Abstract
To achieve accurate spatiotemporal patterns of gene expression, RNA-binding proteins (RBPs) guide nuclear processing, intracellular trafficking, and local translation of target mRNAs. In neurons, RBPs direct transport of target mRNAs to sites of translation in remote axons and dendrites. However, it is not known whether an individual RBP coordinately regulates multiple mRNAs within these morphologically complex cells. Here we identify SFPQ (Splicing factor, proline-glutamine rich) as an RBP that binds and regulates multiple mRNAs in dorsal root ganglion (DRG) sensory neurons and thereby promotes neurotrophin-dependent axonal viability. SFPQ acts within nuclei, cytoplasm and axons to regulate multiple functionally related mRNAs essential for axon survival. Notably SFPQ is required for co-assembly of laminb2 and bclw within RNA granules and for axonal trafficking of these mRNAs. Together these data demonstrate that SFPQ orchestrates spatial gene expression of a newly identified RNA regulon essential for axonal viability.
Regulated intracellular trafficking of mRNAs and of ribosomal machinery allows localized translation, and so provides a mechanism for spatiotemporal control of protein expression. Precise mRNA localization is mediated by an array of RNA-binding proteins (RBPs)1. Individual RBPs interact with newly transcribed mRNAs within the nucleus and enable nuclear export2. Subsequently, mRNAs destined for remote subcellular locations are assembled together with interacting RBPs into RNA granules, which are transported by motor proteins along microtubules to sites of local translation1,2. Intriguingly an individual RBP can bind multiple functionally related mRNAs and also coordinate sequential steps of mRNA processing3,4. In this way, RBPs can synchronize post-transcriptional expression of a set of related mRNAs as an RNA regulon3,4. RNA regulons may be particularly important in highly polarized neurons, where localized mRNAs are translated in axons and dendrites at great distances from the nucleus5,6.
The DBHS (Drosophila Behavior Human Splicing) family member SFPQ7 is a multi-functional RBP with roles in RNA transcription, splicing and 3′ end processing8–12. SFPQ is highly expressed in the developing and mature nervous system and is required for neuronal survival and normal brain development13,14. Within neurons, SFPQ is a critical component of cytoplasmic RNA transport granules in dendrites15,16. However, it is not known whether SFPQ is required for post-transcriptional regulation of mRNAs that are localized to axons. Recent studies have characterized preferred SFPQ binding motifs, providing a resource for identifying mRNAs that might interact with SFPQ17. Here we demonstrate that SFPQ binds and regulates multiple mRNAs that are localized to the axons of DRG sensory neurons and encode proteins that promote axon survival. We show that SFPQ is required for colocalization of laminb2 and bclw mRNA within cytoplasmic RNA granules and for coordinated axonal trafficking of these mRNAs. Together these studies identify an SFPQ-dependent RNA regulon that coordinates the localization of mRNAs to promote axon survival.
RESULTS
SFPQ binds axonal mRNAs
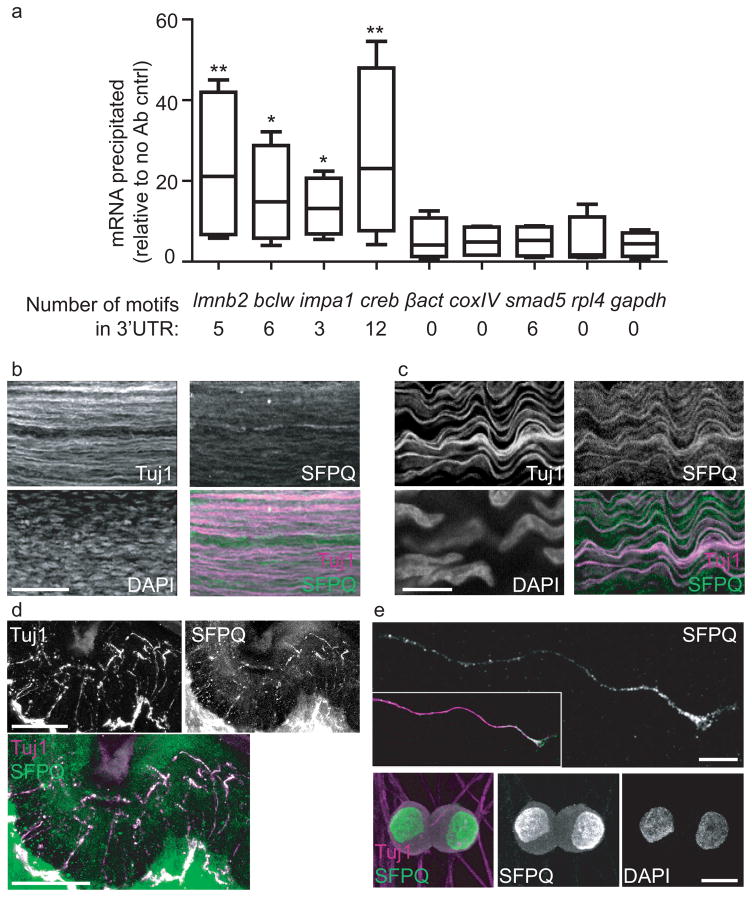
To determine whether SFPQ coordinates spatial regulation of mRNAs in neurons, we searched for putative SFPQ binding motifs17 within mRNAs that have been demonstrated to localize to axons18. We find numerous SFPQ binding motifs within axonal mRNAs encoding proteins that promote neurotrophin-dependent neuronal survival, including laminb219, bclw20, impa121, and creb22 (Fig. 1a and Supplementary Fig. 1a). These motifs are enriched within the 3′UTRs, which are often critical for proper localization of mRNA1,2 (Fig. 1a and Supplementary Fig. 1a). We tested whether SFPQ binds these functionally related mRNAs by formaldehyde crosslinking and RNA-immunoprecipitation (Supplementary Fig. 1b). Endogenous SFPQ co-precipitates with laminb2, bclw, impa1 and creb mRNA, but not with other functionally distinct axonal mRNAs (Fig. 1a). Furthermore, binding of SFPQ to laminb2, bclw, impa1 and creb is regulated by stimulation with high levels of NGF (Supplementary Fig. 1c). These data demonstrate that SFPQ binds multiple axonal mRNAs that contain SFPQ motifs and are involved in promoting neurotrophin-dependent survival.
Figure 1. SFPQ binds axonal mRNAs.
(a) qRT-PCR analysis of SFPQ-precipitated mRNAs from Trk-PC12 cells following formaldehyde crosslinking. Number of SFPQ-binding motifs present within the 3′UTR of each mRNA is shown in the black box. Data, normalized to no antibody control, shows mean ± SEM, n = 4 crosslinking experiments; **P < 0.005, *P < 0.05 (Z-test; P = 0.00073 for laminb2, P = 0.013 for bclw, P = 0.036 for impa1, P = 0.00016 for creb, P = 0.26 for β-actin, P = 0.28 for cox4, P = 0.25 for smad, P = 0.27 for rpl4, P = 0.32 for gapdh). (b, c) Representative SFPQ immunostaining of wholemount P0 peripheral nerve from the fourth lumbar (L4) DRG (n = 3 individual wholemount DRGs). Scale bar, 100 μm (b) and 10 μm (c). (d) Representative SFPQ immunostaining of P0 footpads with Tuj1 showing axons innervating the skin (n = 15 slices from 4 individual footpads). Scale bar, 25 μm. (e) Representative SFPQ immunostaining of cultured sensory neurons in the distal axon and growth cone, and cell bodies and nuclei (n = 3 individual neuronal cultures). Scale bar, 10 μm.
SFPQ promotes axonal localization of mRNAs
We examined the subcellular localization of SFPQ in DRG sensory neurons, as these cells extend long peripheral axons and depend on target-derived neurotrophins for survival23. We detect SFPQ in the peripheral axons that extend towards neurotrophin-producing target tissues (Fig. 1b, c) and in the nerve endings that innervate the skin (Fig. 1d). As an RBP with roles in nuclear RNA processing, SFPQ is also present in nuclei as well as cytoplasm of sensory neurons (Supplementary Fig. 1d). A similar pattern of expression is observed in cultured sensory neurons, where SFPQ localizes to distal axons and growth cones, nuclei and surrounding cytoplasm (Fig. 1e and Supplementary Fig. 1e, f, g). The presence of SFPQ in axons of sensory neurons both in vivo and in vitro suggests a potential role for SFPQ in localization of mRNAs to axons. To test this, we used sensory neurons in compartmented cultures that recapitulate the separation of cell bodies and distal axons observed in vivo. In this system, neurotrophin stimulation of distal axons leads to an increase in laminb2, bclw, impa1, creb, and β-actin mRNA levels in axons (Fig. 2a). We find that shRNA-mediated knockdown of SFPQ (Fig. 2e, Supplementary Fig. 2a) inhibits neurotrophin-dependent increases in multiple axonal mRNAs that are bound by SFPQ (Fig. 2a). Importantly, knockdown of SFPQ does not alter the neurotrophin-dependent increase in axonal β-actin mRNA24, which is not bound by SFPQ (Fig. 2a). Collectively these data demonstrate that SFPQ is required for neurotrophin-dependent axonal localization of multiple functionally related genes.
Figure 2. SFPQ regulates localization of axonal mRNAs.
(a) qRT-PCR analysis of mRNA levels of distal axons from sensory neurons grown in compartmented cultures expressing SFPQ shRNA following 2h neurotrophin stimulation of distal axons (DA). Data shows mean ± SEM, n = 8 experiments from cultures grown from 8 pregnant rats; **P < 0.005, *P < 0.05 (Unpaired two-tailed t-test between shCntrl and shSFPQ; P = 0.0076, t(16) = 3.05 for laminb2, P = 0.0014, t(16) = 3.85 for bclw, P = 0.017, t(12) = 2.78 for impa1, P = 0.13, t(14) = 1.64 for creb, P = 0.55, t(14) = 0.62 for β-actin, P = 0.72, t(14) = 0.36 for cox4, P = 0.6, t(14) = 0.54 for smad and P = 0.33, t(14) = 1.01 for rpl4). (b) Representative single-molecule FISH for laminb2, bclw and β-actin mRNA in DA from sensory neurons grown in microfluidic cultures (n = 3 individual microfluidic cultures). Scale bar, 10 μm. (c) Number of single-molecule FISH mRNA puncta per μm2 in DA following 2h neurotrophin stimulation of DA. Data shows mean ± SEM, n = 3 individual microfluidic cultures; *P < 0.05 (Unpaired two-tailed t-test between Cn and NT; P = 0.012, t(79) = 2.57 for laminb2 and P < 0.0001, t(79) = 5.79 for bclw). (d) Number of single-molecule FISH mRNA puncta per μm2 in DA of sensory neurons expressing SFPQ shRNA. Data shows mean ± SEM, n = 3 individual microfluidic cultures; *P < 0.05 (Unpaired two-tailed t-test between shCntrl and shSFPQ; P = 0.0014, t(68) = 3.32 for laminb2 and P < 0.0001, t(68) = 5.99 for bclw). (e) Western blot of cultured sensory neurons expressing control or SFPQ shRNA with SFPQ protein quantification; data shows mean ± SEM, n = 3 experiments from cultures grown from 3 pregnant rats; *P = 0.01, t(4) = 4.26 (Unpaired two-tailed t-test). Full-length blots are presented in Supplementary Figure 7.
Both laminb2 and bclw mRNA are bound by SFPQ and loss of either component leads to a similar phenotype of selective axon degeneration in vivo19,25. Furthermore, while LaminB2 was initially identified as a nuclear intermediate filament, and Bclw was discovered as an anti-apoptotic Bcl2 family member, both laminb2 and bclw are locally translated within axons, where the resultant protein products localize to mitochondria19,20. Therefore, in the following experiments, we focused on these two functionally related axonal mRNAs. To examine the role of SFPQ in regulating neurotrophin-dependent axonal localization of laminb2 and bclw mRNA, we performed single-molecule fluorescent in situ hybridization (FISH) after validating the specificity of laminb2 and bclw probes (Supplementary Fig. 2b, c). Puncta of laminb2 and bclw mRNA are evident in distal axons of neurons grown in microfluidic cultures (Fig. 2b). Consistent with qRT-PCR data, laminb2 and bclw mRNA puncta are less numerous than β-actin puncta, (Fig. 2b and Supplementary Fig. 1b) and the density of laminb2 and bclw mRNA puncta in axons increases with neurotrophin stimulation (Fig. 2c and Supplementary Fig. 2d). Furthermore, axonal laminb2 and bclw mRNA puncta decrease following knockdown of SFPQ, while β-actin mRNA puncta in axons are not affected (Fig. 2d and Supplementary Fig. 2e). Together these results demonstrate a specific role for SFPQ in neurotrophin-dependent axonal localization of laminb2 and bclw mRNA.
Since SFPQ is a component of RNA transport granules13,14, we asked whether impaired localization of laminb2 and bclw mRNA to axons (Fig. 2a) is associated with corresponding changes of mRNA levels within cell bodies. We carried out subcellular fractionation and isolated mRNAs from nuclei, cytoplasm, and distal axons (Fig. 3a, e). Following knockdown of SFPQ (Supplementary Fig. 2a), laminb2 mRNA accumulates in the cytoplasmic fraction (Fig. 3b), suggesting that SFPQ is required for axonal transport of laminb2 mRNA. In contrast, bclw mRNA accumulates in the nuclear fraction following knockdown of SFPQ (Fig. 3c), suggesting SFPQ may be required for both nuclear export and subsequent axonal transport of bclw mRNA. Importantly, subcellular localization of β-actin mRNA is not altered by SFPQ knockdown (Fig. 3d). Together these results indicate that SFPQ functions throughout the neuron to enable axonal trafficking of interacting mRNAs.
Figure 3. SFPQ is required for axonal trafficking of laminb2 and bclw mRNA.

(a) Subcellular fractionation of mRNA from nuclei (Nuc), cytoplasm (Cyt) and distal axons (DA) from sensory neurons grown in compartmented cultures. (b–d) qRT-PCR analysis of mRNA levels from Nuc, Cyt and DA from sensory neurons expressing SFPQ shRNA following 2h neurotrophin stimulation of DA. Data shows mean ± SEM, n = 8 experiments from cultures grown from 8 pregnant rats; *P < 0.05, ***P < 0.01 (two-way ANOVA with Bonferroni correction; (b) P < 0.0001, F(2,44) = 11.46 for laminb2, (c) P < 0.0001, F(2,44) = 14.10 for bclw, (d) P = 0.49, F(2,44) = 0.722 for β-actin). (e) Subcellular fractionation confirmed by PCR of U6 snRNA for nuclear mRNA and S14 ribosomal RNA for cytoplasmic mRNA. Representative image of 3 individual experiments; full length gel shown in Supplementary Figure 6.
SFPQ is required for mRNA coassembly in transport granules
It is not known whether distinct mRNAs that bind the same RBP are co-assembled into an individual RNA transport granule. Electron microscopy studies estimate RNA granules to be 100–250 nm in diameter26. To determine whether laminb2 and bclw mRNA are packaged together, we performed single-molecule FISH and looked for colocalization of laminb2 and bclw mRNA in the cell body, where RNA transport granules are packaged for axonal transport (Fig. 4a). Using super-resolution quantitative colocalization analysis, we observe an enrichment of laminb2 and bclw mRNAs localized within a distance <270 nm from one another (Fig. 4c and Supplementary Fig. 3a). In contrast, this enrichment is not observed for bclw and γ-actin mRNA, a control mRNA that is not localized to axons (Fig. 4b, d and Supplementary Fig. 3a). Together these data suggest that laminb2 and bclw mRNAs can be assembled within a single RNA granule for axonal transport. We used the same approach to analyze localization of laminb2, bclw and γ-actin mRNA following knockdown of SFPQ (Fig. 4d, Supplementary Fig. 3b). Given the variability across cells, we performed a per-cell analysis rather than population-level analysis to determine whether SFPQ is important for colocalization. The percentage of cells in which laminb2 and bclw mRNA puncta are enriched within 270 nm from one another is significantly reduced following knockdown of SFPQ (Fig. 4e). Importantly, the percentage of cells with colocalization of bclw and γ-actin is very low and is not affected by SFPQ knockdown. Intriguingly we also observe occasional colocalization of laminb2 and bclw mRNA puncta in distal axons (Fig. 4f), while these mRNAs never colocalize with β-actin puncta. Together these data indicate that SFPQ promotes colocalization of laminb2 and bclw mRNA and may therefore facilitate co-assembly and co-trafficking of laminb2 and bclw mRNA within individual RNA transport granules that travel into distal axons.
Figure 4. SFPQ is required for co-assembly of bclw and laminb2 mRNA within RNA transport granules.
(a, b) Representative single-molecule FISH for bclw and laminb2 mRNA (a) and bclw and γ-actin (b) of sensory neurons grown in microfluidic cultures, with DAPI (n = 3 individual microfluidic cultures). Scale bar, 5 μm. Zoom-in (white dotted area) shows colocalization of bclw and laminb2 mRNA (white arrow). Scale bar, 1 μm. (c) Super-resolution quantitative colocalization analysis of adjacent bclw and laminb2 mRNAs (blue; n = 131 cells) and adjacent bclw and γ-actin mRNAs (gray; n = 143 cells) within neuronal cell bodies in control conditions from 3 experiments. Normalized density is frequency of adjacent mRNAs at each distance on x-axis normalized to the frequency expected by chance. Shaded region shows 99% bootstrapped confidence intervals and red line is at y=1 (equal to chance) and x=270 nm. Colocalization of laminb2 and bclw mRNAs occurs 2x to 5x more often than would be expected by chance. (d) Super-resolution quantitative colocalization analysis (as shown in c) in neuronal cell bodies expressing SFPQ shRNA; n = 112 cells from 3 experiments. (e) Percentage of cells exhibiting significant colocalization of bclw and laminb2 or bclw and γ-actin within 270 nm in control and shSFPQ conditions; n = 131 cells for Cntrl bclw and laminb2, n = 143 cells for Cntrl bclw and γ-actin, n = 112 cells for shSFPQ bclw and laminb2 or γ-actin from 3 experiments, *P < 0.05 (Fisher’s exact test). Dotted line represents percentage of cells expected to show colocalization by chance. (f) Representative single-molecule FISH for laminb2, bclw and β-actin mRNA in distal axons of sensory neurons grown in microfluidic cultures (n = 3 individual microfluidic cultures). Scale bar, 10 μm. Zoom-in (white dotted area) shows colocalization of laminb2 and bclw mRNA (white arrow), but no colocalization with β-actin mRNA (arrowhead). Scale bar, 2.5 μm.
SFPQ binds critical motifs for axonal transport
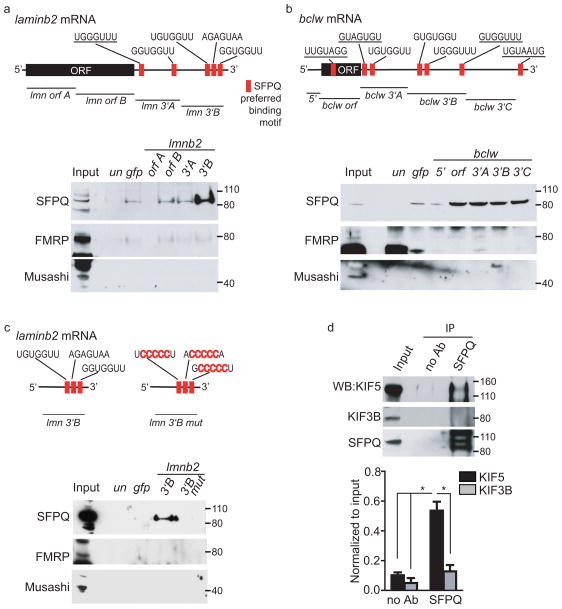
The putative SFPQ binding motifs have not yet been rigorously assessed in biological assays. To validate the putative SFPQ binding motifs within the 3′UTR of laminb2 and bclw mRNA, we used in vitro synthesized, biotinylated regions of laminb2 and bclw mRNAs, or a gfp control, to pull-down interacting RBPs from lysates of sensory neurons (Fig. 5a, b). RNA corresponding to regions of laminb2 and bclw that contain SFPQ motifs19 successfully co-precipitate SFPQ, but not the control neuronal RBPs, Fragile X Mental Retardation Protein (FMRP)27 or Musashi28 (Fig. 5a, b). These data indicate that SFPQ binds motifs within the 3′UTR of laminb2 and bclw mRNA. To determine whether the putative SFPQ motifs are required for binding, we carried out site-directed mutagenesis. We mutated the core region within each of the three motifs of the laminb2 3′UTR (3′B) (Fig. 5c). The mutant laminb2 3′B mRNA lacking consensus SFPQ sites no longer co-precipitates with SFPQ, indicating that these predicted sites within the 3′UTR are essential for mRNA binding to SFPQ (Fig. 5c). Interestingly, the 3′UTR of bclw is also important for neurotrophin-dependent axonal localization (Supplementary Fig. 4a), indicating that 3′UTR sites may be generally critical for SFPQ-dependent intracellular trafficking.
Figure 5. SFPQ binds motifs within the 3′UTRs of bclw and laminb2.
(a, b) Schematic of rat laminb2 (a) and bclw (b) mRNA with SFPQ binding motifs13. Underlined motifs are conserved among rat, mouse and human. Western blot of proteins eluted from unbiotinylated mRNA (un), biotinylated gfp RNA (gfp), biotinylated 5′UTR (5′), open reading frame (orf) or 3′UTR (3′) mRNA pull-down from sensory neuron protein lysates probed for SFPQ, FMRP and Musashi; data representative of 3 experiments. (c) Schematic of rat laminb2 3′UTR (3′B) with mutated SFPQ binding motifs19. Western blot of proteins eluted from unbiotinylated mRNA (un), biotinylated gfp mRNA (gfp), biotinylated 3′UTR (3′B) or biotinylated mutated 3′UTR (3′B mut) pull-down from sensory neuron protein lysates probed for SFPQ, FMRP and Musashi; data representative of 3 experiments. (d) Western blot following immunoprecipitation with α-SFPQ from sensory neuron protein lysate. Quantification of pull-down relative to input; data shows mean ± SEM, n = 3 experiments from cultures grown from 3 pregnant rats; *P = 0.0001, F(3,8) = 28.9 to noAb or KIF3B pull-down (one-way ANOVA with Bonferroni correction). Full-length blots are presented in Supplementary Figure 7.
Axonal transport of RNA granules containing RBPs and associated mRNAs is mediated by microtubule-dependent molecular motors13,29,30. In sensory neurons, SFPQ co-precipitates with the kinesin KIF5, but not KIF3B, suggesting this motor protein transports RNA granules containing SFPQ, laminb2 and bclw along microtubules of sensory axons (Fig. 5d).
SFPQ is required for axonal survival
As laminb2 and bclw mRNAs are transported to axons where they are locally translated19,20, we asked whether SFPQ also regulates axonal levels of LaminB2 and Bclw protein. We used shRNA to knockdown SFPQ in compartmented cultures and find that SFPQ is required for neurotrophin-dependent increases of LaminB2 and Bclw protein levels in axons (Fig. 6a). Within axons of sensory neurons, SFPQ localizes near mitochondria and ribosomes (Fig. 6b). Thus SFPQ may deliver laminb2 and bclw mRNA to sites of local translation adjacent to mitochondria where their protein products function to promote axon viability31. To determine whether SFPQ is therefore required for axon survival, we used compartmented cultures wherein survival of sensory axons depends on local application of neurotrophins (Fig. 6c)20,32. We find that acute knockdown of SFPQ specifically abrogates neurotrophin-dependent axonal survival (Fig. 6c, d and Supplementary Fig. 5a). These findings are consistent with in vivo data showing that mutations in SFPQ lead to neuronal apoptosis and axonal degeneration16. Similarly, loss of LaminB219 (Fig. 6c, d and Supplementary Fig. 5b) or Bclw20,25, like SFPQ, each result in axonal degeneration suggesting that they function in a common pathway. Introduction of recombinant Bclw protein into axons rescues axon degeneration due to knockdown of SFPQ or LaminB2, indicating that Bclw acts downstream of SFPQ to promote axon health (Fig. 6d and Supplementary Fig. 5c). Together these data suggest that SFPQ-dependent regulation of Bclw and LaminB2 protein levels in axons contributes to axon viability.
Figure 6. SFPQ regulates functionally related genes to promote axonal viability.
(a) Western blot of protein from distal axon (DA) lysate from sensory neurons grown in compartmented cultures expressing control or SFPQ shRNA following 8h neurotrophin stimulation of DA. Quantification of LaminB2 and Bclw protein levels normalized to GAPDH; data shows mean ± SEM, n = 3; *P ≤ 0.05 (Paired two-tailed t-test; P = 0.05, t(2) = 2.84 for LaminB2 and P = 0.04, t(2) =3.29 for Bclw). Full-length blots are presented in Supplementary Figure 7. (b) Representative immunostaining of sensory neuron axons with SFPQ, RPL17 and mitotracker (n = 3 individual neuronal cultures). Scale bar, 10 μm. (c) Representative binarized Tuj1-labeled axons in compartmented cultures expressing control (n = 56 axon tracts), SFPQ (n = 24 axon tracts) or LaminB2 (n = 32 axon tracts) shRNA in the absence (-NT) or presence (+NT) of neurotrophins (NGF+BDNF) from 3 individual experiments. Scale bar, 40 μm. (d) Quantification of axon degeneration; data shows mean values ± SEM, n = 25 axon tracts for shSFPQ+Bclw and n = 16 axon tracts for shLmnB2+Bclw from 3 experiments; *P < 0.0001, F(9,301) = 10.55 (one-way ANOVA with Bonferroni correction). Following introduction of Bclw protein to DA; P < 0.0001, F(9,301) = 10.55 by one-way ANOVA with Dunnett’s multiple comparison test to shCntrl -NT condition.
DISCUSSION
It has become evident that the coordination of gene expression relies heavily on post-transcriptional events regulated by RBPs. The RNA regulon model proposes that an individual RBP propels multiple functionally related mRNAs along a coordinated pathway of RNA processing, from splicing to nuclear export, stability, and translation3,4. The best evidence for this type of coordinated regulation comes from yeast; where the RBP Puf3p interacts selectively with nuclear-encoded mRNAs for mitochondrial proteins that localize to mitochondria for translation33,34. In the brain, the splicing factor Nova binds a subset of mRNAs that encode protein with synaptic function35, suggesting that RNA regulons may also be important for neuronal functions. Here we identify an SFPQ-dependent regulon critical for axonal viability, with SFPQ coordinating nuclear export and axonal transport of functionally related target mRNAs (Fig. 6e). Our studies demonstrate that SFPQ is required for neurotrophin-dependent axon survival through direct regulation of multiple mRNAs, including laminb2 and bclw mRNA.
Previous studies using single-molecule FISH have not detected colocalization of distinct mRNAs within neuronal dendrites. These studies suggested that different mRNAs are transported individually36. Our data demonstrates colocalization of laminb2 and bclw within cell bodies and axons, indicating that distinct axonal mRNAs can be transported within single RNA granules. By nearest neighbor analysis, we find 8.8% of laminb2 and bclw puncta colocalize. As Bclw and LaminB2 protein are expressed and functional within cell bodies as well as within axons, this number may reflect the fraction of mRNA transported to axons for local translation. Further work will be needed to define the complete composition of SFPQ-containing mRNA granules, and how they are directed along the post-transcriptional pathway.
Since SFPQ functions as a multimer37, numerous SFPQ-binding sites along mRNAs may be required for assembly of SFPQ-containing RNA granules. We find that mRNAs bound and regulated by SFPQ contain multiple SFPQ-binding motifs, suggesting that more than one site may be required for efficient mRNA localization. We show that mutagenesis of multiple motifs blocks binding of SFPQ to a target mRNA. The binding motifs of RBPs are still being defined and investigated functionally, thus additional mutagenesis studies will be required to establish the precise nature and number of motifs required for RNA binding and axonal regulation.
Our studies demonstrate that SFPQ functions in mRNA localization, and that SFPQ is required for neurotrophin-dependent regulation of LaminB2 and Bclw protein in axons. Decreases in axonal LaminB2 and Bclw proteins levels may be due to a reduction in the pool of localized mRNAs available for local translation following loss of SFPQ. However, data that NGF regulates SFPQ binding to laminb2 and bclw mRNA (Supplementary Fig. 1c) suggests SFPQ may also control stimulus-induced translation of target mRNAs, as release of mRNA from RBPs often enables translation initiation38. Phosphorylation of SFPQ can modulate binding to mRNAs39, providing a potential mechanism by which target-derived neurotrophins locally regulate the fate of mRNA once they reach the axon.
Together, the findings presented here demonstrate that SFPQ coordinately regulates mRNAs that are locally translated and promotes neurotrophin-dependent axon viability: together these studies define an RNA regulon for axon survival. Changes in SFPQ activity, expression or localization have been implicated in Alzheimer’s disease, dyslexia and bipolar disorder40–42, suggesting that this newly defined SFPQ RNA regulon may be critical for diverse developmental and degenerative disorders.
METHODS
SFPQ binding motif analysis
bclw: UUGUAGG, GUAGUGU, UGUGGUU, GUGUGGU, UGGGUUU, GUGGUUU, UGUAAUG; laminb2: UGGGUUU, GGUGGUU, UGUGGUU, AGAGUAA, GGUGGUU; creb: UUGUAAA, GU/GUGGU/GU, GGUGGUU, G/UGGUGU/U, UGGUGUU, UGGGUUU, G/UGGUGU/U, UUGUAAA, AGAGUAA, AGAGUAA, UGGUGUU, U/UGGGUU/U; impa1: UUGUAAA, UUGUAGG, G/UGGUGU/U, UUGUAAA; par-3: UGUUGGU, UUGUAAA; smad5: UGGUGUU, UGGGUUU, UUGUAAA, UGGUGUU, UGUAAUG, UUGUAAA, UGUUGGU, GUAAUGG; smad1: UGUUGGU, UUGUAAA, UGGGUUU, UGGGUUU; rhoA: UGUUGGU; rpl4: GGUGGUU, AGAGUAA, GGUGGUU; stat3: UGGUGUU, UGGGUUU, UGUGGUU, UGUGGUU, UGGGUUU; importinβ: UUGUAAA, U/GUAAUG/C; nf-l: UGUUGGU. Underlined motifs are conserved between rat, mouse and human,/represents overlapping motifs.
Formaldehyde crosslinking
Trk PC1243 cells were differentiated in low serum media and 50 ng/mL NGF for 24 hours before starvation in DMEM alone for 2 hours, followed by NGF (100 ng/mL) stimulation or vehicle control (BSA 100 ng/mL) for 2 hours. Cells were collected in HBSS, before crosslinking with 1% wt/vol formaldehyde (AR grade, 37% wt/wt) in PBS for 10 minutes at RT. Crosslinking reaction was stopped with 1 M glycine to a final concentration of 0.25 M glycine for 5 minutes at RT and washed 2 times with ice-cold PBS. Cells were resuspended in RIPA buffer (50 mM Tris-HCl pH 7.4, 1% NP-40, 0.5% Na-deoxycholate, 0.05% SDS, 1 mM EDTA, 150 mM NaCl, protease inhibitors, RNasin inhibitors), and sonicated 5 times using a 3 second burst. Lysate was centrifuged at 13,000 rpm to remove insoluble material and incubated overnight in antibody (αSFPQ 1:100, abcam) or no-antibody control at 4°C. Beads (A/G plus-agarose, Pierce) were added for 4 hours at 4°C and collected by centrifugation at 2500 rpm and washed 4 times with RIPA buffer. Beads were resuspended in 50 mM Tris-HCl pH7, 5 mM EDTA, 10 mM DTT, 1% SDS and incubated at 70°C for 45 minutes to reverse crosslinks. Immunoprecipitated RNAs were isolated with Trizol (Invitrogen) and analyzed by qRT-PCR.
qRT-PCR
Dorsal root ganglia (DRG) sensory neuron RNA from a minimum of four compartmented cultures was isolated with Trizol (Invitrogen). Reverse transcription was performed using the cDNA archive kit (Applied Biosystems) according to the manufacturer’s specifications. qRT-PCR was performed using TaqMan gene expression assays (Applied Biosystems) to assess the RNA levels of rat bclw (Rn00821025_g1), laminB2 (Rn01408653_g1), β-actin (Rn00667869_m1), impa1 (Rn00583189_m1), creb (Rn00578829_g1), coxIV (Rn00665001_g1), smad5 (Rn00572484_m1), rpl4 (Rn00821091_g1) and sfpq (Rn01179807_m1). For each sample, average ct values were normalized to the level of rat gapdh (Applied Biosystems).
Animal Use
All experimental procedures were done in accordance with the National Institutes of Health guidelines and were approved by the Dana-Farber Cancer Institutional Animal Care and Use Committee. Timed-pregnant Sprague-Dawley rats were purchased from Charles River. Wild-type 129/SvJae mice were purchased from The Jackson Laboratory. bclw−/− mice on a C57BL/6J genetic background were a generous gift from Grant MacGregor44. Animals are kept in a 12:12 hour light/dark cycle and housed with 2 rats or 2–3 mice per cage.
Whole-mount immunostaining
Whole DRG with peripheral nerves were dissected from P0 rats of either sex and fixed overnight in 4% paraformaldehyde. DRGs were washed in PBS, permeabilized in 0.5% Triton X-100 for 1 hour and blocked in 5% BSA and 0.5% Triton X-100 for 4 hours. DRGs were incubated for 48 hours in primary antibody at 4°C and washed overnight in PBS. DRGs were then incubated in secondary antibody (Alexa-Fluor; 1:1000) at RT for 2 hours and counterstained with DAPI (1:1000). Antibodies used are listed in Supplementary Table 1. Images were acquired using Nikon C2 Si laser-scanning confocal microscope with 20x air and 60x oil objective.
Footpad immunostaining
P14 C57Bl6/J WT mice were killed with isoflurane, and then footpad tissue from hindpaws was removed, fixed overnight in Zamboni’s fixative at 4°C. Footpads were frozen, and 30 μm sections were prepared. Tissue sections were blocked in 10% normal goat serum and 0.1% Triton X-100 in PBS for 1 hour at room temperature and then incubated in primary antibody overnight at 4°C. Sections were then incubated in secondary antibody (Alexa-Fluor; 1:1000) for 2 hours at room temperature, counterstained with DAPI (1:1000) and mounted on gelatin-coated slides. Antibodies used are listed in Supplementary Table 1. Images were acquired using Nikon C2 Si laser-scanning confocal microscope with 60x oil objective.
Neuronal cultures
DRGs were dissected from embryonic day 15 (E15) rats and dissociated in trypsin. Cultures were maintained in Neurobasal media with 2% B-27, 1% glutamax, 1% penicillin-streptomycin, 0.08% glucose, 0.3 μM AraC, and neurotrophins (NGF+BDNF). Compartmented (Campenot) cultures were prepared as previously described45 with 120,000 neurons plated in the center of a Teflon divider (Camp10; Tyler Research) attached to a P35 tissue culture plate coated with matrigel (Invitrogen). NGF+BDNF were added to the cell body compartment at a concentration of 10 ng/ml and to the axon compartment at a concentration of 100 ng/ml. Microfluidic cultures (Xona Microfluidics) were prepared as previously described46, with 100,000 neurons plated into the channel of the microfluidic chamber attached to cover glass coated with poly-M-lysine/laminin. NGF+BDNF were added to the cell body compartment at a concentration of 10 ng/ml and to the axon compartment at a concentration of 100 ng/ml. Mass cultures were grown on matrigel-coated (Invitrogen) P35 tissue culture plates in 100 ng/ml NGF+BDNF. At 8 DIV neurons were starved of neurotrophins for 2 hours in Neurobasal media, followed by stimulation of axon compartments with NGF+BDNF (100 ng/ml) or BSA (100 ng/ml) vehicle control for 2 hours. Compartmented cultures were selected for experiments based on whether axons had successfully grown through the divider with no cell body leakage into the axonal compartment. Each week, cultures were randomly assigned to conditions for each experiment. Experiments that showed no response to neurotrophins, as measured by cfos induction, were excluded (this occurred in <2% of experiments). Due to high variability among compartmented cultures, experiments were typically repeated 5–8 times providing a sample size similar to those reported in previous publications47.
Immunocytochemistry
Neuronal cultures were fixed with 4% paraformaldehyde, permeabilized in 0.5% Triton 100-X, and blocked in 3% BSA and 0.1% Triton 100-X. Cultures were incubated overnight at 4°C in primary antibody followed by incubation in secondary antibody (AlexaFluor, 1:1000) for 1 hour at room temperature, and counterstained with DAPI (1:1000). Antibodies used are listed in Supplementary Table 1. Images were acquired using Nikon C2 Si laser-scanning confocal microscope with 40x oil objective.
Lentiviral shRNA knockdown
Lentiviral shRNA constructs were obtained from the Broad institute RNAi Consortium (TRC). Virus conditioned media was collected from 293T packaging cells transfected using Fugene6 reagent (Promega) in DMEM with 10% fetal bovine serum. E15 rat DRG sensory neurons were grown in compartmented cultures for 1 DIV, infected with concentrated shRNA lentivirus in the cell body compartment for 24 hours, and then selected with 0.5 μg/mL puromycin and cultured for a further 7 DIV before RNA or protein collection.
Western Blotting
DRG sensory neuron protein lysate from a minimum of six compartmented cultures was collected in lysis buffer (20 mM Tris-HCl pH 7.4, 140 mM NaCl, 10% glycerol, 1% Triton X-100) with protease inhibitor. Lysates were separated by 4–12% Bis-Tris SDS-PAGE and protein transferred to PVDF membrane and blotted with primary antibodies. Bands were visualized with secondary antibodies conjugated to HRP and SuperSignal chemiluminescent substrate kit. Antibodies used are listed in Supplementary Table 1. Protein levels were quantified using National Institutes of Health ImageJ software.
Fluorescent in situ hybridization (FISH)
DRG sensory neurons in microfluidic cultures were fixed in 3.7% formaldehyde, dehydrated, and stored in ethanol at −20°C. Cultures were rehydrated, permeabilized, and RNAscope (Advanced Cell Diagnostics) kit was used to perform FISH with custom probes designed for bclw, laminb2, β-actin, and γ-actin mRNAs. Cells were incubated with protease pre-treatment 3 (1:15) for 10 minutes at 40°C. Probe incubation was performed for 2 hours at 40°C followed by washing and amplification steps according to manufacturer’s protocol. Microfluidic coverslips were washed in beakers with wash buffer ~10 times between steps. Blinded images were acquired using Nikon C2 Si laser-scanning confocal microscope and PlanApo 60x (NA 1.4) oil objective with 2.29 zoom to obtain a pixel size of 0.09 μm.
Axonal FISH analysis
Images of axons were binarized so that axonal areas were converted to black and background areas were converted to white. Axonal area was selected using the threshold Huang function of National Institutes of Health ImageJ software to measure the total axonal area. A blinded researcher manually counted RNA FISH puncta and the mRNA per μm2 was calculated as the ratio of RNA puncta over the total axon area.
Colocalization analysis
Manual per-cell region-of-interest selection was combined with thresholding and morphological post-processing to segment cell areas. Robust point source detection and Gaussian PSF-model fitting were then performed within the cell area to identify puncta with a localization precision ranging from approximately 15–70 nm48. A distance-based colocalization measure was then calculated as described previously49,50. Briefly, closest distances between points in the two image channels were calculated, and a frequency vs. search radius curve generated. This curve was then normalized to the mean frequency of interpoint distances observed in 1000 rounds of randomly generated point positions with the same number of points and within the same cell area. These 1000 rounds of randomization were repeated twice: once holding points from image channel one (bclw) fixed while randomizing channel two (laminb2 or γ-actin), and a second time randomizing channel one while holding channel two fixed. Alternately randomizing the points in the two channels controls for the fact that spatial patterns in the points in one channel can potentially induce spurious indications of colocalization in the other channel. In our data, no difference was seen between these alternate randomizations (Supplementary Fig. 3a). Significance of colocalization at the single-cell level was determined by comparing the measured density vs. distance to the 99th percentile of the density seen in the 1000 randomizations in that cell. For population level analysis, the normalized density curves for each cell were averaged and a 99% confidence interval was then calculated at each distance by 5000 bootstrap repetitions, by sampling from the individual cell curves with replacement. Significance was determined by comparing these confidence intervals to 1, the normalized frequency at any given distance, which would be expected purely by chance. Nearest-neighbor distances were also calculated between points to determine the fraction of points within the 270 nm cutoff from one another.
Code availability
The colocalization code is available upon request from the Image and Data Analysis Core at Harvard Medical School (http://idac.hms.harvard.edu/).
RNA pull-downs
Rat mRNA sequences for bclw, laminb2 and β-actin were amplified by PCR to add a T7 promoter to the 5′ end of the PCR product. DNA bands were purified using QIAquick Gel Extraction Kit (Qiagen) and confirmed by sequencing. The laminb2 mutant cDNA was synthesized (Genewiz) to contain five cysteines within the core of the SFPQ motifs. In vitro transcription with T7 RNA polymerase (Promega) was used to generate RNA from PCR products according to manufacturer’s protocol and was purified using the RNeasy MinElute Cleanup Kit (Qiagen). RNA was biotinylated using the RNA 3′ End Biotinylation Kit (Pierce) and extracted with Trizol (Invitrogen). DRG sensory neuron lysate was collected from mass cultures in polysome extraction buffer (20 mM TrisHCl pH7.5, 100 mM KCl, 5 mM MgCl2 and 0.5% NP-40) with protease inhibitor, RNasin (1:100), 0.2 mM PMSF, 2 mM NaOVa, and 10 mM NaF. Protein lysate was pre-cleared with MyOne streptavidin C1 beads (Invitrogen), yeast tRNA, and in vitro transcribed GFP RNA. GFP, bclw and laminb2 RNAs were conjugated to streptavidin beads in 10 mM Tris-HCl, 1 mM EDTA, and 2 M NaCl. RNA-beads were incubated with protein lysate, washed, eluted by boiling in SDS buffer, and resolved by western blot.
Axon degeneration assay
At 8DIV, DRG sensory neurons in compartmented cultures were maintained in 10 ng/mL NGF+BDNF (cell bodies) and in DMEM alone (distal axons) for 12 hours. Cultures were fixed and stained with mouse anti-Tuj1 (1:400, Covance), goat anti-mouse Alexa-Fluor 488 (1:1000), and counterstained with DAPI (1:1000). Blinded images of distal axons were obtained using a 40X objective and axonal degeneration was quantified as previously described9.
Protein transfection
Recombinant His-tagged Bclw (R&D Systems) was introduced into E15 sensory neurons using the Chariot protein transfection system (Active Motif). Bclw was introduced into the distal axon compartments of compartmented chamber cultures using 2 μl of Chariot reagent. The compartmented culture system allows selective introduction of protein into axons, without altering levels of protein in the cell bodies (see Supplementary Fig. 5c).
Statistical analysis
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those generally employed in the field. Statistical differences were tested using ANOVAs (one-way or two-way) with Bonferroni or Dunnett’s multiple comparison tests using GraphPad Prism, or with two-tailed t-tests (unpaired or paired) or Z-tests using Excel from at least 3 independent experiments. Data distribution was assumed to be normal but this was not formally tested. Two-tailed Fisher’s exact test was performed using the fisher test function in MATLAB.
A Supplementary Methods Checklist is available.
Supplementary Material
Acknowledgments
We thank members of the Segal lab, A. Eisner, M. McClintock, M. Greenberg for discussions, and G. McGregor (University of California, Irvine, CA) for providing bclw−/− mice. This work was supported by grants from the National Institutes of Health (NIH), Grant R01 NS050674 and R01 MH091662 to R.A.S. and Grant F31 NS077620 to S.J.F., and fellowships from the Harvard Mahoney, the Harvard NeuroDiscovery Center and the Alice and Joseph E. Brooks fund to K.E.C., and from the Victoria Quan fund to S.J.F.
Footnotes
AUTHOR CONTRIBUTIONS
K.E.C., S.J.F. and R.A.S. designed experiments and wrote the manuscript. K.E.C., S.J.F. and M.F.P.-M. performed compartmented culture experiments. K.E.C. performed formaldehyde cross-linking experiments. M.F.P.-M. performed axonal degeneration and protein transfection experiments. H.L.E performed analysis of FISH data. K.E.C. and S.J.F. performed all other experiments.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell. 2014;20:127–133. doi: 10.1016/j.molcel.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 3.Blackinton JG, Keene JD. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin Cell Dev Biol. 2014;34:44–54. doi: 10.1016/j.semcdb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keene JD. RNA regulons: coordination of post-transcriptional events. Nature Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 5.Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung H, et al. Remote control of gene function by local translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarosh CA, et al. PSF: nuclear busy-body or nuclear facilitator? Wiley Interdiscip Rev RNA. 2015;6:351–367. doi: 10.1002/wrna.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong X, et al. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol Cell Biol. 2007;27:4863–4875. doi: 10.1128/MCB.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirose T, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patton JG, et al. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 11.Danckwardt S, et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 2007;26:2658–2669. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall-Pogar T, et al. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA. 2007;13:1103–1115. doi: 10.1261/rna.577707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chanas-Sacre G, et al. Identification of PSF, the polypyrimidine tract-binding protein-associated splicing factor, as a developmentally regulated neuronal protein. J Neurosci. 1999;57:62–73. doi: 10.1002/(SICI)1097-4547(19990701)57:1<62::AID-JNR7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 14.Lowery LA, Rubin J, Sive H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev Dyn. 2008;236:1347–1357. doi: 10.1002/dvdy.21132. [DOI] [PubMed] [Google Scholar]
- 15.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 16.Kunde SA, et al. The X-chromosome-linked intellectual disability protein PQBP1 is a component of neuronal RNA granules and regulates the appearance of stress granules. Hum Mol Genet. 2011;20:4916–4931. doi: 10.1093/hmg/ddr430. [DOI] [PubMed] [Google Scholar]
- 17.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deglincerti A, Jaffrey SR. Insights into the roles of local translation from the axonal transcriptome. Open Biol. 2012;2:120079. doi: 10.1098/rsob.120079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon BC, et al. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148:752–764. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cosker KE, et al. Target-derived neurotrophins coordinate transcription and transport of bclw to prevent axonal degeneration. J Neurosci. 2013;33:5195–5207. doi: 10.1523/JNEUROSCI.3862-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreassi C, et al. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- 22.Cox LJ, et al. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snider WD. Functions of the neurotrophins during nervous system development: what the knockouts are teaching us. Cell. 1994;77:627–638. doi: 10.1016/0092-8674(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Courchesne SL, et al. Sensory neuropathy attributable to loss of Bcl-w. J Neurosci. 2011;31:1624–1634. doi: 10.1523/JNEUROSCI.3347-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batish M, et al. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci USA. 2012;109:4645–4650. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Rubei S, Bagni C. Fragile X mental retardation protein control of neuronal mRNA metabolism: Insights into mRNA stability. Mol Cell Neurosci. 2010;43:43–50. doi: 10.1016/j.mcn.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Okano H, et al. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Gagnon JA, Mowry KL. Molecular motors: directing traffic during RNA localization. Crit Rev Biochem Mol Biol. 2011;46:229–239. doi: 10.3109/10409238.2011.572861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirokawa H, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- 31.Spillane M, et al. Mitochondria coordinate sites of axon branching through localized intro-axonal protein synthesis. Cell Rep. 2013;5:1564–1575. doi: 10.1016/j.celrep.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campenot RB. Development of sympathetic neurons in compartmentalized cultures. II Local control of neurite survival by nerve growth factor. Dev Biol. 1982;93:13–21. doi: 10.1016/0012-1606(82)90233-0. [DOI] [PubMed] [Google Scholar]
- 33.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PloS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Rodriguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J Cell Biol. 2007;176:197–207. doi: 10.1083/jcb.200606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 36.Farris S, et al. Selective localization of arc mRNA in dendrites involves activity- and translation-dependent mRNA degradation. J Neurosci. 2014;34:4481–4493. doi: 10.1523/JNEUROSCI.4944-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee M, et al. The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 2015;43:3826–3840. doi: 10.1093/nar/gkv156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huttelmaier S, et al. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 39.Buxade M, et al. The PSF. p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor alpha. J Biol Chem. 2008;283:57–65. doi: 10.1074/jbc.M705286200. [DOI] [PubMed] [Google Scholar]
- 40.Ke Y, et al. Tau-mediated nuclear depletion and cytoplasmic accumulation of SFPQ in Alzheimer’s and Pick’s disease. PLoS One. 2012;7:e35678. doi: 10.1371/journal.pone.0035678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tapia-Paez I, et al. The complex of TFII-I, PARP1, and SFPQ proteins regulates the DYX1C1 gene implicated in neuronal migration and dyslexia. FASEB J. 2008;22:3001–3009. doi: 10.1096/fj.07-104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubota M, et al. Therapeutic implications of down-regulation of cyclophilin D in bipolar disorder. Int J Neuropsychopharmacol. 2010;13:1355–1368. doi: 10.1017/S1461145710000362. [DOI] [PubMed] [Google Scholar]
- 43.Hempstead BL, et al. Overexpression of the trk tyrosine kinase rapidly accelerates nerve growth factor-induced differentiation. Neuron. 1992;9:883–896. doi: 10.1016/0896-6273(92)90241-5. [DOI] [PubMed] [Google Scholar]
- 44.Ross AJ, et al. Testicular degeneration in Bclw-deficient mice. Nat Genet. 1998;18:251–256. doi: 10.1038/ng0398-251. [DOI] [PubMed] [Google Scholar]
- 45.Pazyra-Murphy MF, Segal RA. Preparation and maintenance of dorsal root ganglia neurons in compartmented cultures. J Vis Exp. 2008;20 doi: 10.3791/951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fenstermacher SJ, Pazyra-Murphy MF, Segal RA. Campenot cultures and microfluidics provide complementary platforms for spatial study of dorsal root ganglia neurons. Springer Protocols: Neuromethods. 2015;103:105–124. [Google Scholar]
- 47.Watson FL, et al. Rapid nuclear responses to target-derived neurotrophins require retrograde transport of ligand-receptor complex. Nat Neurosci. 1999;19:7889–7900. doi: 10.1523/JNEUROSCI.19-18-07889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguet F, et al. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev Cell. 2013;26:279–291. doi: 10.1016/j.devcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lachmanovich E, et al. Co-localization analysis of complex formation among membrane proteins by computerized fluorescence microscopy: application to immunofluorescence co-patching studies. J Microsc. 2003;212:122–131. doi: 10.1046/j.1365-2818.2003.01239.x. [DOI] [PubMed] [Google Scholar]
- 50.Mendoza MC, et al. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol Cell. 2011;41:661–671. doi: 10.1016/j.molcel.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.