Regular exercise has cardiovascular and metabolic health benefits resulting from multiorgan physiologic adaptations in response to physical activity. At the cellular level these adaptations entail transcriptional changes that lead to enhanced metabolic and cardiovascular capacities. One major organ undergoing dramatic changes in response to exercise is the skeletal muscle, where training increases oxidative myofibers, mitochondrial volume/size and respiratory capacity, fatty acid oxidation, and glucose metabolism (1, 2). These exercise-driven changes impart endurance, metabolic efficiency, and insulin sensitivity to the skeletal muscle. Conversely, inactivity, aging, and metabolic diseases such as diabetes are generally linked with a decrease in aerobic capacity and performance in the muscle (3, 4). There has been a long-standing interest in delineating the molecular pathways mediating exercise-induced adaptations in the muscle, which in turn can guide therapeutic replication of exercise benefits.
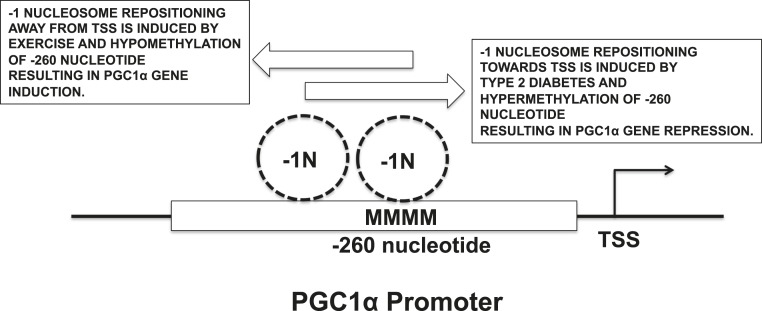
Studies in the last two decades have identified key transcriptional regulators that encode various metabolic and contractile features of the skeletal muscle (1), the most prominent of which is the nuclear receptor coregulator peroxisome proliferator-activated receptor co-activator 1 α (PGC1α). PGC1α transcribes a gene program that enriches the muscle with highly oxidative, mitochondria-rich myofibers, which also express fatigue-resistant slow-contractile proteins (5). Although PGC1α is dynamically regulated by exercise, mechanism of its induction is only starting to be explored. Recent studies identified epigenetic modifications via DNA methylation/demethylation as a molecular means of regulating muscle PGC1α gene expression in exercise and type 2 diabetes (6, 7). PGC1α gene expression (and consequently its target mitochondrial genes and density) is controlled by DNA methyltransferase 3B dependent–methylation of non-CPG nucleotides in the PGC1α promoter, particularly at the −260 position with reference to the transcriptional start site (TSS). Free-fatty acid or tumor necrosis factor-α–stimulated human myotubes, as well as muscle biopsies from patients with type 2 diabetes, exhibit hypermethylation of PGC1α promoter, concomitant with decreased PGC1α transcript levels as well as metabolic genes and mitochondrial density. On the other hand, acute exercise in humans promotes hypomethylation of PGC1α promoter (and other endurance gene promoters), thus increasing PGC1α and metabolic gene expression in the skeletal muscles. In a study published in the current issue, Bajpeyi et al. (8) further unravel the epigenetic mechanism of PGC1α gene regulation (Fig. 1). The highlight of this study is that PGC1α promoter methylation status at −260 nucleotide directly correlates with −1 nucleosome positioning at this location. Hypomethylation at −260 nucleotide in PGC1α promoter correlates with −1 nucleosome repositioning away from the TSS, presumably exposing important cis regulatory elements in the PGC1α promoter. The methylation/nucleosome positioning in PGC1α promoter directly affects the cofactor expression and, in turn, metabolic genes, mitochondrial genes, and exercise responsiveness of serum insulin and intramyocellular lipids.
Figure 1.
Mechanism of PGC1α gene regulation in exercise and diabetes. −1N, −1 nucleosome; M, methylation.
In muscles samples collected pre- and postexercise, the authors first observed that exercise bouts induced repositioning of −1 nucleosome away from the TSS in the PGC1α promoter. Conversely, in primary human myotubes from patients with type 2 diabetes, they detected an increased nucleosome occupancy (at −1 position) compared with the lean individuals, which is reversible by treatment with exercise mimetic agents. Therefore, the −1 nucleosome in the PGC1α promoter exhibits positional shifting. Exercise promotes a nucleosome shift away from the TSS, whereas metabolic syndrome results in a shift toward the TSS. This shift in −1 nucleosome directly correlated with the PGC1α expression, such that repositioning away from TSS and promoter hypomethylation leads to an increase in PGC1α expression.
The authors also identify a potential epigenetic mechanism for interindividual differences typically observed in responsiveness to exercise. They found that the high responders (to exercise) showed a significantly greater decrease in −260 nucleotide methylation in PGC1α promoter compared with the low responders. Consequently, PGC1α transcript is induced more in the high responders. The exercise-induced decrease in insulin levels as well as intramyocellular lipid content is also greater in the high responders compared with low responders, correlating with the extent of epigenetic PGC1α promoter modification.
These findings, although shading new light on epigenetic exercise and metabolic regulation in the skeletal muscle, simultaneously raise many exciting questions for future investigation. What is the temporal link between promoter methylation and nucleosome occupancy? Is methylation and nucleosome repositioning restricted to regulators such as PGC1α and a handful of PGC1α-targeted metabolic genes, and how generalized in the epigenetic regulation to exercise sensitive genes in muscle (or other tissues such as the heart)? Is methylation/nucleosome repositioning by exercise reversible, and therefore a potential mechanism for transient effects of exercise? Along the same lines, are these transcriptional modifications more sustainable with long-term exercise? Is hypermethylation of muscle genes a feature of inactivity and sarcopenia, both of which are related to exercise and metabolic inefficiencies? More importantly, are DNA methyltransferases potential targets for promoting hypomethylation, exercise mimesis, and therefore insulin sensitivity in the skeletal muscle? In summary, the authors report an important link between exercise and PGC1α regulation through promoter methylation and nucleosome repositioning, which have important implications for understanding exercise and metabolic regulation, raising the possibility for exercise mimesis.
Acknowledgments
Acknowledgments
This work was supported by the UTHealth intramural source and The Welch Foundation endowment in Chemistry & Related Science Grant L-AU-0002, as well as American Diabetes Association Grant ADA 1-13-BS-127 and National Institutes of Health/National Heart, Lung and Blood Institute Grant 1 R01 HL129191-01.
Disclosure Summary: The author has nothing to disclose.
Footnotes
- PGC1α
- peroxisome proliferator-activated receptor co-activator 1 α
- TSS
- transcriptional start site.
References
- 1.Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17(2):162–184. [DOI] [PubMed] [Google Scholar]
- 2.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91(4):1447–1531. [DOI] [PubMed] [Google Scholar]
- 3.Thyfault JP, Booth FW. Lack of regular physical exercise or too much inactivity. Curr Opin Clin Nutr Metab Care. 2011;14(4):374–378. [DOI] [PubMed] [Google Scholar]
- 4.Venables MC, Jeukendrup AE. Physical inactivity and obesity: links with insulin resistance and type 2 diabetes mellitus. Diabetes Metab Res Rev. 2009;25(Suppl 1):S18–S23. [DOI] [PubMed] [Google Scholar]
- 5.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. [DOI] [PubMed] [Google Scholar]
- 6.Barrès R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O’Gorman DJ, Zierath JR. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15(3):405–411. [DOI] [PubMed] [Google Scholar]
- 7.Barrès R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10(3):189–198. [DOI] [PubMed] [Google Scholar]
- 8.Bajpeyi S, Covington JD, Taylor EM, Stewart LK, Galgani JE, Henagan TM. Skeletal muscle PGC1α -1 nucleosome position and -260nt DNA methylation determine exercise response and prevent ectopic lipid accumulation in men. Endocrinology. 2017;158(7):2190–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]