Abstract
Induced pluripotent stem cells (iPSCs) are being developed as a source for autologous cell therapies, many of which aim to treat aged patients1–5. To explore the impact of age on iPSC quality, we produced iPSCs from blood cells of 16 donors aged 21–100. We find that while reprogramming resets most of the epigenome, iPSCs retain an epigenetic signature of age that diminishes with passaging. Reprogramming via clonal expansion also exposes somatic mutations present in individual donor cells, which are missed by other methods. We find that exomic mutations in iPSCs increase linearly with age and each iPSC line analyzed carries at least one gene-disrupting mutation, of which several have previously been linked to cancer or dysfunction. Unexpectedly, elderly donors (>90 yrs) harbor fewer mutations than predicted and their distribution suggests that blood in elderly donors derives from a contracted progenitor pool. These studies show that harnessing clonal expansion during reprogramming can uncover age-associated processes relevant to the clinical use of iPSCs.
Pluripotent stem cells are being developed as sources of differentiated cells for autologous cell replacement therapies in applications such as spinal cord injury, Parkinson disease and blindness1–5. Because iPSCs are generated directly from somatic tissues, they can be genetically matched to individuals, reducing the need for immunosuppression. However, this approach also engenders the risk that epigenetic aberrations or mutations in the donor cells will be carried through to iPSCs and their derived tissues, leading to increased tumorigenicity or dysfunction4, 6, 7. Indeed, a recent clinical trial of iPSC-derived tissues in Japan was halted upon discovery of potentially tumorigenic mutations in iPSCs8, 9. While many well-characterized iPSC lines are derived from youthful tissue, iPSCs intended for autologous therapy will in many cases be derived from the somatic cells of aged patients. Therefore, it is important to understand how age influences the epigenomes and genomes of iPSCs.
Aberrant DNA methylation in iPSCs can alter their differentiation capacity and tumorigenicity10–12. Aberrant DNA methylation can arise from stochastic errors in reprogramming or from a failure to reset patterns of methylation characteristic of donor cells. Thus, iPSCs generated from different cell types often retain some lineage-specific epigenetic signatures, termed epigenetic memory, which may influence their function13–17. Recent studies of aging in somatic tissues have identified a strong epigenetic signature of age based on DNA methylation at CpG sites18, 19. This signature is robust enough that DNA methylation patterns at only a small number of sites can accurately predict the chronological age of a donor18–20. Whether reprogramming of elderly donor cells into iPSCs can fully reset this signature has not been examined, although some iPSCs have been reported to have an apparent age of ~ −0.7 years18. To improve iPSC-based disease modeling and establish guidelines for clinical studies, we must determine the influence of age on epigenetic aberrations in iPSCs.
Mutations present in iPSCs contribute to the tumorigenic risk of iPSC-derived cells. Such mutations can be inherited from the original donor cell or acquired during the process of reprogramming and iPSC expansion. Mutations in iPSCs have been investigated in various ways21–23, with most studies concluding that the majority of mutations are derived from pre-existing somatic mutations in the donor cells while a minority arise during reprogramming, although this may reflect technical difficulties in detecting rare mutations associated with later stages of expansion24–26. The influence of age on the chromosomal somatic mutation load in iPSCs has not been studied experimentally, although it has been inferred that the number of somatic mutations in iPSCs would increase with donor age based on parallel increases in somatic mutation in bulk studies of aging tissues27–29. However, it is becoming apparent that individual cells accumulate rare or unique somatic mutations that are invisible in bulk DNA sequencing studies. Because reprogramming clonally expands the genome of a single cell, iPSCs can capture these rare changes. Thus, reprogramming enables high-resolution detection of rare somatic mutations without the errors and noise inherent to single-cell whole-genome sequencing 25,30–33. Here we apply this approach to assess the impact of age on rare exomic mutations in blood cells and iPSC lines derived from them, which has not been systematically reported for any human lineages.
Because age reduces the efficiency of reprogramming fibroblasts34, we elected to reprogram frozen peripheral blood mononuclear cells (PBMCs), which were expanded to eliminate T and B cells and enrich for erythroid progenitor cells, using Yamanaka non-integrated, episomal-based reprogramming35, 36. We modified the protocol of ref. 36 by optimizing parameters for erythroid progenitor cell expansion, survival after electroporation and iPSC culture conditions and applied this method to PBMCs from 5 young (21, 22, 23, 23, and 29 yrs), 5 middle-aged (49, 57, 62, 69 and 79 yrs), and 6 elderly (86, 87, 92, 94, 95 and 100 yrs) individuals (Fig. 1a, Supplementary Fi. 1c). We were able to generate iPSC lines from all donors (at least 6 lines per donor were banked). Donor age did not significantly influence reprogramming efficiency or iPSC quality based on karyotyping and immunostaining for pluripotency markers TRA-1–60 and SSEA4 (Supplementary Fig. 1). Three iPSC lines per donor that were positive for these pluripotency markers, karyotypically normal and that maintained an undifferentiated morphology during passaging were selected for analyses of DNA methylation and exomic mutations, based on comparisons with the original pBMCs obtained from the donors (Fig. 1a).
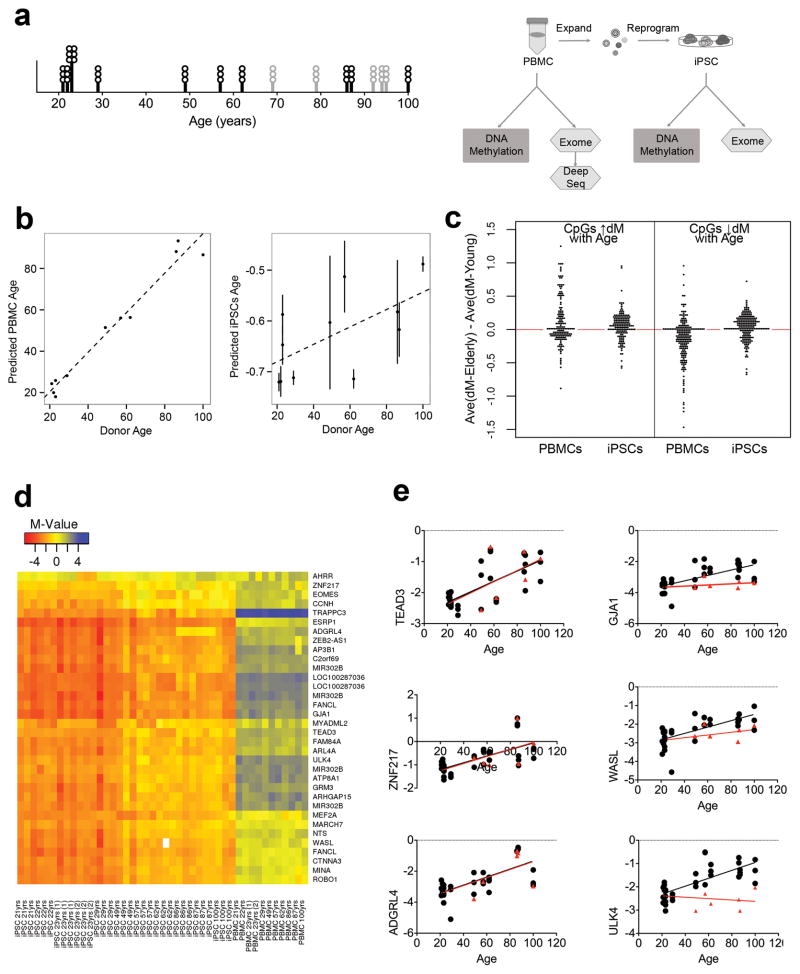
Figure 1. Study overview and epigenetic analyses.
A) Left, graph shows the age distribution of donors. Circles represent the number of iPSC lines generated from each individual. Black bars and circles indicate samples used for both epigenetic and exomic analyses. Gray lines indicate additional samples used for somatic mutation detection. Right, schematic of the study design. PBMCs from donors were isolated, expanded and reprogrammed with episomal reprogramming factors. Three iPSC lines per donor were characterized. Unexpanded PBMCs and iPSCs were subjected to exome sequencing and DNA methylation analysis. B) Donor age is strongly correlated with predicted age based on PBMC methylation (left panel) (R2 = 0.96, p-value = 6.4·10-8). The predicted age of iPSCs is weakly but significantly correlated with donor age (right panel) (R2 = 0.38, p-value = 0.04). Each filled circle represents the average predicted age of three iPSCs per individual; the vertical line is standard error. The dashed line is the linear best fit across all individuals. C) The difference in average methylation levels between elderly and young donors is plotted for CpG sites previously reported to predict age (CpGs that increase in methylation with age on the left; CpGs that decrease in methylation with age on the right). In PBMCs, CpG sites expected to increase in methylation with age (left) show an average increase of methylation in elderly individuals (average of left PBMC plot = 0.09), and CpG sites expected to decrease in methylation with age (right) show an average decrease in of methylation in elderly individuals (average of right PBMC plot = −0.17). On the other hand, in iPSCs, both CpG sites expected to increase (left) and decrease (right) in methylation with age show an average increase of methylation in elderly individuals (average of left iPSC plot = 0.04, average of right iPSC plot = 0.01). Visually, there is a clear dissimilarity in the distribution of the methylation differences at age-related CpG sites in in PBMCs vs iPSCs – with age-related CpG sites displaying two different distributions of methylation differences in PBMCs derived from young vs old donors whereas age-related CpG sites uniformly display increased methylation levels in iPSCs derived from older donors. D) Heat map displaying the methylation status of all CpG sites found to be significantly resistant to demethylation in older individuals compared to younger donors (blue, high, red, low). iPSCs are ordered by the age of donor. CpG sites are clustered based on their methylation pattern across individuals. Genes are labeled on the right side. E) DNA methylation profiles for selected iPSC lines were analyzed at early/intermediate and late passages. The methylation status of six CpG sites is plotted with black circle representing M-value for iPSCs at early passage and red triangle M-value for late passage in selected iPSC lines. The right side shows three CpG sites that correlate with age at early passage, whose methylation is restored after passaging (red linear regression). GJA1 (early passage R2 (eR2)= 0.44 p-value < 0.0001; late passage R2 (lR2)= 0.043 p-value=0.36), WASL (eR2= 0.45 p-value < 0.0001; lR2= 0.094 p-value = 0.17), ULK4 (eR2= 0.53 p-value < 0.0001; lR2= 0.042 p-value = 0.37). The left part show three CpG that maintained a significant correlation with age, independent of passage TEAD3 (eR2= 0.51 p- value < 0.0001; lR2= 0.54 p-value = 0.0001 ), ZNF217 (eR2 = 0.45 p-value<0.0001; (lR2)= 0.45 p-value = 0.0008), ADGRL4 (eR2 = 0.47 p-value<0.0001; (lR2)= 0.45 p-value = 0.0009). Pearson correlation with two-tailed p-value.
We analyzed DNA methylation via Illumina’s Infinium Methylation Assay. As the age of blood cell donors can be predicted based on DNA methylation profiles18, 19, we applied the method of Horvath18 and found that we could accurately predict the age of PBMC donors across all age groups (R2 = 0.96, p-value = 6.4·10−8) (Fig. 1b – left panel). IPSCs exhibit a ‘negative’ predicted age, similar to that of embryonic stem cells, indicating that reprogramming can reset the epigenetic aging clock 18. While the genome-wide changes in DNA methylation we observed predicted a negative age for all of our iPSC lines, we also detected a weak residual signature of the donor age (R2 = 0.38) (Fig. 1b – right panel). Although some variability in predicted methylation age was observed, the relationship was significant when assessed via a linear mixed effects model accounting for repeated measures per individual donor (p-value = 0.04) (see Methods for rationale and description of the model).
During aging, DNA methylation reproducibly increases at some CpG sites and decreases at others. The epigenetic signature of age in blood includes both types of CpG sites, although the former are the best preserved between different tissues and individuals and these are also thought to be most likely to alter gene expression37. To identify the source of the residual age-associated signature in iPSCs, we separately examined the influence of age on methylation levels at these two sets of CpG sites. In the PBMCs, as expected, CpG sites at which methylation has been reported to increase with age exhibit increased DNA methylation in older donors, as shown by plotting the average difference in methylation at each site for the old donors vs. the young donors (Fig. 1c left - average > 0, average = 0.09). In the iPSCs, these same CpG sites similarly exhibited significantly higher methylation in the older donor cells compared to the younger (Fig. 1c left - average > 0, average = 0.04) (generalized Friedman rank sum test with replicated data: p-value = 1.3·10−7). This could result from a residual age-based program of epigenetic regulation or reflect a broader resistance to demethylation in iPSCs from older donors.
To address this we next examined CpG sites reported previously to decline in methylation with age. We found that, as expected, in PBMCs from our cohort the methylation levels at these sites were lower in the cells from the older donors, plotted as average methylation in older donors minus average in younger donors (Fig. 1c right – average < 0, average = -0.17) (generalized Friedman rank sum test with replicated data: p-value < 2.2·10−16). However, in the iPSCs we derived from these PBMCs the average methylation at each CpG site was significantly higher in the lines derived from older compared to the younger donors (Fig. 1c left - points average > 0, average = 0.04) (generalized Friedman rank sum test with replicated data: p-value = 1.3·10−7) (raw data displayed in Supplementary Table 1, Supplementary Fig. 2). These data suggest that the epigenetic signature of age in iPSCs reflects an overall resistance to demethylation during reprogramming at both types of age-associated CpG sites, with a more significant demethylation defect affecting CpG sites whose methylation levels increase with age.
To determine whether this resistance to demethylation during reprogramming of iPSCs from elderly donors is a global phenomenon, we examined DNA methylation at all CpG sites that demethylate significantly during reprogramming (adjusted p-value <0.05) from PBMCs to iPSCs (defined as CpGs with average an M-value > 1 (>67% methylation) in PBMCs and an average M-value < -1 (<33% methylation) in iPSCs). We identified 3,896 reprogramming-associated CpG sites that met these criteria. These reprogramming-associated CpG sites were more methylated in iPSCs derived from elderly vs. young individuals, by ~1/3rd of a standard deviation (Supplementary Fig. 3a), uncovering a small but highly significant global demethylation deficiency in iPSCs from elderly donors (generalized Friedman rank sum test with replicated data, p-value < 2.2·10−16). In absolute terms, this corresponds to a ~5% increase in the global methylation levels in iPSCs from older individuals.
To search for specific genomic regions that might be particularly relevant to understanding the epigenetic variation in iPSCs derived from elderly individuals, we identified CpG sites whose methylation status in iPSCs was significantly associated with donor age (adjusted p-value < 0.05) and whose methylation status displayed a significant and meaningful magnitude of change during reprogramming (an average absolute change in M-value of 1.0 - which corresponds to a 2-fold change in the ratio of methylated to unmethylated CpGs). 109 such CpG sites were identified, including CpG sites that were either defective in demethylation or remethylation during reprogramming (Supplementary Table 2, Supplementary Fig. 4). Of these 109 CpG sites, we focused on 34 sites that did not fully demethylate during reprogramming of iPSCs from elderly individuals( Figure 1d and Supplementary Figure 3b). A number of genes involved in neoplastic processes were identified, for example TEAD3 (Hippo signaling), ADGRL4 (tumor angiogenesis), and AHRR (tumor suppressor).
Epigenetic signatures in iPSCs, such as cell type ‘memory’, can sometimes be reduced with extended passaging. As we found that cells from older donors are significantly different from those of younger donors at passage 84, we tested whether these age-associated changes could be eliminated with even more passaging by expanding one line per donor for an additional 20 passages. All but six of the 34 persistent changes were restored to levels similar to younger donors (Figure 1e, Supplementary Figure 3b).
To investigate mechanisms underlying the resistance to demethylation, we examined the expression of genes involved in active CpG demethylation and methylation, such as the TET (Ten-Eleven Translocation) enzymes38. We found a decrease with age in the expression level of TET1 (R2= 0.21 p-value = 0.0015), TET3 (R2= 0.46 p-value< 0.0001) and TET2A (R2= 0.25 p-value= 0.0005), but not TET2B (R2= 0.04 p-value=0.15) (Supplementary Fig 3c). Thus, lower TET family member expression in the iPSCs may contribute to the observed transient resistance to demethylation.
In contrast to epigenetic aberrations, which can be heterogeneous even among clonal cells, somatic mutations in the original donor cell are expected to be uniformly persistent in iPSCs of the same clonal lineage, yet diverse across iPSCs from separate clonal lineages due to the presumed stochastic nature of somatic mutation. To determine how donor age influences the number, type and potential genic impact of mutations in iPSCs, we performed exome sequencing of the PBMC pool along with at least three iPSC lines from the same donor. Each iPSC line is clonally derived from a single peripheral blood cell, in analogous manner to the derivation of the major clone of a tumor from a single normal cell. Therefore, we identified candidate somatic mutations in iPSC lines by comparing them to the bulk original blood sample and applying standard filters used to identify somatic mutations in tumor-normal sequencing studies – i.e. filtration of variants detected by the MuTect algorithm (See Online methods)39. Exomic mutations in the iPSCs derived from somatic exomic mutations in the starting donor cell are present at 50% allelic frequency in the iPSC population. Similarly, mutations induced during reprogramming of the initial donor cell will be present at 50% allelic frequency in the iPSC population. Thus, we restricted our analysis to the most likely to be valid somatic mutations, as opposed to false positive candidate somatic mutations due to errors in sample preparation, sequencing and/or variant calling, using a variant allele frequency filter (See Online methods). This approach identified somatic point mutations in all iPSC lines (Supplementary Table 3). No somatic CNVs were detected in any line based on read-depth analysis, although this method has limited sensitivity.
Indeed, exomic mutations in iPSCs derived from individual PBMCs increased with donor age, doubling, on average, from 20 to 90 years of age (Fig. 2a). This correlation was highly significant as assessed via a linear mixed effects model accounting for repeated measures per individual donor (R2 = 0.38, p-value = 0.0011) (Fig. 2a – dashed line). Notably, this relationship was independent of variation in coverage of our iPSC samples (R2 = 0.002, p-value = 0.38) (Fig. 2c, Supplementary Table 4). While some rare reprogramming-associated mutations may be included in these analyses, due to noise in low coverage regions, the linear increase we observed with age is difficult to explain as anything other than somatic mutation.
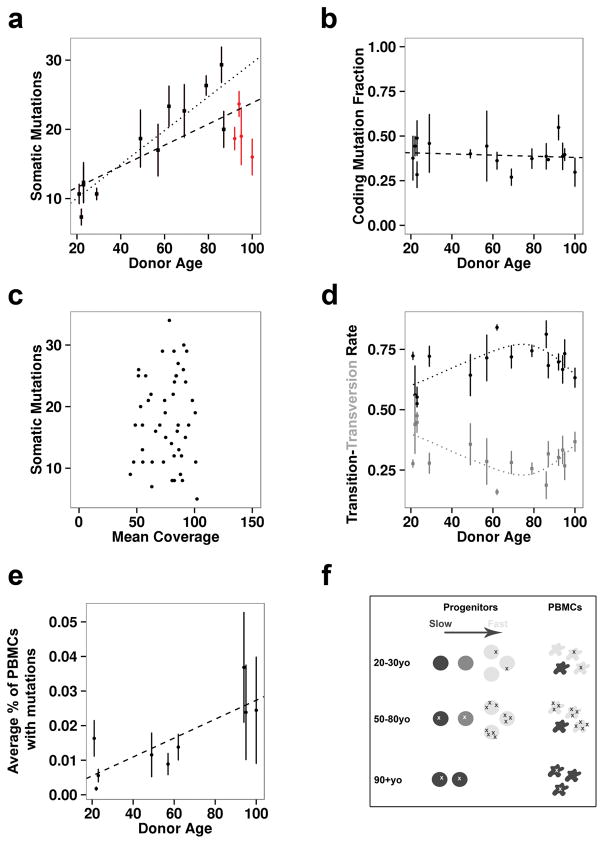
Figure 2. Somatic Mutations in iPSCs.
A) The number of somatic mutations per iPSC line per individual is plotted vs. donor age. Each point represents the average number of somatic ?7 mutations observed in three iPSC lines from one individual. A standard error bar intersects each point. The dashed line represents a linear best fit across all individuals (R2 = 0.38, p-value = 0.0011) and the dotted line represents a linear best fit across all individuals younger than 90 yrs (R2 = 0.63, p-value < 0.0001). Lines from donors over 90 years old are red. B) The fraction of all identified somatic mutations that were either missense or nonsense is plotted against donor age. Each data point represents the average fraction of somatic coding mutations observed in three iPSC lines per individual. A standard error bar intersects each point. The dashed line represents a linear best fit across all individuals (R2 = 0.004, p-value = 0.34). C) The number of somatic mutations is plotted against the coverage per iPSC line. No relationship was observed (R2 = 0.002, p-value = 0.38). D) The proportion of somatic mutations that are nucleotide transitions (black circles) vs. transversions (grey squares) is plotted relative to donor age. Each data point represents the average fraction of somatic mutations that are transitions or transversions in the three iPSC lines per individual. A standard error bar intersects each point, representing the variation in somatic mutations per iPSC line in each individual. The dotted lines displays a LOESS curve fit across transition and transversion proportion averages. E) The average level of somatic mosaicism in PBMC samples as determined by deep amplicon sequencing of randomly selected somatic mutations observed in the derived iPSC samples is plotted against donor age. Each data point represents the average level of somatic mosaicism observed per individual based on three somatic mutations per donor. A standard error bar intersects each point, representing the variation in the level of somatic mosaicism per donor. The dashed line represents a linear best fit across all individuals (R2 = 0.66, p-value = 0.028). F) Schematic of a model that can explain the apparent reduction in somatic mutations in iPSCs derived from the oldest donors (>90 years old). In this model, few mutations (x’s) are present in all cells of younger donors. Then in middle age the rapidly dividing cells (light grey) accumulate mutations. At very old ages, these cells disappear and smaller number of slowly dividing progenitors (darkest grey) with fewer mutations contribute to the PBMC population.
Of the 326 somatic coding mutations we identified, 24 were nonsense mutations affecting genes in which loss of function mutations are known to impair cellular function or lead to tumorigenesis (Table 1). For example, we observed nonsense mutations in MYO7A (c.5899C>T, p.R1967X) and KIRREL3 (c.553C>T, p.R185X). Loss of function mutations in one copy of either of these genes cause Usher syndrome, type 1B (OMIM:276903) and autosomal dominant mental retardation (OMIM:607761), respectively. These observations predict an elevated risk for dysfunctional tissues derived from iPSCs of older donors. We also observed a nonsense mutation in TET2 found in somatic myelodysplastic syndromes, and another mutation implicated in cancer (PPM1D). All observed mutations, their coding consequences, and coverage/counts in iPSCs vs. donor PBMCs are provided in Supplementary Table 3. Despite this, we did not detect any signatures of positive or negative selection of these mutations based on two lines of evidence. First, the proportion of observed missense and nonsense variants to total coding variants did not significantly deviate from expectations (expected 77% nonsynonymous, observed 75% ± 5%; 95% confidence interval). Second, the fraction of somatic mutations that were coding was also not correlated with age (R2 = 0.004, p-value = 0.34) (Fig. 2b). These data also argue that the majority of mutations we identified are of somatic origin.
Table 1. Potential loss of function mutations.
Each gene disrupting mutation found in iPSCs is listed with alongside the donor of origin and the phenotypic consequence of this mutation in humans. Importantly, many potential deleterious mutations may have unknown consequence so this is likely an underestimate.
| Donor | Gene | cDNA Change | Protein Change | OMIM Disease |
|---|---|---|---|---|
| iPSC 23 yrs | MYO7A | c.5899C>T | p.R1967X | Deafness, AD |
| iPSC 23 yrs | OR5B17 | c.602C>A | p.S201X | -- |
| iPSC 29 yrs | NLRP13 | c.590G>A | p.W197X | -- |
| iPSC 29 yrs | PACSIN2 | c.703C>T | p.Q235X | -- |
| iPSC 62 yrs | KIRREL3 | c.553C>T | p.R185X | Mental retardation, AD |
| iPSC 62 yrs | SIRT1 | c.322C>T | p.Q108X | -- |
| iPSC 62 yrs | C11orf1 | c.13C>T | p.Q5X | -- |
| iPSC 62 yrs | STAB2 | c.5353C>T | p.Q1785X | -- |
| iPSC 62 yrs | PACRG | c.1A>T | p.M1L | -- |
| iPSC 69 yrs | HS6ST3 | c.1081C>T | p.Q361X | -- |
| iPSC 69 yrs | NOBOX | c.280G>T | p.E94X | Premature ovarian failure 5, AD |
| iPSC 69 yrs | ATRNL1 | c.2799G>A | p.W933X | -- |
| iPSC 69 yrs | KMT2B | c.3247C>T | p.R1083X | -- |
| iPSC 86 yrs | TMPRSS2 | c.135C>G | p.Y45X | -- |
| iPSC 86 yrs | RSAD2 | c.1083G>A | p.W361X | -- |
| iPSC 86 yrs | NINL | c.2641C>T | p.Q881X | -- |
| iPSC 86 yrs | LY75 | c.3G>A | p.M1I | -- |
| iPSC 87 yrs | PPM1D | c.1375G>T | p.E459X | Breast cancer, AD |
| iPSC 92 yrs | CCNB1 | c.169C>T | p.Q57X | -- |
| iPSC 92 yrs | CCNB1 | c.169C>T | p.Q57X | -- |
| iPSC 94 yrs | TRMT10A | c.397C>T | p.R133X | Microcephaly, short stature, and impaired glucose metabolism, AR |
| iPSC 94 yrs | PPFIA2 | c.3591G>A | p.W1197X | -- |
| iPSC 95 yrs | TET2 | c.2185C>T | p.Q729X | Myelodysplastic syndrome, somatic |
| iPSC 100 yrs | SCG2 | c.196C>T | p.R66X | -- |
AD = autosomal dominant, AR = autosomal recessive
Unexpectedly, iPSCs from extremely elderly donors (~90 years or older) harbored significantly fewer exomic mutations than predicted by the linear increase in somatic mutations observed in the overall population (Fig. 2a – red points). To determine whether this apparent decline in somatic mutation rate was significant, we compared the goodness of fit for somatic mutations by age for individuals from 20 – 90 years of age (R2 = 0.63, p-value < 0.0001) (Fig. 2a – dotted line) vs. 20 – 100 years of age (R2 = 0.38, p-value = 0.0011) (Fig. 2a – dashed line). These results confirmed a strong linear relationship between age and somatic mutation count between the ages of 20 to 90, which begins to degrade at extreme age. A linear mixed effects model with the addition of a parameter to indicate individuals over the age of 90 (extreme age indicator), and an interaction term between age and this extreme age indicator, confirmed a statistically significant effect of extreme age on the number of somatic mutations per iPSC line (AIC = 297 vs. 310 for model with and without extreme age parameter respectively, difference in fit p-value = 0.0003 via ANOVA). Thus, iPSC lines produced from donors of extreme age harbored fewer mutations than expected and in some cases, mutational loads were similar to donors one half to two thirds of their age.
To investigate the potential mechanism behind this shift in mutations, we examined the nucleotide context of the mutations in each iPSC line. Cell division correlates with the number of mutations in a cell and with the ratio of transition mutations to transversions. As expected, the ratio of transitions to transversions increases from 20 to ~90 years of age (Fig. 2d). However, in cells from extremely elderly donors, this ratio begins to revert back to resemble that of young donors (Fig. 2d). These results suggest that the PBMC progenitor cells in the extremely elderly donors have undergone fewer divisions than would be expected.
One possible explanation for this is that extremely elderly individuals have a restricted pool of hematopoetic stem cells that may be slowly dividing and thus longer lived than other progenitors. To address this question, we reasoned that if extremely elderly individuals have smaller progenitor pools, the proportion of cells in the PBMCs that carry a particular somatic mutation should be higher than in younger donors. Thus, we randomly selected mutations observed in iPSCs and deeply sequenced the corresponding donor PBMC populations in order to determine the number of cells that were likely to carry these mutations, in other words, the level of somatic mosaicism at these sites. As predicted, we observed a strong linear relationship between donor age and the degree of detectable somatic mosaicism for these mutations (R2 = 0.66, p-value = 0.028) (Fig. 2e, Supplementary Table 5). This result is consistent with models in which PBMCs from older individuals derive from a restricted and/or genomically protected hematopoetic progenitor pool that has undergone fewer cell divisions than progenitors supplying PBMCs to middle-aged donors (Fig. 2f).
Together, our results demonstrate that donor age poses a risk to the safety of iPSCs because the likelihood of both epigenetic and genetic aberrations in iPSCs increases with age. First, we show that the previously described epigenetic signature of advanced biological age is evident in blood cells derived from our donors. Next we show that demethylation during reprogramming of these cells into iPSCs is less complete in elderly donors than younger donors, leading to an epigenetic signature of age in the iPSCs. This signature correlated with decreased expression of several TET family genes and also impacted genes linked to cancer. However, we find that extended passaging can erase most of the residual aberrant DNA methylation, suggesting that improved reprogramming techniques or increased monitoring of iPSC methylation can reduce the potential for aberrant DNA methylation to negatively impact iPSC safety or quality in clinical and research settings.
In contrast, improved reprograming techniques cannot reduce the burden of mutations derived from somatic cells. Encouragingly, we saw no evidence for positive selection of somatic mutations in iPSCs and observed a large degree of heterogeneity in the somatic mutations identified between lines derived from the same individual, suggesting that genome sequencing would be an effective means to screen and select iPSC lines least prone to dysfunction. Computational means to assess the pathogenicity of somatic mutations will be important to identify mutations that promote tumorigenicity and dysfunction of the derived tissue. Given our source of donor material (PBMCs), our study could not differentiate between inborn germline mutations and subsequent mutations that underwent positive selection and rose to high abundance, as has been observed in elderly individuals27–29. To avoid this possibility, we suggest that, especially in elderly individuals, a second tissue should be used as a reference for germline mutations.
Somatic mosaicism may increase with age not only through selection of growth-promoting mutations but also through contraction of the cellular population available for reprogramming at advanced age. This has been difficult to assess because bulk sequencing studies cannot identify rare mutations, and single-cell whole-genome sequencing suffers from a high degree of noise due to amplification and loss of individual DNA molecules. Our results show that amplification of the genomes of blood cells from extremely elderly individuals by reprogramming facilitates assessment of the burden of rare mutations. We observed an overall decrease in the number of somatic mutations per line from donors older than ~90. Our observation of the reversion of transition vs transversion rates in extremely elderly individuals, and the age-associated increase in allelic frequency of rare somatic mutations in the overall PBMC population, suggests that a reduction in the progenitor cell population for PBMCs may explain the overall decline in the number of somatic mutations observed in extremely elderly individuals—a phenomena previously observed in a single supercentenarian with only two hematopoietic lineages remaining41. We cannot definitively differentiate between this possibility and some enhancement of DNA repair in long-lived individuals42. However, if such ‘less mutated’ donor cells are present also in donors <90 years old, it would be useful to perhaps identify markers for these cells or use genome sequencing to identify iPSCs derived from them for clinical use.
Our systematic study of the effect of age and advanced age on the genetic and epigenetic dynamics of iPSCs was made possible by our efficient technique for reprogramming PBMCs of elderly individuals. Biomaterials from elderly individuals are not typically used during the development of techniques for cell therapies, although these individuals are most likely to receive clinical benefit. This work highlights some important considerations for clinical translation of reprogramming techniques to the clinic and offers an approach for investigating the impact of age on the epigenetic and genetic features of iPSCs and somatic cells.
Online Methods
Peripheral blood mononuclear cell isolation and culture
Study participants were enrolled and informed consent obtained under study IRB-11-5676 approved by the Scripps Institutional Review Board. PBMC isolation from blood in Heparin vacutainers was performed using Ficoll-Paque Premium (GE Healthcare, cat #17-5442-03). Blood was diluted with 10 ml 1X PBS, layered over the top of 20 ml Ficoll and then centrifuged at 750xg for 35 min with the acceleration at its lowest setting. The white interphase (buffy coat) between the plasma and Ficoll fractions was transferred into 35 ml 1X PBS and centrifuged for 10 min at 350 xg. The cells were then resuspended in 10 ml 1X PBS and centrifuged for 10 min at 250 xg. Cells were resuspended in 10% DMSO in heat-inactivated FBS (Life Technologies, cat # 10082139), frozen at −80C and then transferred to liquid nitrogen for long-term storage.
PBMC reprogramming to IPSCs
iPSCs were generated from 5 young (21, 22, 23, 23, and 29 yrs), 5 middle-aged (49, 57, 62, 69 and 79 yrs), and 6 elderly (86, 87, 92, 94, 95 and 100 yrs) individuals via Yamanaka episomal- based reprogramming of peripheral blood mononuclear cells (PBMCs). We selected normal healthy blood donors based simply on their age. The methods used were optimized from previously described protocols (Chou et al., 2011). Below we describe the complete original protocol from Chou et al including the additional changes we applied. Changes applied in our protocol will be highlighted with underlined text. Isolated and cryopreserved PBMCs were thawed and cultured for 12–14 days in Mononuclear Cell (MNC) complete medium (MNC basal medium (IMDM (Gibco Cat#21056023), Hams F12 (Gibco Cat#31765035), Glutamax (Gibco Cat#35050-061), Chemically Defined Lipid Concentrate (Gibco Cat#11905031), ITS-X (Gibco Cat#51500-056), 1- Thioglycerol (Sigma, 435uM), Bovine Serum Albumin (Sigma Cat#A9418, 0.5%w/v), and Ascorbic acid (Sigma Cat#A8960, 50ug/mL)) supplemented with rhEPO (R&D 2U/mL), hSCF (R&D 50ng/mL), rhIGF1 (R&D 40ng/mL), hIL3 (R&D 10ng/mL), Dexamethasone (Sigma Cat#D2915 1uM), and Holo-transferrin (R&D 50ug/mL)). Cell counts were performed on Days 2, 5, 8, and 11 (more frequent feeding than originally described), with cells seeded at 2.5×106 cells/mL for days 2–5, and 1.5×106 cells/mL on days 8 and 11. We proceeded with expansion for 14 days, reporting between 1 and 3 fold by D11.(transfection was performed at D8-9 in Chou et al) 2×106 cells were transfected (Amaxa Nucleofector Technology) with plasmids containing the reprogramming transcription factors (pCXLE- hOCT3/4-shp53; pCXLE-hSK; pCXLE-hUL), 2ug each plasmid (Chou and collegue used a different set of reprogramming plasmid pEB-C5 and pEB-Tg from Linzhao Cheng Laboratory). Human CD34+ cells nucleofector kit (Cat#VPA-1003) was used with program U-008 (instead of T-016, to increase transfection efficiency) on an Amaxa Nucleofector II device. Cells were allowed to rest in MNC complete medium for 2 days, then plated on inactivated MEF feeders (3×104 cells/cm2) in MNC basal medium supplemented with 10% FBS (instead of MEF medium), and transitioned to mTeSR1 medium (Stemcell Tech Cat#05850) over the next 3 days (KOSR medium was used in Chou et al). Cultures were fed with fresh mTeSR1 on days 7, 9, and daily thereafter. From Days 10–20, 50% fresh mTeSR was supplemented with 50% fibroblast conditioned mTeSR (inactivated human foreskin fibroblasts (EMD Millipore Cat#SCC057)). Medium were supplemented with the small molecule enhancer of reprogramming sodium butyrate (0.25mM) as nascent colonies became visible with timing varying on a donor-by-donor basis, typically around Day 13. Colonies were allowed to develop until approximately 22 days post-transfection and then picked to feeder-free conditions on Matrigel (Corning Cat#354277), at which time sodium butyrate supplementation was stopped.
Induced pluripotent stem cells culture and characterization
Induced Pluripotent Stem Cells (iPSCs) were cultured in Matrigel-coated plates (Greiner Bio- one Cat#657160) with mTeSR medium. Cells were passaged every three to four days using 0.5mM EDTA as the dissociation reagent. PBMC-derived iPSCs were characterized after approximately 8–9 passages in culture. Karyotyping analysis was performed by Infinium HumanCore BeadChip. Validation of the iPS cells was performed by immunofluorescence staining and flow cytometry. 250,000 fixed cells per sample were incubated at room temp for 30 minutes in Blocking Buffer (5% heat inactivated FBS in 1XPBS, -Ca, -Mg). SSEA4 (Stemgent, Cat#09-0003, use 1:5) and TRA 1–60 (Millipore, Cat# MAB4360, use 1:100) were added to samples individually and incubated for 30 minutes RT. Three washes were performed with 1% heat inactivated FBS in 1XPBS, -Ca, -Mg. After the final wash, the SSEA4-stained samples were resuspended in 500 ul of Wash Buffer and transferred to FACS tubes. The TRA 1–60 samples were then resuspended in Goat anti-mouse-IgM Alexa 488 (Life Technologies, Cat#A21042, use 1:200) secondary, and incubated 30 minutes RT. Three washes were performed using the wash buffer listed above. The cells were resuspended in 500ul wash buffer and transferred to FACS tubes. The samples were read and analyzed on the LSR II. The LookOut® Mycoplasma PCR Detection Kit (Sigma, Cat. # MP0035) was used to check for Mycoplasma contamination, following the manufacturer’s instructions and using JumpStart Taq DNA Polymerase (Sigma, Cat #D9307).
Genomic DNA extraction
Cell pellets of about 1*106 cells were lysed in 250ul (1 volume) of Lysis Buffer (100mM Tris HCl pH 8; 5 mM EDTA, 200 mM NaCl and 0.2% SDS) with Proteinase K by gently pipetting and incubating at 55 C for approximately 1 hour. RNase A treatment was performed by incubating the lysates for 15 minutes at RT with 4ul of RNase A (100mg/ml Qiagen). One volume of Phenol/Chloroform/Isoamyl alcohol was added and after gentle shaking the samples were centrifuged at 16,000 xg for 20 min at 4 C. The aqueous phase was collected, added to 1/10 volume of 3M sodium acetate, and precipitated by adding 2 volumes of 100% ethanol. Samples were centrifuged for 15 minutes at 16,000 xg at 4 C. The DNA pellet was washed with 1mL of 70% ethanol and centrifuged at 4 C for 10 min. Supernatant was aspirated and the dried pellets were resuspended in 50–200ul TE (pH 8.0, Ambion) by incubating the samples at 50C for one to two hours. Genomic DNA quantification was performed using Nanodrop (Thermo Scientific Nanodrop 1000 device).
RNA extraction, retrotranscription, qPCR
Total RNA extraction from frozen or fresh cells was performed with the Trizol reagent (Cat#15596) and Zymo Direct-zol RNA mini-prep kit according to manufacturer protocols. RNA was eluted in water and treated with Ambion DNA Free (Cat#AM1906) according to manufacturer protocols. cDNA for quantitative PCR was produced from isolated RNA using iScript cDNA Synthesis Kit (Cat#170-8891) in 20uL reaction volumes, according to manufacturer protocol. qPCR reactions were performed in 10uL final volume in 384-well plates, using iTaq Universal SyBr Green Supermix (Bio-Rad Cat#1725121) for detection on a Bio-Rad CFX384 Touch Real-Time PCR machine. Each reaction was performed using 4 ng of cDNA. Relative mRNA expression was determined by the ΔΔ-Ct method with the average of young donors used as control. Gene expression was normalized using PPIA (peptidylprolyl isomerase A) as reference gene. Primer sequences: PPIA (Fwd: 5′ CCGAGGAAAACCGTGTACTATTAG 3′; Rev: 5′ TGCTGTCTTTGGGACCTTG 3′); TET1 (Fwd: 5′ TTCGTCACTGCCAACCTTAG 3′; Rev: 5′ ATGCCTCTTTCACTGGGTG 3′) TET2 isoform A (Fwd: 5′ GGAGCAGGTCCTAATGTGGC 3′; Rev: 5′ ACATCCCTGAGAACTTTTGCCT 3′); TET2 isoform B (Fwd: 5′ CATGGCGTTTATCCAGAATTAGC 3′; Rev: 5′ TCACAAGACACAAGCATCGG 3′); TET3 (Fwd: 5′ GCCCACAAGGACCAGCATAA 3′; Rev: 5′ CGCAGCGATTGTCTTCCTTG 3′).
Identification of Somatic Mutations
Whole exome sequencing (WES) of donor bulk PBMC populations and 3 iPSC lines per donor was performed utilizing Agilent SureSelect exome hybridization followed by barcoding and sequencing of paired 100bp reads on an Illumina HiSeq2500 instrument (Illumina Inc., San Diego, CA). Somatic mosaicism in donor PBMC populations was performed by amplicon sequencing. Read mapping and initial variant calling and quality filtration was performed using the standard BWA-GATK6, 43 best practices variant quality score recalibration approach. A mean coverage of 150X for PBMCs and 100X for iPSCs was targeted sample with ~95% of the target exome covered by >10. Somatic point mutations were detected using MUTECT 39. Somatic indels were not interrogated given the false positive somatic variant calls. Somatic variants were then annotated using the SG-ADVISER for further filtration44. Somatic variants present in dbSNP or observed at greater than 0.1% frequency in the HapMap45, 1,000 Genomes46, NHLBI GO Exome Sequencing Project47, or 500 genomes from the Scripps Wellderly population, as well as variants in regions known to be prone to false positive somatic calls; segmental duplications, pseudogenes and other repetitive regions were removed. Additional filters were applied to eliminate false positive somatic variant calls, somatic variants must have: 1) alternate allele mapping quality greater than 30, 2) no overlapping indels, 3) at least 50X coverage in the normal PBMC sample, and 4) between 30%–60% population frequency in the iPSC sample. The final filter was necessary to remove systematic errors in variant calling evident by recurrent “mutations” appearing in multiple lines across individuals or observed in somatically re-arranged antibody genes.
Methylation Analysis
PBMC and iPSC methylation was determined via the Infinium Human Methylation 450k array (Illumina Inc., San Diego). Methylation analysis was conducted on the 21, 22, 23 (1), 23 (2), 29, 49, 57, 62, 86, 87, and 100 year old individuals and processed via a pipeline based on Touleimat and Tost 201248. Probes known to overlap with known SNP or CNV sites in any 1000 Genomes population, known to have non-optimal binding, or mapping to sex chromosomes or multiple sites in the genome were removed. Methylation values were determined using the lumi R package49, where color bias was adjusted via simple shift and scaling normalization, background adjusted separately per color, and cross array normalization was performed using via simple shift and scaling normalization. Resultant M-values values were smoothed with a 250bp window size50. The mean overall signal in young iPSC samples was −0.32 vs. −0.34 in iPSC samples from elderly individuals. This difference is non-significant, thus no technical bias exists and, if anything, the trend in bias is towards decreased methylation in iPSCs from elderly individuals – i.e. in the opposite direction of the demethylation defects we observe. Biological age was determined using R code produced by Horvath18.
Statistical Analysis
Linear mixed effects models were utilized to account for repeated measures (i.e. multiple iPSC lines) per donor. In analyses assessing the relationship between donor age on genomic outcomes (predicted iPSC and number of somatic mutations), the age of the donor acts as the fixed effect and the source donor acts as the random effect (e.g. the subject-specific effect). Regression is performed via the generic lme function from the nlme R package – with random intercepts and normality assumed and homogenous variance not assumed. Appropriateness of the normality assumption was confirmed by the distribution of observed residuals.
Differential methylation analysis was conducted similarly utilizing the limma R package with the source donor encoded as a blocking factor and the random effect fit for this blocking factor via the intra-subject correlation between iPSC samples derived from the same source donor51. An empirical Bayes adjustment is applied to moderate the resultant statistics and the Benjamini & Hochberg method is applied for multiple testing correction. All reported p-values reflect the corrected values. Statistical analysis for Figure 1E, Supplementary Figure 3b and 3c was performed using GraphPad Prism software.
Supplementary Material
Acknowledgments
This work is supported by a Scripps Translational Science Institute pilot award (5 UL1 TR001114) from Scripps Genomic Medicine, a NIH-NCATS Clinical and Translational Science Award (CTSA; 5 UL1 RR025774) to STSI. Further support is provided by the NextGen Consortium NHLBI 5 U01 HL107436 to STSI and TSRI (E.J.T. and K.K.B), and U54GM114833 to STSI (A.T.).
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
Author Contributions
Conceptualization, K.K.B., A.T., E.J.T; Methodology, K.K.B., V.L.S., A.T.; Formal Analysis, A.T., G.A.E.; Investigation, V.L.S., W.F., A.T.; Resources, A.T., K.K.B., E.J.T.; Writing - Original Draft, A.T., K.K.B.; Writing - Review and Editing, K.K.B., V.L.S., W.F., A.T., E.J.T.; Visualization, V.L.S., K.K.B., A.T.; Supervision, A.T., K.K.B., E.J.T; Project Administration, A.T., K.K.B., E.J.T.; Funding Acquisition, A.T., K.K.B., E.J.T
References
- 1.Barker RA, Drouin-Ouellet J, Parmar M. Cell-based therapies for Parkinson disease-past insights and future potential. Nature reviews Neurology. 2015;11:492–503. doi: 10.1038/nrneurol.2015.123. [DOI] [PubMed] [Google Scholar]
- 2.Keirstead HS, et al. Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants remyelinate and restore locomotion after spinal cord injury. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:4694–4705. doi: 10.1523/JNEUROSCI.0311-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz SD, et al. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379:713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 4.Daley GQ. The promise and perils of stem cell therapeutics. Cell stem cell. 2012;10:740–749. doi: 10.1016/j.stem.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz SD, et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet. 2015;385:509–516. doi: 10.1016/S0140-6736(14)61376-3. [DOI] [PubMed] [Google Scholar]
- 6.Amariglio N, et al. Donor-derived brain tumor following neural stem cell transplantation in an ataxia telangiectasia patient. PLoS medicine. 2009;6:e1000029. doi: 10.1371/journal.pmed.1000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nature reviews Cancer. 2011;11:268–277. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 8.Kamao H, et al. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem cell reports. 2014;2:205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coghlan A. Mutation alert halts stem-cell trial to cure blindness. New Scientist. 2015 [Google Scholar]
- 10.Polo JM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohi Y, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell stem cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Kim K, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiler D, et al. Quantification of Retinogenesis in 3D Cultures Reveals Epigenetic Memory and Higher Efficiency in iPSCs Derived from Rod Photoreceptors. Cell stem cell. 2015;17:101–115. doi: 10.1016/j.stem.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S. DNA methylation age of human tissues and cell types. Genome biology. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyn H, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji J, et al. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–440. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- 22.Laurent LC, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell stem cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugiura M, et al. Induced pluripotent stem cell generation-associated point mutations arise during the initial stages of the conversion of these cells. Stem cell reports. 2014;2:52–63. doi: 10.1016/j.stemcr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng L, et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell stem cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan AR, et al. Genome sequencing of mouse induced pluripotent stem cells reveals retroelement stability and infrequent DNA rearrangement during reprogramming. Cell stem cell. 2011;9:366–373. doi: 10.1016/j.stem.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young MA, et al. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell stem cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs KB, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurie CC, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biesecker LG, Spinner NB. A genomic view of mosaicism and human disease. Nat Rev Genet. 2013;14:307–320. doi: 10.1038/nrg3424. [DOI] [PubMed] [Google Scholar]
- 30.Evrony GD, Lee E, Park PJ, Walsh CA. Resolving rates of mutation in the brain using single-neuron genomics. Elife. 2016;5 doi: 10.7554/eLife.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lodato MA, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350:94–98. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazen JL, et al. The Complete Genome Sequences, Unique Mutational Spectra, and Developmental Potency of Adult Neurons Revealed by Cloning. Neuron. 2016;89:1223–1236. doi: 10.1016/j.neuron.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abyzov A, et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahmoudi S, Brunet A. Aging and reprogramming: a two-way street. Curr Opin Cell Biol. 2012;24:744–756. doi: 10.1016/j.ceb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okita K, et al. A more efficient method to generate integration-free human iPS cells. Nature methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 36.Chou BK, et al. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21:518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roadmap Epigenomics C et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross AM, et al. Methylome-wide Analysis of Chronic HIV Infection Reveals Five-Year Increase in Biological Age and Epigenetic Targeting of HLA. Molecular cell. 2016;62:157–168. doi: 10.1016/j.molcel.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holstege H, et al. Somatic mutations found in the healthy blood compartment of a 115- yr-old woman demonstrate oligoclonal hematopoiesis. Genome Res. 2014;24:733–742. doi: 10.1101/gr.162131.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willcox DC, Willcox BJ, Hsueh WC, Suzuki M. Genetic determinants of exceptional human longevity: insights from the Okinawa Centenarian Study. Age (Dordr) 2006;28:313–332. doi: 10.1007/s11357-006-9020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DePristo MA, et al. A framework for variation discovery and genotyping using next- generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pham PH, Shipman WJ, Erikson GA, Schork NJ, Torkamani A. Scripps Genome ADVISER: Annotation and Distributed Variant Interpretation SERver. PLoS One. 2015;10:e0116815. doi: 10.1371/journal.pone.0116815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genomes Project C et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Touleimat N, Tost J. Complete pipeline for Infinium((R)) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 49.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 50.Du P, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.