Abstract
Context:
Recent studies showed that transcription of the MYC gene is driven by the interaction of bromodomain and extraterminal domain (BET) proteins with acetylated histones on chromatin. JQ1, a potent inhibitor that effectively disrupts the interaction of BET proteins with acetylated histones, preferentially suppresses transcription of the MYC gene. We recently reported that JQ1 decreased thyroid tumor growth and improved survival in a mouse model of anaplastic thyroid cancer (ATC) by targeting MYC transcription. The role of MYC in human ATC and whether JQ1 can effectively target MYC as a treatment modality have not been elucidated.
Objective:
To understand the underlying molecular mechanisms of JQ1, we evaluated its efficacy in human ATC cell lines and xenograft models.
Design:
We determined the effects of JQ1 on proliferation and invasion in cell lines and xenograft tumors. We identified key regulators critical for JQ1-affected proliferation and invasion of tumor cells.
Results:
JQ1 markedly inhibited proliferation of four ATC cell lines by suppression of MYC and elevation of p21and p27 to decrease phosphorylated Rb and delay cell cycle progression from the G0/G1 phase to the S phase. JQ1 blocked cell invasion by attenuating epithelial-mesenchymal transition signals. These cell-based studies were further confirmed in xenograft studies in which the size and rate of tumor growth were inhibited by JQ1 via inhibition of p21-cyclin/cyclin-dependent kinase-Rb-E2F signaling.
Conclusions:
These results suggest targeting of the MYC protein could be a potential treatment modality for human ATC for which effective treatment options are limited.
Inhibition of MYC expression by the BET protein inhibitor JQ1 resulted in suppression of proliferation and invasion of ATC cells. BET inhibitors could be used as a treatment modality.
Anaplastic thyroid cancer (ATC) is one of the most aggressive cancers in humans. Studies have shown that human ATC derives from complex and heterogeneous genetic changes (1), making effective treatment a major challenge. Although well-differentiated thyroid cancer responds well to radioiodine therapy and usually has a favorable therapeutic outcome, it is rare for a patient with an anaplastic thyroid tumor to survive beyond 1 year. Intensive efforts have been undertaken in the search for effective ways to treat ATC (2).
Preclinical studies and clinical trials have demonstrated that targeting epigenetic alterations could be effective for cancer treatment. Epigenetic modifications through histone acetylation are key steps in the regulation of the gene expression in both normal and tumor cells (3). Bromodomain and extraterminal domain (BET) proteins interact with acetylated histones to regulate gene transcription (4). Specific inhibitors, such as JQ1, have been shown to block the interaction of BET proteins (e.g., BRD4) with acetylated histones to affect transcriptional events (5, 6). JQ1 has been reported to exhibit inhibitory effects on lung and prostate cancers (7–9). JQ1 has also been shown to suppress cell proliferation and tumor growth of both differentiated and undifferentiated thyroid cancer cell lines (10, 11).
We recently created a mutant mouse, expressing both mutated thyroid hormone receptor β (TRβPV) and KrasG12D mutants (ThrbPV/PVKrasG12D mice). These mice spontaneously developed metastatic undifferentiated thyroid cancer mimicking ATC (12). We found that the highly elevated expression of MYC at the messenger RNA (mRNA) and protein levels propels the aggressive growth of thyroid tumors of ThrbPV/PVKrasG12D mice. Concurrent with the increased Myc expression is the suppressed expression of thyroid differentiation transcription factors, paired box gene 8 (PAX8) and NKX2-1. Recent studies have shown that the MYC transcription program is particularly sensitive to the inhibitory effect of JQ1. Indeed, we found that treatment of ThrbPV/PVKrasG12D mice with JQ1 markedly reduced thyroid tumor growth and prolonged survival. These preclinical studies supported the idea that epigenetic modifications through blocking the interaction of BET proteins with acetylated chromatin by JQ1 and its analogues could be a potential treatment modality of human ATC.
To test the feasibility of this idea, we evaluated the efficacy of JQ1 in four human cell lines, designated THJ-11T, THJ-16T, THJ-21T, and THJ-29T, established from human primary ATC tumors (13). They were shown to harbor complex genetic alterations. In addition to copy number gains and losses in various genes, THJ-11T cells expressed KRASG12V mutation; THJ-16T cells expressed PI3KE454K, TP53, and Rb mutations; THJ-21T cells expressed BRAFV600E, TP53, and Rb mutations; and THJ-29T cells expressed Rb mutations (13). These authenticated cell lines have been used by investigators as model cell lines to interrogate the functional consequences of these mutations and to identify potential molecular targets for treatment (14, 15).
In the present studies, we found JQ1 treatment was effective in suppressing the proliferation and invasion of tumor cells in cell-based studies and a mouse xenograft model. Consistent with preclinical studies using ThrbPV/PVKrasG12D mice, the expression of the MYC gene was sensitive to the inhibitory effect of JQ1, leading to the upregulation of cyclin-dependent kinase inhibitor 1 (p21Cip1) to arrest the cell cycle progression. Moreover, the expression of regulators of epithelial-mesenchymal transition (EMT) were decreased by JQ1 to attenuate tumor cell invasion. These results suggest that JQ1 could be considered favorably for treatment of human ATC.
Materials and Methods
Cell culture
The human ATC cell lines (i.e., THJ-11T, THJ-16T, THJ-21T, and THJ-29T) were obtained from Dr. John A. Copland III at the Mayo Foundation for Medical Education and Research. All patient tissues used in this study were de-identified. This study was approved by the Mayo Institutional Review Board. Four human ATC cell lines, designated THJ-11T, THJ-16T, THJ-21T, and THJ-29T, were established from human primary ATC tumors and were extensively characterized and authenticated using DNA short tandem repeat analysis (13). These four cell lines have different genetic characteristics that were detailed by Marlow et al. (13). The cells were cultured in RPMI-1640 media supplemented with 10% fetal bovine serum, 1% nonessential amino acid, 1% sodium pyruvate, and 1% antibiotic-antimycotic solution in 5% CO2 at 37°C in a humidified incubator. The medium was replaced every 48 hours.
Cell proliferation assays
To evaluate the effect of JQ1 on cell proliferation, cells were cultured in the medium with increasing concentrations of JQ1 (250 nM, 500 nM, or 1 µM) or dimethyl sulfoxide (DMSO) in six-well plates in triplicates. Cell counts were measured every 24 hours for 96 hours using a cell counter (Beckmann Coulter, Indianapolis, IN).
Invasion assay
Invasion assay was performed in 8-μm−pore transwells (6.5 mm; Costar, Corning, NY) in quadruplicate. Transwell filters were layered with 100 μL of Matrigel (BD Biosciences, San Jose, CA, cat. 356230) diluted 1:10 in phosphate-buffered saline (PBS). After rinsing with PBS, cells were plated as described previously. Twenty-four hours later, cells migrating to the bottom of the filter were evaluated after removal of material from the upper side of the filter, with 0.1% crystal violet staining and measurement of solubilized dye at A590 with triplicates.
Western blot analysis
Western blot analyses were carried out as described previously (16). Tumor tissues or cultured cells were washed with PBS and homogenized in a solution with 50 mM Tris buffer, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1% NP40, and proteinase/phosphatase inhibitors. After centrifuge at 14,000 rpm for 5 minutes, lysates were used for Western blot analysis. The antibodies against MYC were used (1:5000 dilution; ab32072); cyclin-dependent kinase 4 (CDK4) (1:2500 dilution; ab75511) from Abcam; p21 (1:200 dilution; sc-6246), E2F-1 (1:200 dilution; sc-251), E2F-3 (1:200 dilution; sc-878), and TWIST1 (1:200 dilution; sc-81417) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); phosphorylated Rb (S780) (1:1000 dilution; #9307), Rb (1:2000 dilution; #9309), p27 (1:1000 dilution; #3686), zinc finger protein SNAIL (Snail; 1:1000 dilution; #3895), and Slug (1:1000 dilution; #9585) from Cell Signaling; α-tubulin (1:5000 dilution; T6199) from Sigma-Aldrich; cyclin D1 (1:2000 dilution; RB9041) from Thermo Fisher Scientific (Fremont, CA); and hexamethylene bis-acetamide inducible 1 (HEXIM1; 1:2000 dilution; A303-113A) and BRD4 (1:5000 dilution; A301-985A) from Bethyl Laboratories, Inc. After being washed, the membranes were incubated with horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) (1:2500 dilution; NA9310 GE Healthcare) or anti-rabbit IgG (1:2500 dilution; NA9340 GE Healthcare) as the secondary antibody and were subsequently detected by means of an ECL system (Western Lightning® Plus-ECL, PerkinElmer, Waltham, MA). For control of protein loading, the blots were striped with Re-Blot Plus (MilliporeSigma, Temecula, CA) and were reprobed with α-tubulin. Band intensities were quantified with ImageJ software (ImageJ 1.48v; Wayne Rasband, National Institutes of Health). Western blot analyses were repeated three times, each with triplicates from independent tissues.
Histological analysis
Slides of 14 normal thyroid tissues and 12 tumor tissues from ATCs from the Mayo Clinic were used to analyze MYC expression in human ATCs. For analysis of proteins in ATC cells, cell pellets were fixed in 10% neutral buffered formalin for 24 hours and were embedded in paraffin for analysis. Immunohistochemistry (IHC) was performed on formalin-fixed paraffin tumor sections as described previously (12). Primary antibodies used were anti–Ki-67 antibody (dilution: 1:300; Thermo Fisher Scientific; #RB-9043-P0) and anti-MYC antibodies (dilution: 1:300; Abcam, ab32072). Staining was developed with diaminobenzidine using the diaminobenzidine substrate kit for peroxidase (Vector Laboratories, Burlingame, CA; SK-4100). Cells positively stained by anti–Ki-67 antibody or anti-MYC antibody were counted (>100 cells/slide in randomly selected three to five fields in each slide with triplicates; repeated twice). The quantitative analysis was carried out for Ki-67– or MYC-positive cells with ImageJ software. IHC was carried out in triplicates.
RNA extraction and real-time reverse transcription polymerase chain reaction analysis
RNA extraction and real-time reverse transcription polymerase chain reaction analyses were carried out as previously described (17): for the MYC gene: forward, 5′-CCTACCCTCTCAACGACAGC-3′; reverse, 5′-CTCTGACCTTTTGCCAGGAG-3′; for the p21 gene: forward, 5′-GGAAGACCATGTGGACCTGT-3′; reverse, 5′-GGGCTCCTCTTGGAGAAGAT-3′; for the glyceraldehyde-3-phosphate dehydrogenase gene: forward, 5′-CGAGATCCCTCCAAAATCAA-3′; reverse, 5′-GGTGCTAAGCAGTTGGTGGT-3′.
The experiments were repeated three times, each with triplicates.
Flow cytometric analysis of cell cycle
Cells were seeded into six-well plates at a density of 1 × 105 cells. After 24 hours, 500 nM JQ1 was treated during 48 hours at 37°C. Control cells were treated with DMSO. After treatment, all harvested cells were washed with ice-cold PBS and then fixed with cold 70% (vol/vol) ethanol and stored at −20°C at least 2 hours. After this, cells were washed in PBS buffer and then incubated with 1 µg/mL 4′,6-diamidino-2-phenylindole solution for 10 minutes before being analyzed. The cell cycle profiles were determined using flow cytometry (LSRFortessa II, BD Bioscience, San Jose, CA) and analyzed with FlowJo (FlowJo LLC, Ashland, OR). All experiments were performed in at least triplicate.
In vivo xenograft tumor assays
All animal experiments were performed under protocols approved by the National Cancer Institute’s Animal Care and Use Committee. The xenograft assays used female athymic nude mice 6 to 8 weeks old. THJ-11T and THJ-16 cells (5 × 106 cells) in 200-μL suspension mixed with Matrigel Basement Membrane Matrix (BD Biosciences, cat. 354234) were inoculated subcutaneously into the right flank of the mice. The tumor volume was calculated as L × W × H × 0.5236. JQ1 or vehicle treatment began when the median tumor size reached ∼100 mm3. Tumor growth rate was calculated using the following equation:
where V2 represents tumor volume at euthanasia; V1 represents tumor volume at JQ1 started; t2 represents the day at euthanasia; and t1 represents the day at JQ1 started.
Before treatment, tumor-bearing mice were randomized into two arms such that the median tumor sizes of each arm were approximately the same (DMSO as vehicle vs JQ1). JQ1 was first dissolved in DMSO and diluted to 20 volumes of β-cyclodextrin (Sigma-Aldrich, cat. H107-5G) in pure water and administered through intraperitoneal injection. JQ1 was administered at 50 mg/kg/mouse twice per day (THJ-11T) and once per day (THJ-16T). Tumor size was measured with calipers every day (THJ-11T) or every other day (THJ-16T) until it reached 2 cm in diameter at the longest dimension. Then, the mice were euthanized and the tumor tissues were collected for further analysis.
Statistical analysis
All data are expressed as mean ± standard deviation. All tests were two-sided, and P < 0.05 was considered significant. GraphPad Prism version 5.0 for Mac OS X was used to perform analyses of variance.
Results
Human ATC cells aberrantly express elevated levels of MYC
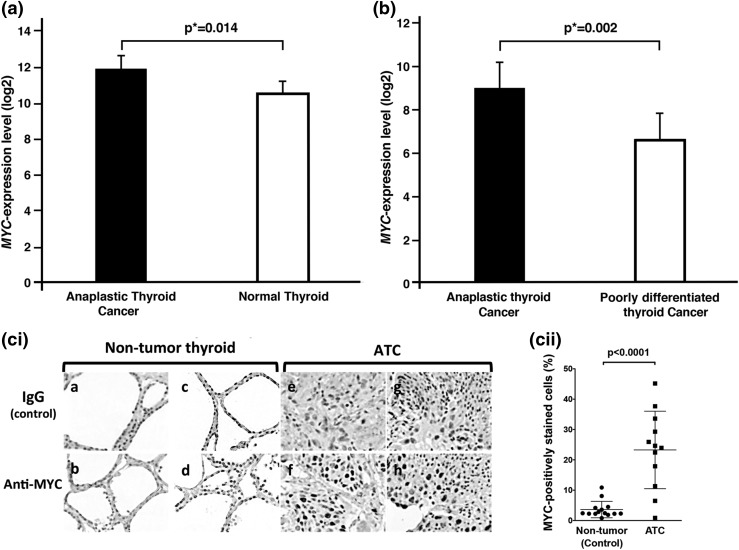
Using gene expression microarray analysis, von Roemeling et al. (18) found that MYC mRNA expression was elevated 2.83-fold in ATC tumors (n = 12) compared with normal thyroid tissues (n = 13; adjusted P value = 0.0014) [Fig. 1(a); GSE65144 data set by von Roemeling et al. (18)]. Recently, in a different study, Landa et al. (1) also reported that MYC expression was 5.06-fold higher in human ATC (n = 20) than in poorly differentiated thyroid cancer (n = 17) [Fig. 1(b); GSE76039 data set from Landa et al. (1)]. It is important to note that Fig. 1(a) and 1(b) were from two different data sets of two different unrelated studies using two different ranges of (log2) scale in the y-axis.
Figure 1.
MYC overexpressed in ATC. (a) and (b) show elevated expression of MYC in ATC by two data sets deposited by two different independent studies in the National Center for Biotechnology Information Gene Expression Omnibus database. (a) MYC expression was significantly upregulated in anaplastic thyroid tumors compared with normal thyroid tissue [from data set GSE65144 deposited by von Roemeling et al. (18)] and (b) was also higher in ATC than in poorly differentiated thyroid cancer [from data set GSE76039 deposited by Landa et al. (1)]. The microarray data were reanalyzed with limma, Bioconductor packages, and false discovery rate was used to calculate adjusted P values (*). (ci) Immunohistochemical analysis was carried out using antibody against MYC protein for 14 nontumor thyroid tissues and 12 ATC tissues. Representative images are shown for two normal thyroids as controls (panels a to d) and two ATC tissues (panels e to h). (cii) The anaplastic thyroid tumor tissues had significantly higher numbers of cells stained for MYC than did the control nontumor thyroid tissues (P < 0.0001).
These findings prompted us to carry out IHC analysis to examine whether MYC protein levels were increased in the human ATC tumors. Figure 1(ci) shows clearly that MYC–positively labeled cells were strongly detected in the nuclei of tumor cells (panels f and h), whereas only a few cells were weakly stained with anti-MYC antibodies in nontumor thyroid (panels b and d). Panels a, c, e, and g were the respective negative controls. Quantitative data showed that MYC–positively stained cells were 8.4-fold higher in tumors than in nontumor thyroid [Fig. 1(cii)]. We further analyzed the MYC abundance in a normal human thyroid cell line (116N) and human ATC cell lines THJ-11T, THJ-16T, THJ-21T, and THJ-29T. We found that MYC was not detectable in the 116N cell line (panel I, lane 1, Supplemental Fig. A (271KB, tif) ) but was highly abundant in the four ATC cell lines (panel I, lanes 2 to 5, Supplemental Fig. A (271KB, tif) ). Moreover, MYC was not detectable in human follicular thyroid cancer cells (FTC133 and FTC236, lanes 5 to 8, panel II, Supplemental Fig. A (271KB, tif) ) or in rat pCCL3 cells (lanes 9 and 10, panel II, Supplemental Fig. A (271KB, tif) ). Taken together, these results suggest that MYC could be a molecular target in human ATC.
JQ1 inhibits cell proliferation in human ATC cell lines
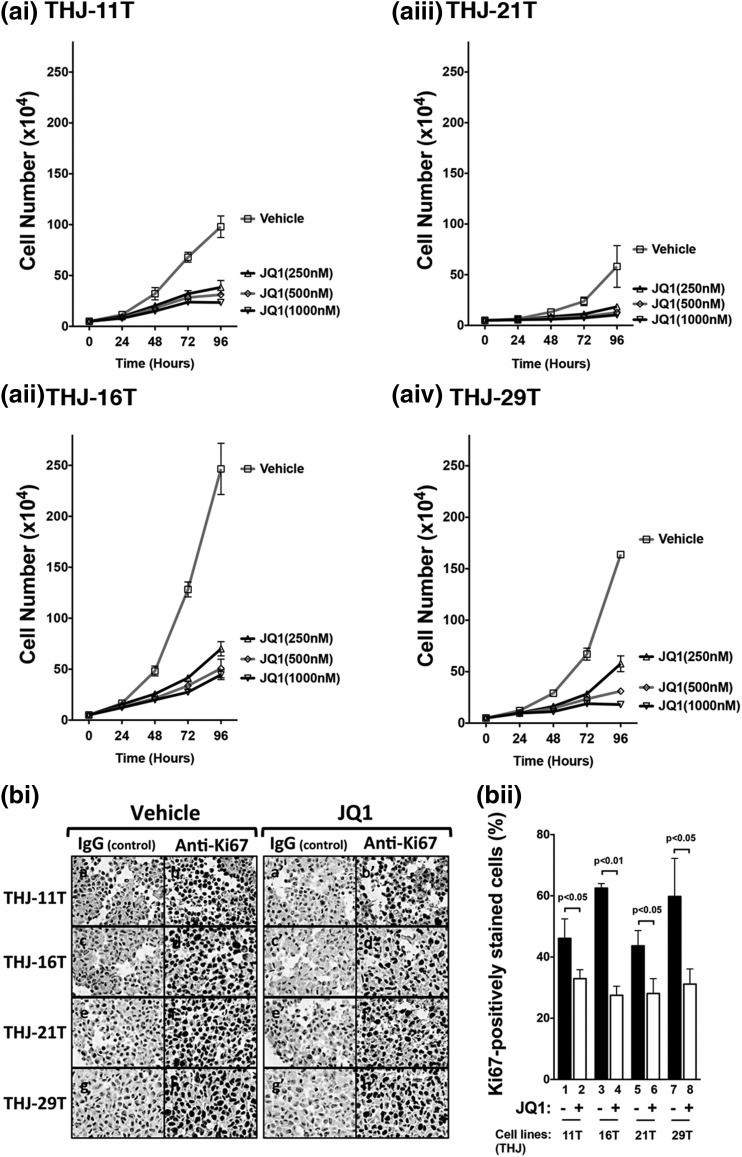
Recent studies have shown that JQ1 is a strong inhibitor in suppressing MYC transcription activity. JQ1 has been shown to induce cell differentiation of NUT midline carcinoma and to attenuate growth of myeloid tumors (e.g., acute myeloid leukemia and multiple myeloma) (5, 19–21). To test whether JQ1 was effective in inhibiting MYC expression in human ATC, we evaluated the effect of JQ1 on the proliferation of these four human ATC cell lines: THJ-11T, THJ-16T, THJ-21T, and THJ-29T. We found the rank of proliferation rates was THJ-16T > THJ-21T > THJ-11T > THJ-29T [Fig. 2(ai–aiv)]. However, JQ1 inhibited cell proliferation in a concentration-dependent manner in these cell lines. At the concentration of 250 nM, JQ1 inhibited 72%, 68%, 60%, and 65% in THJ-16T, THJ-21T, THJ-11T, and THJ-29T cells, respectively. At the highest concentration of 1000 nM, 80% to 90% of cell proliferation was inhibited in these four cell lines [Fig. 2(ai–aiv)]. Of note, JQ1 was similarly effective in inhibiting cell proliferation irrespective of the genetic background of these four cell lines.
Figure 2.
JQ1 inhibited cell proliferation in human ATC cell lines. (ai–aiv) Four cells lines were treated with vehicle (DMSO) or with JQ1 at increasing doses of 250 nM, 500 nM, or 1000 nM for 24, 48, 72, or 96 hours. Cells were harvested, and cell numbers were counted. (bi) Immunohistochemical analysis was carried out to detect the cell proliferation marker Ki67 after cells were treated with vehicle (DMSO) or JQ1 (500 nM) for 48 hours. (bii) Quantification of Ki67-positive cells. After treatment with JQ1, the cell lines of the THJ-11T (bar 2), THJ-16T (bar 4), THJ-21T (bar 6), and THJ-29T (bar 8) cells had significantly lower numbers of cells stained for Ki67 than those treated with DMSO as controls.
We performed further IHC analysis to assess the extent of JQ1-induced inhibition of tumor cell proliferation using the cell proliferation marker Ki67. Figure 2(bi) compares the number of cells positively stained for Ki67 in tumor cells. JQ1 treatment decreased the number of Ki67-positive cells as shown in Figure 2(bi) (compare panels b′, d′, f′, and h′ with b, d, f, and h, respectively. Quantitative analysis showed that consistent with proliferation assays shown in Fig. 2(ai–aiv), fewer Ki67–positively stained cells were found in JQ1-treated cells than in the vehicle-treated cells [In Fig. 2(bii), compare bars 2, 4, 6, and 8 with 1, 3, 5, and 7, respectively]. Taken together, these data indicate that JQ1 was effective in inhibiting cell proliferation in these four cell lines.
JQ1 suppresses MYC and key cell cycle regulators in human ATC cell lines
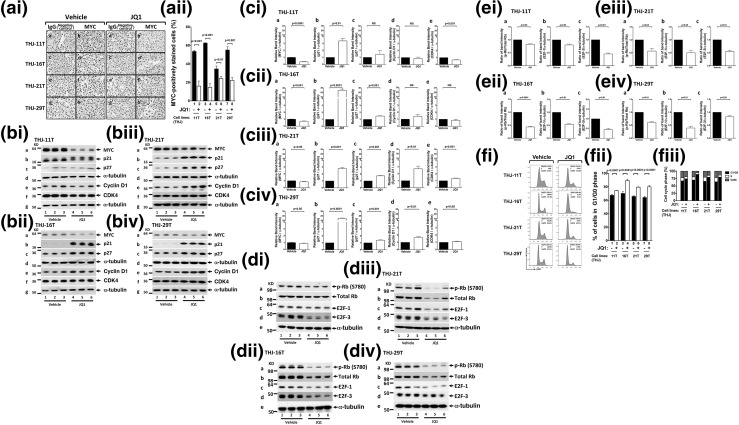
That JQ1 inhibited cell proliferation of the four anaplastic cancer cell lines prompted us to examine whether the MYC protein abundance was affected by JQ1. We first assessed the MYC protein abundance in these four cell lines by IHC analyses. Figure 3(ai) shows that in vehicle-treated cells, cells positively stained with MYC were apparent (panels b, d, f, and h for THJ-11T, THJ-16T, THJ-21T, and THJ-29T cells, respectively). Panels a, c, e, and g were the respective negative controls in which IgG was used. In JQ1-treated cells, however, the number of cells positively stained with MYC was markedly reduced [Fig. 3(ai), panels b′, d′, f′, and h′]. MYC–positively stained cells were counted, and the quantitative data expressed as percentage of positively stained cells to total cells are shown in Fig. 3(aii). The number of JQ1-treated tumor cells stained with MYC was reduced 69% for THJ-11T (bar 2), 78% for THJ-16T (bar 4), 35% for THJ-21T (bar 6), and 62% for THJ-29T (bar 8), indicating that JQ1 could suppress MYC expression in these cell lines. But the extent of inhibition in the protein levels varied in the different cell lines.
Figure 3.
JQ1 decreased MYC and elevated p21 protein abundance in human ATC cell lines. (ai) Immunohistochemical analysis for MYC protein was carried out after the THJ-11T, THJ-16T, THJ-21T, and THJ-29T cell lines were treated with vehicle or with JQ1 (500 nM) for 48 hours. (aii) Quantification of MYC-positive cells indicated reduction of MYC protein by JQ1. (bi–biv) Cell lysates were prepared from the THJ-11T, THJ-16T, THJ-21T, and THJ-29T cell lines. The blots were probed with the antibodies against MYC, p21, p27, cyclin D1, and CDK4 and the loading control α-tubulin, as described in the Materials and Methods section. (ci–civ) Band intensities in (b) were quantified, allowing a comparison between vehicle-treated and JQ1-treated cells. The P values are shown. (di–div) Cell lysates were prepared from the THJ-11T, THJ-16T, THJ-21T, and THJ-29T cell lines. The blots were probed with the antibodies against p-Rb, total Rb, E2F-1, and E2F-3 and the loading control α-tubulin, as described in the Materials and Methods section. (ei–eiv) Band intensities in (di–div) were quantified, allowing a comparison between vehicle-treated and JQ1-treated cells. The P values are shown. (fi–fiii) JQ1 inhibited cell cycle progression of ATC cell lines. (fi) Cell cycle profiles in THJ cell lines. The percentages of cell populations of different cell cycle phases are shown in the upper-right corner. After JQ1 treatments, all THJ cell lines were blocked in the G1/G0 phase, and there was reduction in the S and G2/M phases. (fii) JQ1 treatment lengthened the G1/G0 phase in ATC cells. (fiii) JQ1 treatment affected the distribution of cell cycle. Different cell cycle phases were quantified by 4′,6-diamidino-2-phenylindole staining, followed by fluorescence-activated cell sorting analysis in THJ cell lines.
The findings from IHC analyses were further confirmed in four cell lines by Western blotting [Fig. 3(bi–biv)]. Consistent with findings by IHC analysis [Fig. 3(ai)], the abundance of MYC was clearly lower in JQ1-treated than in vehicle-treated cell lines [Fig. 3(bi–biv), lanes 4 to 6 vs lanes 1 to 3, in panel a], albeit with varied extent of inhibition of MYC protein levels by JQ1 depending on the cell lines. The quantitative differences were apparent in that THJ-11T was most sensitive to the inhibitory effect of JQ1 [∼65% inhibition, Fig. 3(ci-a)], followed by THJ-16T [28%; Fig. 3(cii-a)], with THJ-21T and THJ-29T being the least sensitive [5% to 10%; Fig. 3(ciii-a) and 3(civ-a)].
p21Cip1, also known as p21Waf1 and hereafter referred to as p21, is a cyclin-dependent kinase inhibitor. The p21 protein binds to and inhibits the activity of cyclin-CDK complexes to regulate cell cycle progression at the G1 and S phases. MYC directly represses p21 transcription (22), thereby driving cell cycle progression. We therefore evaluated levels of p21, p27, cyclin D1, and CDK4 proteins by Western blot analysis [Fig. 3(bi–biv)]. Concomitant with decreased MYC protein levels in JQ1-treated cells, p21 protein levels were highly elevated [panel b in Fig. 3(bi–biv)]. Levels of cyclin-dependent kinase inhibitor 1B (p27Kip1), another CDK inhibitor, were also elevated (panel c), though less affected by JQ1 than p21 was. The protein levels of cyclin D1 and CDK4 were not significantly affected by JQ1 [panels e and f, respectively, in Fig. 3(bi–biv); the quantitative JQ1-induced changes in p21, p27, cyclin D1, and CDK4 are shown in panels b, c, d, and e, respectively, of Fig. 3(ci–civ) for the four cell lines].
The effects of elevated p21 and p27 were apparent in that phosphorylated Rb (p-Rb) was decreased (panel a in Fig. 3(di–div); see also quantitative data in the ratios of p-Rb vs total Rb, Fig. 3(ei–eiv), panel a in ei–eiv for the respective cell lines). In addition, E2F-1 and E2F-3 were also decreased [panels c and d in Fig. 3(di–div); see also quantitative data in panels b and c in Fig. 3(ei–eiv) for the respective cell lines]. These results indicate that together with decreased MYC, JQ1-decreased cell proliferation was mediated via the upregulation of CDK inhibitors p21 and p27.
The inhibitory action of p21 and p27 on cell proliferation was further elucidated by cell cycle analysis. Figure 3(fi–fiii) shows that JQ1-induced elevated p21 and p27 delayed the entry of cells from the G1 phase to the S phase. It was clear that cells in the G1/G0 phase were significantly prolonged in all four cell lines [Fig. 3(fii) and 3(fiii)]. The subsequent cycles of the S and G2/M phases were also delayed by JQ1 treatment [Fig. 3(fii) and 3(fiii)]. Thus, the cell inhibitory effect by JQ1 was mainly mediated by decreases in cell cycle progression.
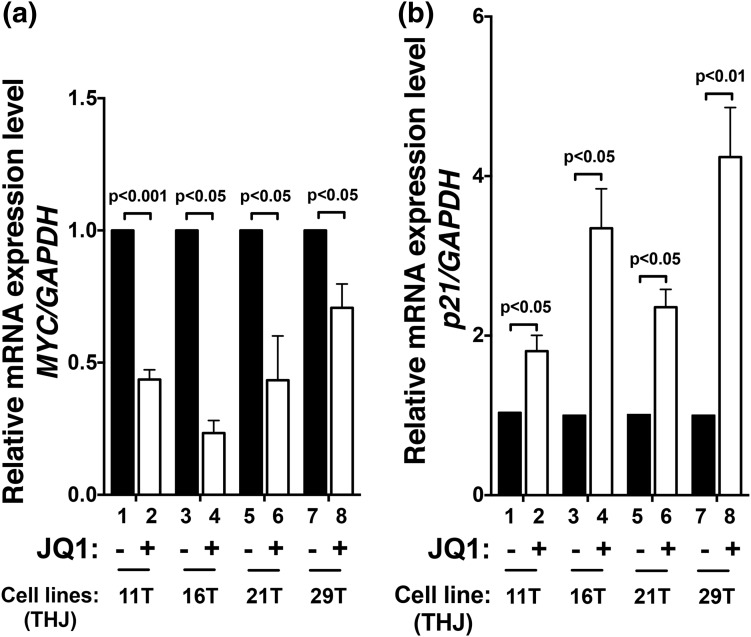
That MYC protein levels were decreased by JQ1 treatment [Fig. 3(ai–aii) and 3(bi–biv)] led us to ascertain the effect of JQ1 at the mRNA levels. Through inhibition of the binding of BRD4 to acetyl-lysine on chromatins, JQ1 is known to selectively inhibit transcription of the MYC gene in many tumors (6, 19, 23). Using a mouse model of ATC, we recently showed that Myc expression was suppressed in thyroid tumors of JQ1-treated mice, resulting in improved survival and inhibition of tumor growth (17). Consistent with these findings, we found that the expression of MYC mRNA in these four cell lines was inhibited by JQ1, ranging from 77% [THJ-16T cells, bar 4, Fig. 4(a)] to 29% [THJ-29T, bar 8, Fig. 4(a)]. Because p21 is directly negatively regulated by MYC (24) and is MYC’s downstream effector, we further ascertained the mRNA expression of the p21 gene, which is a downstream effector of MYC. We found that concurrently with the suppression of MYC by JQ1, p21 mRNA was markedly elevated by JQ1 [Fig. 4(b)]. These data indicated that the concordant changes of the MYC and p21 genes by JQ1 inhibited tumor cell proliferation. It should be pointed out that JQ1 could not suppress the overexpressed MYC by lentiviral transduction (Supplemental Fig. B (280.4KB, tif) , lanes 10 to 12 vs lanes 7 to 9, panel a). Suppression of MYC led to the elevation of p21 and p27 in the control cells (only control lentivirus was used; lanes 4 to 6 vs lanes 1 to 3, panels b and c, respectively), whereas highly overexpressed MYC led to the suppression of p21 and p27 (lanes 10 to 12, panels b and c, respectively). These data clearly indicate that overexpression of MYC overcame the JQ1 effect and further support our conclusions that MYC plays a critical role in the proliferation of ATC tumor cells.
Figure 4.
JQ1 suppressed MYC and p21 transcription. Total RNA was extracted from four THJ cell lines treated with vehicle or JQ1 (500 nM) for 24 hours. The expression of (a) MYC and (b) p21 mRNA was analyzed by quantitative real-time reverse transcription polymerase chain reaction (RT-PCR). Triplicate samples were analyzed. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as a reference. The P values are indicated.
JQ1 inhibits cell motility by suppressing EMT signals
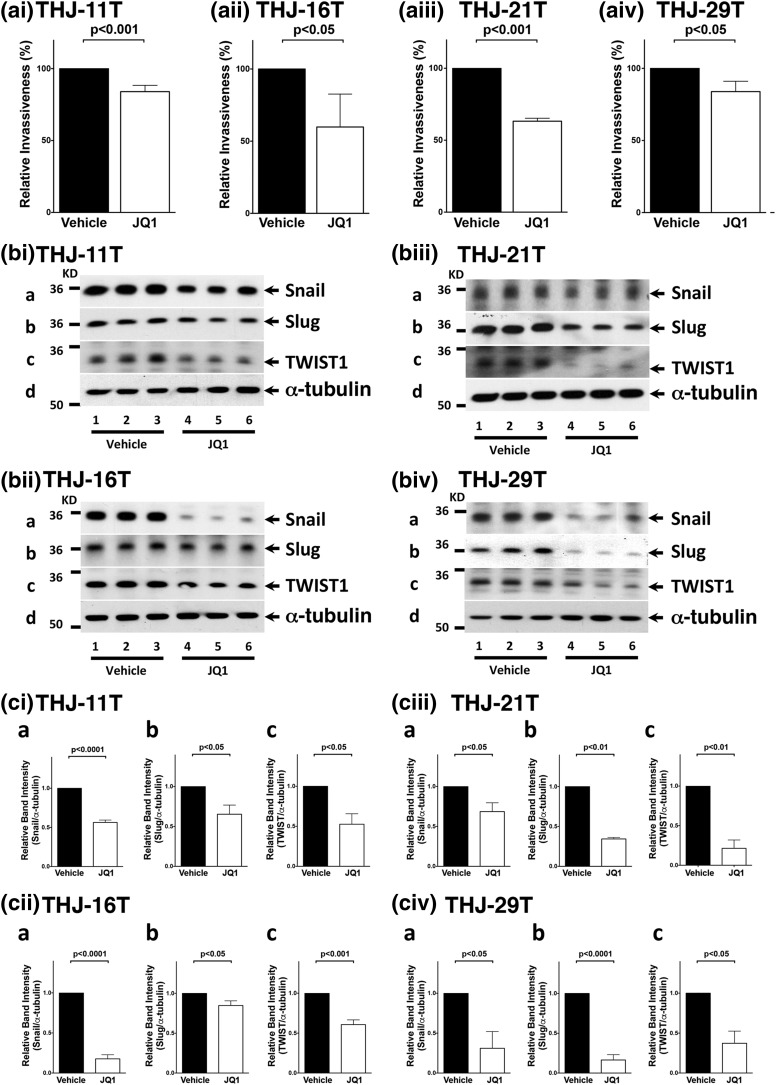
MYC also regulates genes important for tumor cell motility and invasion. Overexpression of MYC induces EMT in mammary epithelial cells. To investigate whether JQ1 inhibited invasion of cells, we carried out a cell invasion assay. Cell invasiveness was significantly decreased in JQ1-treated cell lines compared with vehicle-treated cells [reduction: 16%, 40%, 37%, and 16% in the THJ-11T, -16T, -21T, and -29T cells, respectively; Fig. 5(civ-a)]. We further identified the regulators in EMT affected by JQ1. Snail is a transcription factor that represses the expression of the adhesion molecule E-cadherin to promote EMT. We found that Snail protein levels were lower in JQ1-treated cells [panel a in Fig. 5(bi–biv)]. The quantitative data show that the decreases ranged from 82% in JQ1-treated THJ-16T [Fig. 5(cii-a)] to 30% in THJ-21T cells [Fig. 5(ciii-a)]. The other two EMT regulators, Slug and TWIST1, were also decreased in JQ1-treated cells [panels b and c, respectively, in Fig. 5(bi–biv)]. The quantitative data show that the decreases ranged from 84% (THJ-29T) to 15% (THJ-16T) for Slug and from 79% (THJ-21T) to 39% (THJ-16T) for TWIST1 [Fig. 5(ci–civ), panels b and c, respectively]. Taken together, these data indicate that JQ1 inhibited tumor cell invasion by suppression of EMT.
Figure 5.
JQ1 inhibited ATC invasion and regulated EMT expression. (ai–aiv) JQ1 inhibited tumor cell invasion. Four cell lines treated with vehicle or JQ1 were examined using an in vitro invasion assay as described in the Materials and Methods section. Data were averaged from three independent experiments. (bi–biv) JQ1 altered EMT key regulators. Four cell lines were treated with vehicle or JQ1 (500 nM) for 48 hours. The blots were probed with antibodies against Snail (panel a), Slug (panel b), or TWIST1 (panel c). α-tubulin was used as the loading control. (ci–civ) Band intensities of Snail, Slug, and TWIST1 proteins were quantified. P values are shown.
Inhibition of MYC expression by JQ1 via collaboration with epigenetic regulators
We further ascertained the effects of JQ1 on the protein levels of BRD4. We found that JQ1 did not decrease the protein levels of BRD4 (panel a of Supplemental Fig. C-I–IV (1.6MB, tif) ) but slightly elevated its protein levels (panel a in Supplemental Fig. C-V–VIII (1.6MB, tif) ). Previously, we showed that in addition to BRD4, HEXIM1 protein also contributed to the inhibition of Myc expression to suppress thyroid tumor growth in a mouse model of ATC (17). HEXIM1 is a transcription regulator that associates with and inhibits the positive transcription elongation factor b (p-TEFb), a key regulator of RNA polymerase II (RNAPII) (25). We found that HEXIM1 was significantly elevated in JQ1-treated cells (panels b, Supplemental Fig. C-I–IV (1.6MB, tif) ). These changes can be seen more clearly in the quantitative analysis shown in Supplemental Fig. C (1.6MB, tif) (panel b in V–VIII). The increases ranged from 1.4- to 2.1-fold for HEXIM1. These data indicated that the effects of JQ1 were not due to a decrease in BRD4 and that the increase of HEXIM1, an inhibitor of RNAPII transcription, further coordinately contributed to the suppression of the MYC transcription.
JQ1 inhibits ATC-induced tumor growth in mouse xenograft models
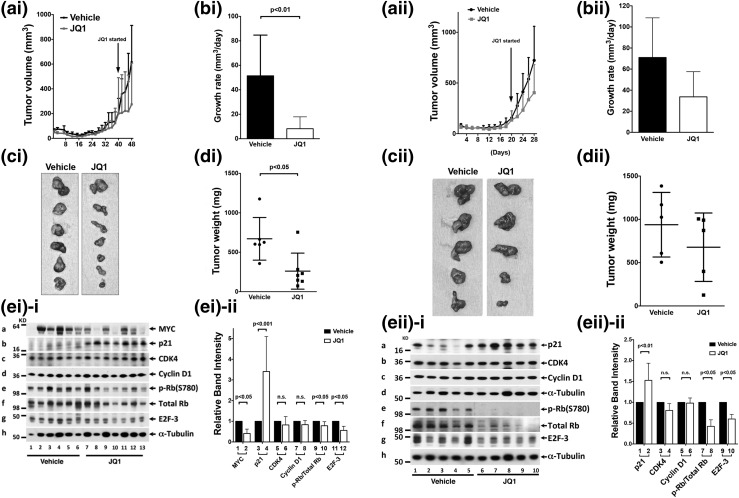
That JQ1 inhibited proliferation and suppressed invasion of cultured ATC cells prompted us to assess the effects of JQ1 in vivo using mouse xenograft models. We first studied the induction of tumor growth by THJ-16T cells in athymic mice. JQ1 treatment suppressed the tumor growth [Figure 6(ai)] and decreased the growth rate of xenograft tumors [Fig. 6(bi)]. JQ1 treatment also reduced the tumor size [Fig. 6(ci)] and the tumor weight [Fig. 6(di)]. Western blot analysis showed that MYC protein levels were lower in JQ1-treated athymic mice than in vehicle-treated mice [compare lanes 7 to 13 with lanes 1 to 6, Fig. 6(ei-i-a); see also quantitative data, bar 2 vs bar 1 in Fig. 6(ei-ii)]. Furthermore, p21 was significantly higher in tumors from JQ1-treated than vehicle-treated mice [compare lanes 7 to 13 with 1 to 6; Fig. 6(ei-i-b); also see Fig. 6 (ei-ii), bar 4 vs bar 3]. Consistent with cell-based studies, no significant changes in protein levels of CDK4 and cyclin D1 were detected in tumors from JQ1-treated mice [Fig. 6(ei-i-c) and 6(ei-i-d); also see Fig. 6(ei-ii), bars 5 to 8]. Importantly, p-Rb (panel e) and E2F-3 (panel g, Fig. 6(ei-i)] and the ratios of p-Rb vs total Rb [bar 10 vs bar 9, Fig. 6(ei-ii)] were also decreased, indicating the inhibiting effect of p21 on CDK to deter the cell cycle progression of tumor cells from the G1 phase to the S phase.
Figure 6.
(ai–di) JQ1 suppressed the growth of tumors derived from THJ-16T ATC cells in a xenograft mouse model. An equal number of THJ-16T cells (5 × 106) were injected into the flanks of mice before treatment. When tumors started to develop (average tumor size reached 100 mm3), JQ1 or vehicle was administered intraperitoneally on day 40 (50 mg/kg/d). JQ1 treatment decreased (ai) tumor growth, (bi) growth rate, (ci) tumor size, and (di) tumor weight. (ei) JQ1 treatment altered the protein abundance of key cell regulators. (ei-i) Western blot analysis was carried out as described in the Materials and Methods section. The proteins analyzed were marked. (ei-ii) The band intensities were determined, and the quantitative data indicated that JQ1 treatment decreased MYC and elevated p21 protein levels to decrease p-Rb/total Rb ratios and E2F-3 without significantly affecting CDK4 and cyclin D1. Six tumors derived from vehicle-treated mice and seven tumors derived from JQ1-treated mice were used in the Western blot analysis for the vehicle- and JQ1-treated mice. P values are indicated. (aii-dii) JQ1 suppressed the growth of tumors derived from ATC THJ-11T cells in a xenograft mouse model. An equal number of THJ-11T cells (5 × 106) were injected into the flanks of mice before treatment. When tumors started to develop (average tumor size reached 100 mm3), JQ1 or vehicle was administered intraperitoneally on day 20 (50 mg/kg per mouse twice per day). JQ1 treatment decreased (aii) tumor growth, (bii) growth rate, (cii) tumor size, and (dii) tumor weight. (eii) JQ1 treatment altered the protein abundance of key cell regulators. (eii-i) Western blot analysis was carried out as described in the Materials and Methods section. The proteins analyzed were marked. (eii-ii) The band intensities from the Western blots shown in eii-i were determined. The quantitative data indicated that JQ1 elevated p21 to decrease p-Rb/total Rb ratios and E2F-3 without significantly affecting CDK4 and cyclin D1. Five tumors each from vehicle- and JQ1-treated mice were used in the Western blot analysis. P values are shown.
We further assessed the effect of JQ1 on tumor growth induced by THJ-11T cells. JQ1 treatment inhibited the tumor growth [Fig. 6(aii)] and growth rates of xenograft tumors [Fig. 6(bii)]. JQ1 treatment also reduced the tumor size [Fig. 6(cii)] and the tumor weight [Fig. 6(dii)]. Western blot analysis of key cell regulators showed that p21 was significantly higher in tumors from JQ1-treated mice than in vehicle-treated mice [compare lanes 6 to 10 with 1 to 5, Fig. 6(eii-i-a); also see Fig. 6(eii-ii), bar 2 vs bar 1]. Consistent with cell-based studies, no significant changes in protein levels of CDK4 and cyclin D1 were detected in tumors from JQ1-treated mice [Fig. 6(eii-i-b) and 6(eii-i-c); also see Fig. 6(eii-ii), bars 3 to 6]. It is important to note that p-Rb and E2F-3 protein levels were decreased [panels e and g in Fig. 6(eii-i)]; see also that the ratio of p-Rb/total Rb was decreased in bar 8 vs bar 7, Fig. 6(eii-ii). These results indicate the inhibiting effect of p21 on CDK to deter cell cycle progression of tumor cells and inhibit tumor cell proliferation.
Discussion
Effective treatment of human ATC has long been a challenge for clinical oncologists. Many approaches, including tyrosine kinase inhibitor–based targeting therapies, failed to improve the outcome of patients with ATC (26, 27). One possible difficulty is the complexity and heterogeneity in the genetic abnormalities underlying ATC. The four human ATC cell lines used in the present studies harbor complex genetic alterations (13). Remarkably, JQ1 treatment was effective in suppressing proliferation and reducing invasion of tumor cells in these four cell lines, albeit with minor quantitative differences in the extent of responses. In the two representative xenograft studies, we also found that JQ1 was effective in inhibiting tumor growth in vivo. These results suggest that targeting chromatin regulators such as BET proteins with JQ1 in these ATC tumor cell lines is effective. These findings raise the possibility that JQ1 and its next-generation analogues could be beneficial for ATC patients.
The inhibitory action of JQ1 on the chromatin regulators, such as BRD4, have been shown to selectively inhibit transcription of the oncogenic driver MYC (6, 28, 29). Indeed, we previously showed that JQ1 treatment of anaplastic thyroid tumors of ThrbPV/PVKrasG12D mice resulted in the suppression of Myc expression to decrease tumor growth (17). Consistent with these findings, the present studies showed that JQ1 suppressed the expression of the MYC gene in the four cell lines, accompanied by concurrent elevated expression of a MYC downstream target gene, p21 [Fig. 3(bi–biv) and 3(ci–civ)]. Although JQ1 selectively suppressed the MYC transcription program, resulting in the inhibiton of tumor growth, MYC-independent effects were also found in the present studies. We showed that JQ1 also acted to increase the expression of HEXIM1 (Supplemental Fig. C (1.6MB, tif) ). It was reported that HEXIM1 inhibited p-TEFb by binding to cyclin T1 and sequestering p-TEFb into an inhibitory complex. Multiple p-TEFb units bind to a HEXIM1 multimer. By sequestering and inhibiting p-TEFb and in turn RNAPII, HEXIM1could be mechanistically involved in mediating JQ1-induced growth inhibition and differentiation of tumor cells (30). The MYC-independent elevated HEXIM1 further collaboratively suppressed the transcription of MYC. The dual actions of JQ1 shown in the present studies were also found in OTX015, an analogous inhibitor of BET proteins in acute leukemia cells (19). It is important to note that despite the different genetic mutations driving tumor cell proliferation in these four cell lines (13), JQ1 was effective in suppressing MYC expression and increasing p21 expression to deter cell cycle progression from the G1/G0 to the S phase [Fig. 3(fi–fiii)]. These results suggest that the MYC-p21 link is critical in the inhibition of tumor growth of ATC (see Supplemental Fig. D (91.7KB, tif) ). This inhibition could effectively counteract the aberrant growth driven by overactivation of other signaling pathways by oncogenic mutations of BRAF, KRAS, TP53, and/or PI3KCA in these cell lines.
Recently the effects of JQ1 and a related BET inhibitor (I-BET762) on thyroid cancer cell lines have been reported (10, 11). Gao et al. (10) reported that treatment of two papillary thyroid cancer lines, BCPAP and K1, with JQ1 resulted in the inhibition of cell proliferation and enhancement in I-131 uptake in vitro and the suppression of tumor growth in vivo. Mio et al. (11) demonstrated that JQ1 and I-BET762 were effective in the inhibition of proliferation of two human ATC cell lines, FRO and SW1736. Consistent with these findings, the present studies showed that JQ1 inhibited cell proliferation in vitro and tumor growth in vivo of four human ATC cells. Although we did not know whether JQ1 inhibited the invasion capacity of FRO, SW1736, BCPAP, and K1 cells studied by Gao et al. (10) and Mio et al. (11), the present studies showed that JQ1 suppressed tumor cell invasion [Fig. 5(ai–aiv)] by inhibiting key regulators to attenuate EMT [Fig. 5(bi–biv) and 5(ci–civ)]. One hallmark of human ATC is aggressive metastasis. The finding that JQ1 was effective in suppressing invasion of tumor cells further highlighted its beneficial effects as a potential therapy for ATC.
The transcriptomic hallmarks of ATC were recently reported from analysis of 33 ATC tumors (1). Many genetic changes were identified in these tumors, including mutations of BRAF, RAS, PI3CA, PTEN, telomerase reverse transcription (TERT) promoters, TP53, genes in the components of the SWI/SNF chromatin-remodeling complex, and genes encoding epigenetic modifiers (1). These complex and diverse genetic changes would make the choice of an effective treatment modality for ATC patients challenging because multi-aberrantly activated pathways would need to be considered. Adding to the complexity are the progressive dynamic changes of genetic alterations during carcinogenesis. Recent studies have documented that the approach of targeting pathway-by-pathway in thyroid cancer treatment may be less effective because of the ultimate development of drug resistance (26, 27, 31, 32).
The present studies suggest that epigenetic modification of chromatin by inhibitors of BET proteins to suppress the gene transcription of oncogenic drivers could be an effective treatment in ATC. In humans, there are 46 bromodomain-containing proteins. Despite highly conserved structural features, they recognize diverse natural ligands. The discovery in 2010 of JQ1 and I-BET762 as potent inhibitors of BET proteins sparked an intense search for additional antagonists for bromodomain proteins for therapeutic applications. The efficacy of JQ1 in attenuating tumor growth has been shown in a number of myeloid-derived tumors such as acute myeloid leukemia and multiple myeloma (19–21) and thyroid anaplastic tumors in mouse models (17). The present studies showed that JQ1 was effective in reducing tumor cell growth in cell-based studies and xenograft mouse models. Moreover, BET inhibitors structurally similar to JQ1 are being tested in clinical trials for a variety of cancers including NUT midline carcinoma (https://www.cinicaltrials.gov/ct2/results?term=bet+inhibitors&search). It is tempting to speculate that other oncogenic drivers, such as TERT, in addition to MYC may be inhibited at the transcriptional levels via epigenetic modifications of bromodomain proteins or other components of chromatin-remodeling complexes. Determining whether such antagonists can be developed for potential treatment of ATC awaits future studies.
Acknowledgments
This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health. K.E. was supported by a grant from the Mochida Memorial Foundation for Medical and Pharmaceutical Research, Japan.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ATC
- anaplastic thyroid cancer
- BET
- bromodomain and extraterminal domain
- CDK4
- cyclin-dependent kinase 4
- DMSO
- dimethyl sulfoxide
- EMT
- epithelial-mesenchymal transition
- HEXIM1
- hexamethylene bis-acetamide inducible 1
- IgG
- immunoglobulin G
- IHC
- immunohistochemistry
- mRNA
- messenger RNA
- PBS
- phosphate-buffered saline
- p-Rb
- phosphorylated Rb
- p-TEFb
- positive transcription elongation factor b
- RNAPII
- RNA polymerase II.
References
- 1.Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho JC, Krishnamoorthy GP, Xu B, Schultz N, Berger MF, Sander C, Taylor BS, Ghossein R, Ganly I, Fagin JA. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126(3):1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee HJ, Yun HJ, Kim S. Lenvatinib in radioiodine-refractory thyroid cancer. N Engl J Med. 2015;372(19):1868. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. [DOI] [PubMed] [Google Scholar]
- 4.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149(1):214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA. 2012;109(47):19408–19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, Gao Y, Cheng KA, Cohoon TJ, Qi J, Akbay E, Kimmelman AC, Kung AL, Bradner JE, Wong KK. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin Cancer Res. 2013;19(22):6183–6192. [DOI] [PMC free article] [PubMed]
- 9.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, Iyer MK, Jing X, Wu YM, Cao X, Qin ZS, Wang S, Feng FY, Chinnaiyan AM. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao X, Wu X, Zhang X, Hua W, Zhang Y, Maimaiti Y, Gao Z, Zhang Y. Inhibition of BRD4 suppresses tumor growth and enhances iodine uptake in thyroid cancer. Biochem Biophys Res Commun. 2016;469(3):679–685. [DOI] [PubMed] [Google Scholar]
- 11.Mio C, Lavarone E, Conzatti K, Baldan F, Toffoletto B, Puppin C, Filetti S, Durante C, Russo D, Orlacchio A, Di Cristofano A, Di Loreto C, Damante G. MCM5 as a target of BET inhibitors in thyroid cancer cells. Endocr Relat Cancer. 2016;23(4):335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Zhao L, Park JW, Willingham MC, Cheng SY. Synergistic signaling of KRAS and thyroid hormone receptor β mutants promotes undifferentiated thyroid cancer through MYC up-regulation. Neoplasia. 2014;16(9):757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marlow LA, D’Innocenzi J, Zhang Y, Rohl SD, Cooper SJ, Sebo T, Grant C, McIver B, Kasperbauer JL, Wadsworth JT, Casler JD, Kennedy PW, Highsmith WE, Clark O, Milosevic D, Netzel B, Cradic K, Arora S, Beaudry C, Grebe SK, Silverberg ML, Azorsa DO, Smallridge RC, Copland JA. Detailed molecular fingerprinting of four new anaplastic thyroid carcinoma cell lines and their use for verification of RhoB as a molecular therapeutic target. J Clin Endocrinol Metab. 2010;95(12):5338–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeb AN, Li W, Lin RY. Bioluminescent human thyrospheres allow noninvasive detection of anaplastic thyroid cancer growth and metastases in vivo. Thyroid. 2014;24(7):1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marlow LA, Bok I, Smallridge RC, Copland JA. RhoB upregulation leads to either apoptosis or cytostasis through differential target selection. Endocr Relat Cancer. 2015;22(5):777–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furumoto H, Ying H, Chandramouli GV, Zhao L, Walker RL, Meltzer PS, Willingham MC, Cheng SY. An unliganded thyroid hormone beta receptor activates the cyclin D1/cyclin-dependent kinase/retinoblastoma/E2F pathway and induces pituitary tumorigenesis. Mol Cell Biol. 2005;25(1):124–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X, Enomoto K, Zhao L, Zhu YJ, Willingham MC, Meltzer PS, Qi J, Cheng SY. Bromodomain and extraterminal protein inhibitor JQ1 suppresses thyroid tumor growth in a mouse model. Clin Cancer Res. 2017;23(2):430–440. [DOI] [PMC free article] [PubMed]
- 18.von Roemeling CA, Marlow LA, Pinkerton AB, Crist A, Miller J, Tun HW, Smallridge RC, Copland JA. Aberrant lipid metabolism in anaplastic thyroid carcinoma reveals stearoyl CoA desaturase 1 as a novel therapeutic target. J Clin Endocrinol Metab. 2015;100(5):E697–E709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coudé MM, Braun T, Berrou J, Dupont M, Bertrand S, Masse A, Raffoux E, Itzykson R, Delord M, Riveiro ME, Herait P, Baruchel A, Dombret H, Gardin C. BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget. 2015;6(19):17698–17712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, Rodig SJ, Kung AL, Bradner JE, Weinstock DM. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120(14):2843–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaidos A, Caputo V, Gouvedenou K, Liu B, Marigo I, Chaudhry MS, Rotolo A, Tough DF, Smithers NN, Bassil AK, Chapman TD, Harker NR, Barbash O, Tummino P, Al-Mahdi N, Haynes AC, Cutler L, Le B, Rahemtulla A, Roberts I, Kleijnen M, Witherington JJ, Parr NJ, Prinjha RK, Karadimitris A. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood. 2014;123(5):697–705. [DOI] [PubMed] [Google Scholar]
- 22.Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, Tyner AL. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci USA. 2001;98(8):4510–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ III. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108(40):16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65(10):3980–3985. [DOI] [PubMed] [Google Scholar]
- 25.Dey A, Chao SH, Lane DP. HEXIM1 and the control of transcription elongation: from cancer and inflammation to AIDS and cardiac hypertrophy. Cell Cycle. 2007;6(15):1856–1863. [DOI] [PubMed] [Google Scholar]
- 26.Antonelli A, Fallahi P, Ulisse S, Ferrari SM, Minuto M, Saraceno G, Santini F, Mazzi V, D’Armiento M, Miccoli P. New targeted therapies for anaplastic thyroid cancer. Anticancer Agents Med Chem. 2012;12(1):87–93. [DOI] [PubMed] [Google Scholar]
- 27.Deshpande HA, Roman S, Sosa JA. New targeted therapies and other advances in the management of anaplastic thyroid cancer. Curr Opin Oncol. 2013;25(1):44–49. [DOI] [PubMed] [Google Scholar]
- 28.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, Huthmacher C, Gudgin E, Lugo D, Beinke S, Chapman TD, Roberts EJ, Soden PE, Auger KR, Mirguet O, Doehner K, Delwel R, Burnett AK, Jeffrey P, Drewes G, Lee K, Huntly BJ, Kouzarides T. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devaraj SG, Fiskus W, Shah B, Qi J, Sun B, Iyer SP, Sharma S, Bradner JE, Bhalla KN. HEXIM1 induction is mechanistically involved in mediating anti-AML activity of BET protein bromodomain antagonist. Leukemia. 2016;30(2):504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viola D, Valerio L, Molinaro E, Agate L, Bottici V, Biagini A, Lorusso L, Cappagli V, Pieruzzi L, Giani C, Sabini E, Passannati P, Puleo L, Matrone A, Pontillo-Contillo B, Battaglia V, Mazzeo S, Vitti P, Elisei R. Treatment of advanced thyroid cancer with targeted therapies: ten years of experience. Endocr Relat Cancer. 2016;23(4):R185–R205. [DOI] [PubMed] [Google Scholar]
- 32.Hsu KT, Yu XM, Audhya AW, Jaume JC, Lloyd RV, Miyamoto S, Prolla TA, Chen H. Novel approaches in anaplastic thyroid cancer therapy. Oncologist. 2014;19(11):1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]