (B); oxalic, citric and acetic acids (C) of Lb. rhamnosus GG in in MRS-added of sodium oxalate with decreasing glucose concentrations (10, 5, 0 g/L) during 72 h of incubation.
The chromatographic analysis confirmed a higher oxalate consumption in presence of the maximum glucose concentration. Citric acid was fully metabolized in the medium without glucose and its consumption decreased with the increase of glucose concentration.
Discussion
In the present study, a screening for oxalates degrading activity was carried out by considering 79 LAB strains known for probiotic features or isolated from foods and evaluated for functional capabilities during previous surveys (Aponte et al., 2008, 2012; Blaiotta et al., 2012; Ricciardi et al., 2014). Strains exhibiting a better growth on MRS agar plates enriched in oxalates were further investigated. According to outcomes, most of the strains showed a low oxalate degrading capacity. Among the tested strains, Lb. rhamnosus GG exhibited the highest percentage of oxalate consumption, above all in the presence of glucose. To authors’ knowledge, this is the first time that this popular probiotic strain has been investigated for such feature. Results even proved a correlation between the presence of sugars and the rate of oxalate consumption, thus underlining that the availability of glucose might provide an input for the oxalate metabolism as already pointed out by Ren et al. (2011). Without glucose, both LbGG and Ec. faecalis 59, isolated from Pecorino di Carmasciano cheese (Ricciardi et al., 2014) fully metabolized citrate and, above all, utilized oxalate at the same level. In other words, LbGG proved to degrade less efficiently the salt, while Ec. faecalis, that was chosen for its inability to degrade oxalate, exhibited an attitude comparable to that recorded for LbGG.
Conclusions
Many reports are currently focused on oxalates degrading capacity of O. Formigenes (Knight et al., 2013). However, its safety issue and health benefits are not as well established as in LAB. The research of different microorganisms able to express the same degrading-capacities led to investigate on several LAB strains mainly referable to Enterococcus spp and Lactobacillus spp (Hokama et al., 2000; Turroni et al., 2007). Bifidobacterium and Lactobacillus strains are commonly used in dairy and pharmaceutical probiotic preparation and are generally recognised as safe for human consumption (Fao, 2002; Iqbal et al., 2014). However, all studies highlighted in LAB cultures a minor oxalate degrading capacity if compared O. formigenes performances, for two main reasons: LAB prefer other substrates to obtain energy (Ren et al., 2011), and the genes encoding for the synthesis of oxalate degrading enzymes are less efficient than those reported in O. formigenes (Turroni et al., 2007). Basing on outcomes obtained in the present survey, Lb. rhamnosus GG, never evaluated before for this feature, may express an interesting potential for the degradation of oxalate. Since LbGG is well known for its ability to survive and to tolerate adverse storage conditions in dairy products (Alampreese et al., 2005), fresh-cut apple (Alegre et al., 2011), maple sap (Khalf et al., 2010), chestnut purees and mousses (Blaiotta et al., 2012; Romano et al., 2014), and emmer beverages (Coda et al., 2011), new perspectives may be hypothetised for the production of probiotic food, with particular emphasis on fermented vegetable foods, with reduced oxalates content and endowed with LbGG, thus likely to promote an oxalate reduction in the human organism, beyond the beneficial and therapeutic effects commonly associated to the ingestion of such probiotic strain.
References
- Alamprese C, Foschino R, Rossi M, Pompei C, Corti S, 2005. Effects of Lactobacillus rhamnosus GG addition in ice cream. Int J Dairy Technol 58:200-6. [Google Scholar]
- Alegre I, Viñas I, Usall J, Anguera M, Abadias M, 2011. Microbiological and physico-chemical quality of fresh-cut apple enriched with the probiotic strain Lactobacillus rhamnosus GG. Food Microbiol 28:59-66. [DOI] [PubMed] [Google Scholar]
- Anbazhagan K, Sasikumar P, Gomathi S, Priya HP, Selvam GS, 2013. In vitro degradation of oxalate by recombinant Lactobacillus plantarum expressing heterologous oxalate decarboxylase. J Appl Microbiol 115:880-7. [DOI] [PubMed] [Google Scholar]
- Aponte M, Blaiotta G, La Croce F, Mazzaglia A, Farina V, Settanni L, Moschetti G, 2012. Use of selected autochthonous lactic acid bacteria for Spanish-style table olive fermentation. Food Microbiol 30:8-16. [DOI] [PubMed] [Google Scholar]
- Aponte M, Fusco V, Andolfi R, Coppola S, 2008. Lactic acid bacteria during manufacture and ripening of Provolone del Monaco cheese: detection by different analytical approaches. Int Dairy J 18:403-13. [Google Scholar]
- Blaiotta G, Di Capua M, Coppola R, Aponte M, 2012. Production of fermented chestnut purees by lactic acid bacteria. Int J Food Microbiol 158:195-202. [DOI] [PubMed] [Google Scholar]
- Coda R, Rizzello CG, Trani A, Gobbetti M, 2011. Manufacture and characterization of functional emmer beverages fermented by selected lactic acid bacteria. Food Microbiol 28:526-36. [DOI] [PubMed] [Google Scholar]
- FAO, 2002. Guidelines for evaluation of probiotics in food. Food and Agriculture Organization of the United Nations, Rome, Italy. [Google Scholar]
- Giardina S, Scilironi C, Michelotti A, Samuele A, Borella F, Daglia M, Marzatico F, 2014. In vitro anti-inflammatory activity of selected oxalatedegrading probiotic bacteria: potential applications in the prevention and treatment of hyperoxaluria. J Food Sci 79:384-90. [DOI] [PubMed] [Google Scholar]
- Gnanandarajah JS, Johnson TJ, Kim HB, Abrahante JE, Lulich JP, Murtaugh MP, 2012. Comparative faecal microbiota of dogs with and without calcium oxalate stones. J Appl Microbiol 113:745-56. [DOI] [PubMed] [Google Scholar]
- Hatch M, Gjymishka A, Salido EC, Allison MJ, Freel RW, 2011. Enteric oxalate elimination is induced and oxalate is normalized in a mouse model of primary hyperoxaluria following intestinal colonization with Oxalobacter. Am J Physiol Gastrointest Liver Physiol 300:461-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson A, 1977. Oxalate content of foods and nutrition. Hodgkinson A. (ed.) Oxalic acid in biology and medicine. Academic Press, London, UK, pp. 193-212. [Google Scholar]
- Hokama S, Honma Y, Toma C, Ogawa Y, 2000. Oxalate-degrading Enterococcus faecalis. Microbiol Immunol 44:235-40. [DOI] [PubMed] [Google Scholar]
- Hoppe B, Von Unruh G, Laube N, Hesse A, Sidhu H, 2005. Oxalate degrading bacteria: new treatment option for patients with primary and secondary hyperoxaluria? Urol Res 33:372-5. [DOI] [PubMed] [Google Scholar]
- Iqbal MZ, Qadir MI, Hussain T, Janbaz KH, Khan YH, Ahmad B. 2014. Review: probiotics and their beneficial effects against various diseases. Pak J Pharm Sci 27:405-15. [PubMed] [Google Scholar]
- Jiang J, Knight J, Easter LH, Neiberg R, Holmes RP, Assimos DG, 2011. Impact of dietary calcium and oxalate, and Oxalobacter formigenes colonization on urinary oxalate excretion. J Urol 186:135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalf M, Dabour N, Kheadr E, Fliss I, 2010. Viability of probiotic bacteria in maple sap products under storage and gastrointestinal conditions. Biores Technol 101:7966-72. [DOI] [PubMed] [Google Scholar]
- Knight J, Deora R, Assimos DG, Holmes RP, 2013. Urolithiasis: the genetic composition of Oxalobacter formigenes and its relationship to colonization and calcium oxalate stone disease. Springer-Verlag 41:187-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieske JC, Goldfarb DS, De Simone C, Regnier C, 2005. Use of a probioitic to decrease enteric hyperoxaluria. Kidney Int 68:1244-9. [DOI] [PubMed] [Google Scholar]
- Murphy C, Murphy S, O’Brien F, O’Donoghue M, Boileau T, Sunvold G, Reinhart G, Kiely B, Shanahan F, O’Mahony L, 2009. Metabolic activity of probiotics - Oxalate degradation. Vet Microbiol 136:100-7. [DOI] [PubMed] [Google Scholar]
- Ren Z, Pan C, Jiang L, Wua C, Liu Y, Zhong Z, Ran L, Ren F, Chen X, Wang Y, Zhu Y, Huang K, 2011. Oxalatedegrading capacities of lactic acid bacteria in canine feces. Vet Microbiol 152:368-73. [DOI] [PubMed] [Google Scholar]
- Ricciardi A, Blaiotta G, Di Cerbo A, Succi M, Aponte M, 2014. Behaviour of lactic acid bacteria populations in Pecorino di Carmasciano cheese samples submitted to environmental conditions prevailing in the gastrointestinal tract: evaluation by means of a polyphasic approach. Int J Food Microbiol 179:64-71. [DOI] [PubMed] [Google Scholar]
- Romano A, Blaiotta G, Di Cerbo A, Coppola R, Masi P, Aponte M, 2014. Spray-dried chestnut extract containing Lactobacillus rhamnosus cells as novel ingredient for a probiotic chestnut mousse. J Appl Microbiol 116:1632-41. [DOI] [PubMed] [Google Scholar]
- Turroni S, Vitali B, Bendazzoli C, Candela M, Gotti R, Federici F, Pirovano F, Brigidi P, 2007. Oxalate consumption by lactobacilli: evaluation of oxalyl-CoA decarboxylase and formyl-CoA transferase activity in Lactobacillus acidophilus. J Appl Microbiol 103:1600-9. [DOI] [PubMed] [Google Scholar]
- Weese JS, Weese HE, Yuricek L, Rousseau J, 2004. Oxalate degradation by intestinal lactic acid bacteria in dogs and cats. Vet Microbiol 101:161-6. [DOI] [PubMed] [Google Scholar]


