The multidrug-resistance transporter MdfA is able to eject a wide variety of compounds from the cytoplasm of E. coli. As it is an integral membrane protein, obtaining diffraction-quality crystals remains a challenge. Here, it is shown that LCP crystallization results in superior crystal packing to vapour diffusion, and that LCP lipid chain-length variation has a marked effect on the morphology and diffraction behaviour of crystals of MdfA in complex with the Fab fragment YN1074.
Keywords: MFS transporter, multidrug resistance, membrane protein, crystallization, antibody fragment, lipidic cubic phase
Abstract
The active efflux of antibiotics by multidrug-resistance (MDR) transporters is a major pathway of drug resistance and complicates the clinical treatment of bacterial infections. MdfA is a member of the major facilitator superfamily (MFS) from Escherichia coli and provides resistance to a wide variety of dissimilar toxic compounds, including neutral, cationic and zwitterionic substances. The 12-transmembrane-helix MdfA was expressed as a GFP-octahistidine fusion protein with a TEV protease cleavage site. Following tag removal, MdfA was purified using two chromatographic steps, complexed with a Fab fragment and further purified using size-exclusion chromatography. MdfA and MdfA–Fab complexes were subjected to both vapour-diffusion and lipidic cubic phase (LCP) crystallization techniques. Vapour-diffusion-grown crystals were of type II, with poor diffraction behaviour and weak crystal contacts. LCP lipid screening resulted in type I crystals that diffracted to 3.4 Å resolution and belonged to the hexagonal space group P6122.
1. Introduction
Active efflux by multidrug-resistance (MDR) transporters is a major cause of bacterial resistance to many classes of antibiotics (Nikaido, 2009 ▸). MDR transporters can be classified into primary transporters [such as ATP-binding cassette (ABC) transporters] that utilize ATP hydrolysis as an energy source, and secondary transporters, which utilize the energy stored in the transmembrane electrochemical gradient. On the basis of similarities in their sequences, the secondary transporters are further categorized into at least four large superfamilies, including the major facilitator superfamily (MFS), the resistance–nodulation–division (RND) family, the multidrug and toxic compounds extrusion (MATE) family and the small multidrug-resistance (SMR) family (Putman et al., 2000 ▸). Proteins belonging to the MFS play a major role in prokaryotic MDR, yet the mechanism of drug transport is not entirely clear. The current accepted paradigm is that MFS transporters utilize a ‘rocker-switch alternating access’ mechanism, whereby the N-terminal and C-terminal six-helix bundles rotate with respect to each other about an axis within the plane of the membrane that passes through the central substrate-binding site. This mechanism requires at least three states: inward open, outward open and a (potentially transient) occluded form that may be further divided into inward and outward occluded states (Yan, 2013 ▸; Quistgaard et al., 2016 ▸).
Sequence analysis of Escherichia coli MdfA suggested the presence of 12 transmembrane (TM) helices, a hallmark of the MFS (Edgar & Bibi, 1997 ▸; Sigal et al., 2006 ▸), which was confirmed by the recent crystal structures of ligand-bound forms of MdfA in the inward-facing state (Heng et al., 2015 ▸; Liu et al., 2016 ▸). MdfA is capable of coupling the efflux of a number of lipophilic cationic, zwitterionic and neutral substrates to the transmembrane proton (H+) or ion chemical gradient, allowing it to translocate antibiotics such as chloramphenicol, erythromycin, ethidium, tetraphenylphosphonium and rhodamine (Edgar & Bibi, 1997 ▸). A second physiological function of MdfA is found in pH regulation owing to its activity as an H+/Na+,K+ antiporter: knockout of MdfA results in bacterial growth restriction under strongly alkaline conditions (Lewinson et al., 2004 ▸).
Although MdfA can transport many structurally unrelated compounds, it has been suggested that similar conformations of the transporter are induced by the different permeant substrates (Fluman et al., 2009 ▸), implying a common transport mechanism within the framework of the rocker-switch model. Two negatively charged residues located in TM helix 1 (Glu26 and Asp34) have been identified as playing critical roles in substrate and proton transport (Edgar & Bibi, 1999 ▸; Flumanet al., 2012 ▸). The postulated transport mechanism involves competition between proton and substrate binding at these two acidic residues in the binding cavity of MdfA. Specifically, Asp34 is proposed to be involved in both proton and substrate binding (supported by the chloramphenicol-bound structure; Heng et al., 2015 ▸), while protonation of Glu26 is thought to shift the conformation of the transporter from the outward open state to the inward open state; interplay between these two sites is thought to drive transport (Fluman et al., 2012 ▸).
For a complete understanding of substrate binding and the transport mechanism, it is essential to identify and visualize additional conformational states of MdfA. Key prerequisites for structural analysis include homogenous and stable MdfA, yet such preparations remain a challenge for membrane proteins, which often suffer from poor expression levels and loss of activity after extraction from their native membranes by detergents. In addition, the resulting detergent micelle surrounding the protein may hamper the protein crystallization process and impact on the diffraction quality of membrane-protein crystals. Co-crystallization of membrane proteins with antibody fragments has been reported to be an effective means of improving the diffraction quality of membrane-protein crystals by limiting intrinsic flexibility. In addition, antibody binding increases the surface area exposed from detergent micelles, which is often thought to be critical for producing crystal contacts (Hino et al., 2013 ▸).
Prior to this study, we expressed and purified the MFS-type MDR transporter MdfA from E. coli to generate and isolate antibody Fab fragments against MdfA, with a view towards using these as potential crystallization chaperones (Hino et al., 2013 ▸). In this way, we were able to identify Fab fragments that stabilize MdfA as measured using the N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM) thermostability assay (Jaenecke et al., 2017 ▸). Here, we show that the Fab fragment YN1074 is also able to suppress pH-dependent stability changes in the transporter. The MdfA–YN1074 complex could be crystallized using both hanging-drop vapour-diffusion and lipidic cubic phase (LCP) methods, and we demonstrate that lipid screening has a significant effect on the quality of crystals grown using LCP. The best crystals grew in LCP using the lipid 1-(8Z-hexadecenoyl)-rac-glycerol (8.8 MAG) and diffracted to a maximum resolution of 3.4 Å.
2. Materials and methods
2.1. Macromolecule production
2.1.1. Materials
All general reagents and materials were purchased from Sigma–Aldrich and Carl Roth, unless otherwise specified. Ni2+–NTA resin was purchased from Qiagen. The detergents n-dodecyl-β-d-maltopyranoside (DDM), n-decyl-β-d-maltopyranoside (DM), n-nonyl-β-d-maltopyranoside (NM) and lauryl maltose neopentyl glycol (LMNG) were obtained from Anatrace (Maumee, Ohio, USA). Monoolein was obtained from Nu-Chek Prep (Elysian, Minnesota, USA) and other MAGs were purchased from Avanti Polar lipids (Alabaster, Alabama, USA). Polyethylene glycols (PEGs) for crystallization were obtained from Molecular Dimensions, whereas other materials for crystallization were obtained from Jena Biosciences, Hampton Research and Rigaku Reagents.
2.1.2. Cloning of MdfA
The mdfA gene (NCBI GenBank accession No. AAC73929.1 for E. coli K-12 substrain MG1655) was amplified from E. coli Top10 cells and cloned upstream of the TEV cleavage-site sequence (TEVcs) of pWaldo-GFPe (Drew et al., 2001 ▸) via the XhoI and KpnI restriction sites, allowing expression of the MdfA-(TEVcs)-GFP-His8 fusion protein. Two nucleotides were introduced between the gene sequences of mdfA and the TEVcs by site-directed mutagenesis in order to ensure the correct reading frame, using the oligonucleotides 5′-TCGCACGAAGGGGGTACCTATGGATCCGAAAACCTGTAC-3′ and 5′-GTACAGGTTTTCGGATCCATAGGTACCCCCTTCGTGCGA-3′. E. coli C43 (DE3) cells were transformed with this plasmid and used for overexpression of the MdfA-(TEVcs)-GFP fusion protein.
2.1.3. MdfA expression and purification
A single colony was inoculated into LB medium containing kanamycin (75 µg ml−1) at 37°C overnight. The overnight culture was diluted (1:100; an OD600 of approximately ∼0.05) in 2× YT medium supplemented with kanamycin and the cells were grown at 37°C to an optical density (OD600) of ∼0.4. The temperature was decreased to 28°C and expression of the protein was induced by the addition of 0.4 mM IPTG. Cells were harvested 6 h after induction by centrifugation at 5000g for 10 min at 4°C.
The cell pellets were resuspended in 20 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA buffer supplemented with 10 µg ml−1 DNAseI, 1 mM PMSF and then disrupted by high-pressure homogenization (APV homogenizers). Cell debris was removed by centrifugation at 10 000g for 15 min, and the membrane fraction was collected by ultracentrifugation at 100 000g for 90 min. Isolated membranes were flash-frozen in liquid nitrogen and stored at −80°C. The membrane fraction was solubilized in 150 ml solubilization buffer (25 mM Tris, 200 mM NaCl pH 7.3) containing 1% DDM; the detergents DM, NM and LMNG were also screened, but only DDM yielded a monodisperse peak in SEC. Insoluble material was removed by centrifugation at 100 000g for 1 h and the solubilized fraction was incubated with 10 ml Ni2+ beads (batch binding) equilibrated in buffer A (20 mM Tris, 150 mM NaCl, 0.02% DDM pH 7.5) for 2 h. MdfA-GFP was purified by immobilized Ni2+-affinity chromatography, with 0.02% DDM added to all buffers. The resin was washed with five column volumes (CV) of buffer A containing 20 mM imidazole, followed by 12 CV of buffer A containing 50 mM imidazole. MdfA-GFP was eluted with buffer A containing 250 mM imidazole, and fractions were pooled and exchanged with buffer A to reduce the concentration of imidazole (to ∼10 mM) before treatment with TEV protease.
MdfA-GFP in the presence of a half-molar ratio of hexahistidine (His6)-tagged TEV protease (Drew et al., 2008 ▸) was dialyzed overnight against buffer A supplemented with 1 mM β-mercaptoethanol at 4°C using a 3 kDa molecular-weight cutoff membrane. After dialysis, the sample was passed through 15 ml Ni2+–NTA resin equilibrated in dialysis buffer to separate the resulting MdfA (flowthrough) from the C-terminally His8-tagged GFP and the His6-tagged TEV protease. The fraction containing MdfA was concentrated and applied onto a Superdex 200 10/300 GL size-exclusion chromatography (SEC) column equilibrated with buffer B (10 mM MES, 20 mM NaCl, 0.02% DDM pH 7.0).
2.1.4. Preparation of Fab fragments
Fab fragments were generated as described previously (Jaenecke et al., 2017 ▸; Supplementary Fig. S1) according to established protocols (Day et al., 2007 ▸). Briefly, a proteoliposome antigen was prepared by reconstituting purified, functional MdfA at high density into phospholipid vesicles that consisted of a 10:1 mixture of egg phosphatidylcholine (Avanti Polar Lipids) and the adjuvant lipid A (Sigma) to facilitate the immune response. BALB/c mice were immunized with the proteoliposome antigen using three injections at two-week intervals. Antibody-producing hybridoma cell lines were generated using a conventional fusion protocol (Köhler & Milstein, 1975 ▸; Pontecorvo, 1976 ▸). Hybridoma clones producing antibodies against MdfA were selected by an enzyme-linked immunosorbent assay on immobilized phospholipid vesicles containing purified MdfA (liposome ELISA), allowing positive selection of those antibodies recognizing the native conformation of MdfA. Additional screening for reduced antibody binding to SDS-denatured MdfA was used to select against linear epitope-recognizing antibodies (negative selection). Whole IgG molecules, collected from large-scale culture supernatant of monoclonal hybridomas and purified using protein G affinity chromatography, were digested with papain (Nacalai) and Fab fragments were isolated using a Superdex 200 gel-filtration column followed by protein A affinity chromatography (Bio-Rad). This procedure resulted in the isolation of four MdfA-specific monoclonal antibodies (YN1006, YN1010, YN1074 and YN1082), the Fab fragments of each of which form a stable complex with the transporter that can be isolated using SEC (Jaenecke et al., 2017 ▸).
2.1.5. Preparation of MdfA–Fab fragment YN1074 complexes
Purified MdfA was incubated with Fab fragment YN1074 in a molar ratio of 1:1.5 for 16 h in buffer B prior to SEC. Peak fractions containing MdfA–YN1074 complexes were concentrated to ∼5 mg ml−1 and used for crystallization. In a second set of experiments, the pH of buffer B during both MdfA–Fab complex formation and subsequent SEC and CPM thermostability assays was modified in the range between pH 5.5 and 7.0.
2.1.6. Thermostability assays of MdfA and the MdfA–Fab complex
CPM thermostability analysis was performed as described by Alexandrov et al. (2008 ▸) with minor modifications. Briefly, 12 µl MdfA or MdfA–Fab complex (2 mg ml−1) was mixed with 45.6 µl buffer B and 2.4 µl CPM dye (at 5 mg ml−1). The reaction mixture was transferred to a clean PCR tube and heated from 25 to 90°C at a rate of 1°C min−1 in a Rotor Gene Q cycler (Qiagen). The fluorescence of the dye (excitation and emission wavelengths of 365 and 460 nm, respectively) was monitored during the heating process. Calculation of the first derivative of the melting curve (performed with the Rotor Gene Q software v.2.1.0) indicates a maximum at the apparent transition temperature/melting temperature T m of the protein.
2.2. Crystallization
MdfA and the MdfA–YN1074 complex were concentrated to 5 and 2.5 mg ml−1, respectively, using a 100 kDa molecular-weight cutoff Amicon (Millipore) prior to crystallization screening. Crystallization trials using the vapour-diffusion method were performed in 96-well sitting-drop plates using a Cartesian MicroSys NQ crystallization robot (Zinsser Analytic) with commercially available screening matrices (MemPlus and MemGold2 from Molecular Dimensions as well as Wizard I, II, III and IV from Rigaku Reagents). Droplets containing equal volumes of reservoir solution (200 nl) and protein solution (200 nl) were incubated against 70 µl of each reservoir solution at 16°C.
Initial LCP crystallization setups were made by mixing MdfA or the MdfA–YN1074 complex at 2.5, 5 or 10 mg ml−1 with monoolein acyl-glycerol (9.9 MAG) in a 2:3 ratio using the two-syringe coupling method (Caffrey & Cherezov, 2009 ▸). Protein-containing LCP (100 nl) was dispensed over each well of the Laminex glass plate (Molecular Dimensions) using a LISSYII robot (Zinsser Analytic) and overlaid with 1 µl precipitant solution from commercially available screening matrices (MemGold, MemGold2, MemStart+MemSys and MemMeso from Molecular Dimensions, JBScreen Membrane and JBScreen Pentaerythritol from Jena Bioscience, Crystal Screen HT, MemFac HT and Index HT from Hampton Research and Wizard 1 & 2 from Rigaku Reagents) for initial screening. Subsequently, the MAG lipids were varied for the MdfA–YN1074 complex using lipid mixing ratios of 1:1 {7.7 MAG [1-(7Z-tetradecenoyl)-rac-glycerol] and 7.8 MAG [1-(7Z-pentadecenoyl)-rac-glycerol]} and 2:3 {7.9 MAG [1-(7Z-hexadecenoyl)-rac-glycerol] and 8.8 MAG [1-(8Z-hexadecenoyl)-rac-glycerol]}. All crystallization trials were performed at 20°C.
2.3. Data collection and processing
Prior to data collection, single crystals of MdfA and the MdfA–YN1074 complex were harvested and flash-cooled directly in liquid nitrogen without additional cryoprotection. All data were collected on beamline PXI (X06SA) at the Swiss Light Source (SLS). For the MdfA and MdfA–YN1074 complex crystals grown using vapour diffusion, diffraction data sets were collected at 100 K using a PILATUS 6M detector, whereas diffraction data for the MdfA–YN1074 complex obtained from LCP were collected using an EIGER 16M detector. Diffraction data were processed and integrated using XDS (Kabsch, 2010 ▸). Molecular replacement was performed with Phaser (McCoy et al., 2007 ▸) to analyse crystal packing using the coordinates of MdfA (PDB entry 4zp0; Heng et al., 2015 ▸) and, where appropriate, a Fab fragment (PDB entry 1ibg; Jeffrey et al., 1995 ▸) as search models (details of the structure solution and analysis will be presented elsewhere). Buried surface areas were calculated using the PISA server (Krissinel & Henrick, 2007 ▸) and crystallographic figures were prepared using PyMOL (Schrödinger).
3. Results and discussion
3.1. Cloning, expression and purification of MdfA
The PCR fragment coding for MdfA was successfully inserted into the XhoI and KpnI sites of pWaldo-GFPe, which was then transformed into E. coli C43 (DE3) cells (Table 1 ▸). The expression level of MdfA-GFP was monitored by GFP fluorescence emission at 512 nm (excitation wavelength of 488 nm) using an ImageQuant LAS 4000 (GE Healthcare). Following isolation of MdfA-GFP by single-step immobilized metal-affinity chromatography (IMAC), untagged MdfA was obtained via TEV cleavage and a subsequent second IMAC step, resulting in >90% purity (Fig. 1 ▸). Approximately 0.3–0.4 mg of purified MdfA was routinely obtained from 1 l of 2× YT medium.
Table 1. MdfA production information.
| Source organism | E. coli |
| DNA source | E. coli Top10 |
| Forward primer | AGGAGACTCGAGATGCAAAATAAATTAGCT |
| Reverse primer | TTTCGGATCCATAGGTACCCCTTCGTGCGAA |
| Expression vector | pWaldo-GFPe |
| Expression host | E. coli C43 (DE3) |
| Complete amino-acid sequence of the construct produced | MQNKLASGARLGRQALLFPLCLVLYEFSTYIGNNMIQPGMLAVVEQYQAGIDWVPTSMTAYLAGGMFLQWLLGPLSDRIGRRPVMLAGVVWFIVTCLAILLAQNIEQFTLLRFLQGISLCFIGAVGYAAIQESFEEAVCIKITALMANVALIAPLLGPLVGAAWIHVLPWEGMFVLFAALAAISFFGLQRAMPETATRIGEKLSLKELGRDYKLVLKNGRFVAGALALGFVSLPLLAWIAQSPIIIITGEQLSSYEYGLLQVPIFGALIAGNLLLARLTSRRTVRSLIIMGGWPIMIGLLVAAAATVISSHAYLWMTAGLSIYAFGIGLANAGLVRLTLFASDMSKGTVSAAMGMLQMLIFTVGIEISKHAWLNGGNGLFNLFNLVNGILWLSLMVIFLKDKQMGNSHEG |
Figure 1.
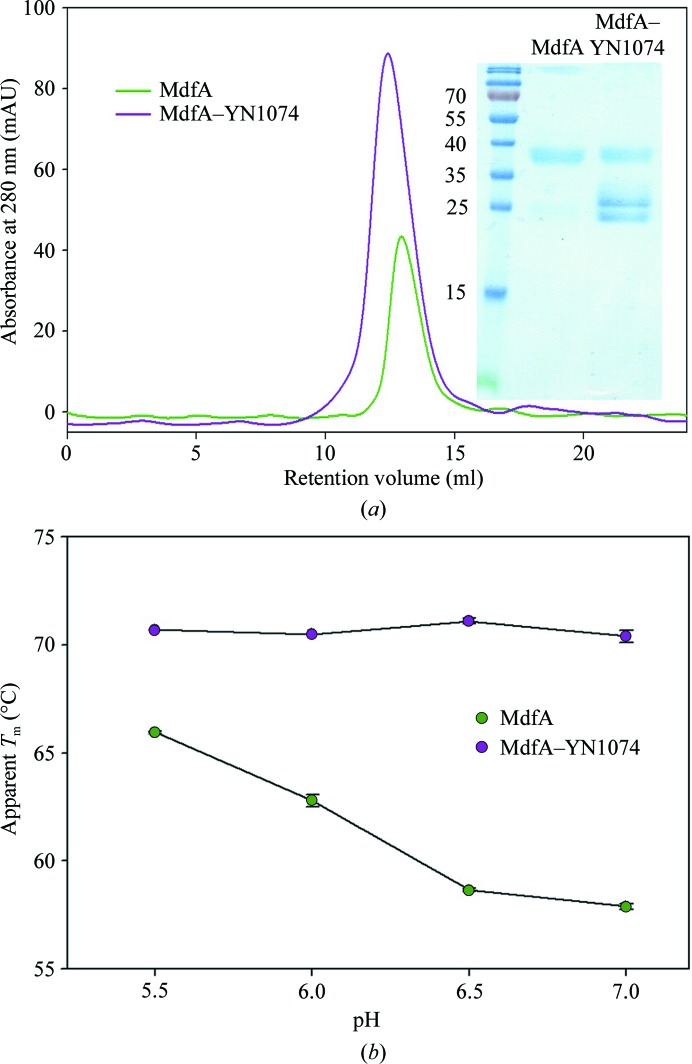
Purification of the MdfA–YN1074 complex and pH-dependent thermostability analyses of MdfA and the MdfA–YN1074 complex. (a) Size-exclusion chromatograms of MdfA (green) and MdfA–YN1074 (purple) on a Superdex 200 10/300 GL size-exclusion column. Inset: SDS–PAGE analysis of the main SEC peaks. (b) Thermostability of MdfA/the MdfA–YN1074 complex as a function of pH was assessed using the CPM thermal denaturation assay. Apparent T m values for MdfA and the MdfA–YN1074 complex were evaluated from the first derivative of the melting curve.
3.2. Effect of Fab fragments on MdfA stability
At pH 7.0, the melting curve of MdfA shows an apparent transition temperature T m of ∼58°C in the CPM assay, which is increased by ∼4°C in the complexes with Fabs YN1006, YN1010 and YN1082 and by ∼12°C in the presence of Fab YN1074 (Jaenecke et al., 2017 ▸). The thermostability of purified MdfA and the isolated MdfA–Fab YN1074 complex were further analysed as a function of pH (Fig. 1 ▸ b). Interestingly, the antiporter exhibits an increased thermostability at lower pH values (T m of ∼66°C at pH 5.5). In contrast, the complex of MdfA with the YN1074 Fab possesses a near-constant T m of ∼71°C at all tested pH values, demonstrating a stabilization of MdfA by the Fab of 5–12°C. The lack of variation of the T m of the complex with pH suggests that YN1074 stabilizes the low-pH form of the antiporter and that MdfA–YN1074 may be suitable for crystallization screening in a wide range of pH conditions.
3.3. Crystallization of MdfA
In the initial crystallization trials, we used commercially available screening kits with the sitting-drop vapour-diffusion method (Supplementary Fig. S1). Microcrystals of MdfA were observed after 1–2 weeks from a number of conditions containing PEG 400 as the precipitant (e.g. 0.1 M MES pH 6.0, 0.2 M Li2SO4, 25–30% PEG 400). These conditions were optimized to improve the crystal morphology using the hanging-drop vapour-diffusion method. The largest crystals were obtained in 100 mM Tris pH 7.5, 100 mM NaCl, 100 mM Li2SO4, 28–30% PEG 400 (MdfA-VD; Fig. 2 ▸ a, Table 2 ▸), which diffracted to resolutions lower than 7 Å (Supplementary Fig. S2a). Processing of the diffraction data demonstrated that the MdfA-VD crystal belonged to the hexagonal space group P6122 or P6522, with unit-cell parameters a = b = 94.5, c = 663.1 Å (Table 3 ▸).
Figure 2.
Crystals of MdfA and MdfA–YN1074. Crystals of (a) MdfA and (b) the MdfA–YN1074 complex grown by the hanging-drop vapour-diffusion method, as well as of the MdfA–YN1074 complex grown using the LCP method with various host lipids: (c) 9.9 MAG, (d) 7.7 MAG, (e) 7.8 MAG, (f) 7.9 MAG and (g) 8.8 MAG.
Table 2. Crystallization of MdfA and MdfA–YN1074.
| Protein | MdfA | MdfA–YN1074 | MdfA–YN1074 |
|---|---|---|---|
| Method | Hanging-drop vapour diffusion | Hanging-drop vapour diffusion | Lipidic cubic phase |
| Temperature (K) | 289 | 289 | 293 |
| Protein concentration (mg ml−1) | 5 | 2.5 | 2.5 |
| Buffer composition of protein solution | 10 mM MES, 20 mM NaCl, 0.02% DDM pH 7.0 | 10 mM MES, 20 mM NaCl, 0.02% DDM pH 7.0 | 10 mM MES, 20 mM NaCl, 0.02% DDM pH 7.0 |
| Composition of reservoir solution | 100 mM Tris pH 7.5, 100 mM NaCl, 100 mM Li2SO4, 28–30% PEG 400 | 100 mM HEPES pH 7, 100 mM CdCl2, 100 mM LiCl, 24–28% PEG 400 | 100 mM ADA pH 6.5, 100 mM NaCl, 100 mM Li2SO4, 32–36% PEG 300 |
| Volume and ratio of drop | 2 µl (1:1) | 2 µl (1:1) | 100 nl |
| Volume of reservoir | 1 ml | 1 ml | 1 µl |
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Protein | MdfA-VD (hanging-drop vapour diffusion) | MdfA–YN1074-VD (hanging-drop vapour diffusion) | MdfA–YN1074-LCP (lipidic cubic phase method) |
|---|---|---|---|
| Diffraction source | PXI (X06SA), SLS | PXI (X06SA), SLS | PXI (X06SA), SLS |
| Wavelength (Å) | 1.000 | 1.000 | 1.000 |
| Temperature (K) | 100 | 100 | 100 |
| Detector | PILATUS 6M | PILATUS 6M | EIGER 16M |
| Space group | P6522 | P212121 | P6122 |
| a, b, c (Å) | 94.5, 94.5, 663.1 | 76.6, 141.6, 296.6 | 73.2, 73.2, 927.9 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 | 90, 90, 120 |
| Resolution range (Å) | 50–7.82 (8.28–7.82) | 50–7.06 (7.48–7.06) | 49–3.40 (3.61–3.40) |
| Total No. of reflections | 39028 | 32684 | 384466 |
| No. of unique reflections | 3376 | 9469 | 22224 |
| CC1/2 (%) | 100 (69.0) | 99.9 (76.5) | 100 (58.3) |
| R meas (%) | 9.7 (122.2) | 6.0 (61.4) | 26.2 (172.4) |
| 〈I/σ(I)〉 | 9.04 (1.65) | 11.49 (2.01) | 11.57 (1.59)† |
| Completeness (%) | 99.1 (98.4) | 98.3 (94.3) | 99.9 (99.9) |
| Multiplicity | 9.97 (9.77) | 3.45 (3.44) | 17.3 (16.03) |
| Mosaicity (°) | 0.229 | 0.286 | 0.097 |
| Solvent content (%) | 74.5 | 71.6 | 68.1 |
| No. of molecules/complexes per asymmetric unit | 2 | 2 | 1 |
The I/σ(I) falls below 2.0 at 3.4 Å resolution. The resolution cutoff was determined by the CC1/2 value (Karplus & Diederichs, 2012 ▸), which is 58.3% (our cutoff value is below 50%).
In parallel, crystallization conditions for MdfA in complex with the four Fabs were screened using vapour diffusion, and (with the exception of Fab YN1010) crystals were obtained (Supplementary Fig. S1). Nevertheless, the crystals of MdfA–YN1006 and MdfA–YN1082 diffracted poorly, with a maximum resolution of ∼30 Å. Only those of the MdfA–YN1074 complex, obtained in 100 mM HEPES pH 7, 100 mM CdCl2, 100 mM LiCl, 24–28% PEG 400 within 3–5 d (MdfA–YN1074-VD; Fig. 2 ▸ b, Table 2 ▸), diffracted to a maximal resolution between 6 and 7 Å (Supplementary Fig. S2b). The MdfA–YN1074-VD crystal belonged to the orthorhombic space group P212121, with unit-cell parameters a = 76.6, b = 141.6, c = 296.6 Å (Table 3 ▸).
The limitations in obtaining diffraction-quality crystals via the vapour-diffusion method prompted us to explore the lipidic cubic phase (LCP) technique. The LCP medium is ubiquitously used as an alternative to detergent micelles during the crystallization of membrane proteins (Caffrey, 2015 ▸). Needle-shaped crystals of the MdfA–YN1074 complex appeared in LCP using 9.9 MAG (the most frequently used host lipid in initial LCP trials; Caffrey, 2015 ▸) in 100 mM Tris pH 7.0, 100 mM NaCl, 100 mM Li2SO4, 40–44% PEG 300 (Fig. 2 ▸ c). These crystals, which belonged to the hexagonal space group P6122 or P6522, with unit-cell parameters a = b = 73.3, c = 950.1 Å, showed weak diffraction to 8 Å resolution.
We then screened the alkyl-chain length of the host lipid between 14-C and 18-C, which is thought to improve the partitioning of the membrane protein into the lipid and to influence the curvature of the bicontinuous lipidic bilayer to optimize the size of the aqueous channels to accommodate the bound Fab (Li et al., 2013 ▸). Selection of lipids was informed empirically by reported membrane-protein structures grown by the LCP method (Caffrey, 2015 ▸). Crystallization in 7.7, 7.8 and 7.9 MAG generated small hexagonal crystals (<50 µm) in 100 mM ADA pH 6.5, 100 mM NaCl, 100 mM Li2SO4, 24–26% PEG 300 (7.7 MAG), 100 mM ADA pH 6, 100 mM NaCl, 100 mM Li2SO4, 28–30% PEG 300 (7.8 MAG) or 100 mM ADA pH 6.5, 100 mM NaCl, 100 mM Li2SO4, 32–36% PEG 300 (7.9 MAG) (Figs. 2 ▸ d, 2 ▸ e and 2 ▸ f), yet the diffraction quality remained limited.
Crystals grown in 8.8 MAG as a host lipid appeared within one week in 100 mM ADA pH 6.5, 100 mM NaCl, 100 mM Li2SO4, 32–36% PEG 300 and matured to full size within between five and seven weeks, when they were harvested (MdfA–YN1074-LCP; Fig. 2 ▸ g, Table 2 ▸). The morphology of these larger crystals was hexagonal, and their maximal size was 80–100 µm. These crystals diffracted to a resolution of slightly over 3.0 Å (Supplementary Fig. S2c). Owing to the presence of a very long c axis, the crystals were mounted perpendicular to the beam with a slight tilt to best resolve the closely spaced diffraction spots. Anisotropy of the diffraction data restricted the resolution of the data set to 3.4 Å. The MdfA–YN1074-LCP crystal belonged to the hexagonal space group P6122 or P6522, with unit-cell parameters a = b = 73.2, c = 927.9 Å (i.e. related to those obtained using 9.9 MAG but with a 22 Å shorter c axis). Data-collection and processing statistics are summarized in Table 3 ▸.
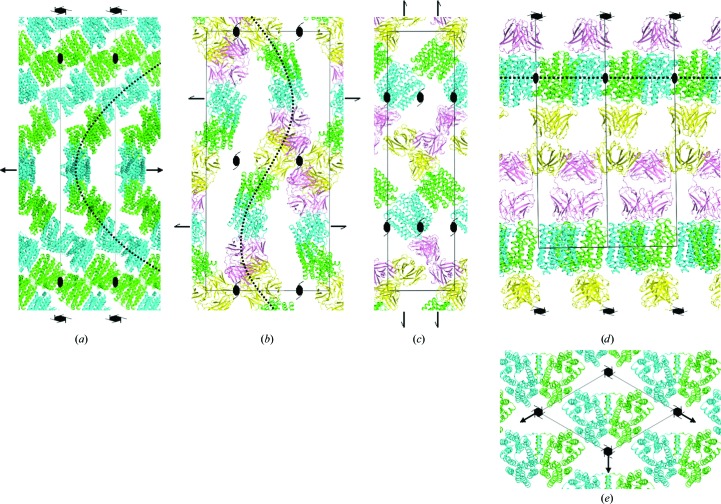
The crystal packing in three of the crystal forms was analysed following molecular replacement (details of the structure solution and analysis will be presented elsewhere). Two MdfA molecules (solvent content of 74.5%) could be located in the asymmetric unit of the MdfA-VD crystal (Fig. 3 ▸ a), two transporter–Fab complexes (solvent content of 71.6%) in that of MdfA–YN1074-VD (Figs. 3 ▸ b and 3 ▸ c) and one complex (solvent content 68.1%) in the asymmetric unit of MdfA–YN1074-LCP (Figs. 3 ▸ d and 3 ▸ e). In each crystal form, individual MdfA molecules associate laterally via their transmembrane regions (although the residues that contact each other differ), with adjacent monomers facing in opposite directions (Fig. 3 ▸).
Figure 3.
Crystal packing in MdfA crystals. Packing arrangements of (a) the transporter (MdfA-VD), (b, c) the MdfA–YN1074 complex within vapour-diffusion-grown (MdfA–YN1074-VD) crystals and (d, e) the MdfA–YN1074 membrane-protein complex within lipidic cubic phase-grown crystals (MdfA–YN1074-LCP). Selected symmetry-element symbols are shown for orientation. (a) The two MdfA monomers (green, cyan) within the asymmetric unit of MdfA-VD align to form infinite superhelical chains (dotted line) that are stabilized by lateral hydrophobic contacts. Individual chains contact each other via a small number of hydrophilic contacts. View parallel to the crystallographic a axis. (b) Crystal contacts in MdfA–YN1074-VD are dominated by interactions between the Fabs (yellow, pink) from symmetry-related molecules. Within the lattice, the molecules are arranged in rippled layers (dotted line), with major contacts between the layers provided by the Fabs. The view is parallel to the crystallographic a axis. (c) One layer from (b), viewed parallel to the crystallographic b axis. (d) In the MdfA–YN1074-LCP crystals, MdfA (green; for clarity, twofold symmetry-related molecules are shown in cyan, although these are crystallographically equivalent) is found in a two-dimensional membrane-like array. MdfA layers sandwich those of the Fab YN1074 (yellow; pink), resulting in a favourable segregation of hydrophobic and hydrophilic crystal contacts. The view is parallel to the crystallographic a axis. (e) The MdfA layer from (d), viewed parallel to the crystallographic c axis.
In the MdfA-VD crystal (Fig. 3 ▸ a), hydrophobic TM-helix residues are responsible for most intermolecular contacts, with buried surface areas of 800 Å2 between monomers within the asymmetric unit and 614 Å2 between crystallographically related monomers. This results in the formation of superhelical ‘chains’ of MdfA molecules with their helix axes parallel to the crystallographic sixfold screw axis. The contacts between adjacent chains, involving residues from the hydrophilic cytoplasmic and periplasmic surfaces of the transporter, are weak, burying a surface area of only 97 Å2. The large spaces observed between the chains are presumably occupied by detergent micellar structures.
As expected, the intermolecular contacts in the MdfA–YN1074-VD crystal (Figs. 3 ▸ b and 3 ▸ c) are dominated by multiple Fab–Fab interactions, which bury a total surface area of 1179 Å2. This is comparable to that of the MdfA–YN1074 interface (936 Å2). Lateral hydrophobic contacts between MdfA molecules are restricted to the interface within the asymmetric unit, with a buried surface area of 707 Å2, whereas crystal contacts between the periplasmic faces of MdfA bury 307 Å2. The complexes are arranged in rippled stacks within the crystals, with inter-stack contacts provided by the Fabs (Fig. 3 ▸ c). As in the MdfA-VD crystals, the interlayer spaces between MdfA molecules provide space for (presumably disordered) detergent.
In contrast to the type II membrane-protein crystals formed using vapour diffusion, the MdfA–YN1074-LCP crystal is of type I, with the MdfA molecules forming an infinite two-dimensional array as in a membrane, albeit with alternate facing monomers (Figs. 3 ▸ d and 3 ▸ e). Within this two-dimensional layer, two sets of lateral hydrophobic contacts are made, burying 1189 and 768 Å2. Alternate layers are connected by Fab–Fab interactions that bury a total surface area of 900 Å2. The favourable partitioning of intramembrane and hydrophilic contacts observed in the packing of these crystals is presumably responsible for their superior diffraction qualities.
Although the data presented here by no means present a complete picture of the membrane-protein crystallization process, some conclusions may be drawn. Thus, it seems that the lipidic cubic phase facilitates optimal lateral contacts between membrane-protein molecules, supported by the fact that type I crystal formation appears to be typical for crystals obtained in LCP (Caffrey, 2015 ▸). In contrast, the arrangements in the vapour-diffusion crystals suggest that masking of the hydrophobic membrane-facing surfaces by detergent molecules prevents such two-dimensional arrangements. Coupled with the need to accommodate bulky disordered detergent micellar structures within the lattice, this results in weak crystal contacts and therefore poor diffraction. As observed previously (Hino et al., 2013 ▸), complexation with antibody fragments can (in addition to stabilizing a particular conformation in flexible membrane proteins) increase the likelihood of obtaining three-dimensional crystals. In our case, however, it appears that Fab crystal contacts should be balanced by favourable membrane-protein interactions within the lattice for suitable diffraction properties, as observed in our LCP-grown crystals. Finally, the nature of the host lipid exhibits a pronounced influence both on the morphology and the diffraction quality of the LCP crystals. The fact that we observe a substantial change in the c axis for crystals grown using 8.8 MAG and 9.9 MAG suggests that the different lipids influence the two-dimensional packing of the membrane-protein layer and/or the orientation of the MdfA molecules, in turn influencing the positioning of the Fabs and allowing optimization of the crystal packing and diffraction quality.
Structural analyses of MdfA will be presented elsewhere.
Supplementary Material
Supplementary Figures S1 and S2.. DOI: 10.1107/S2053230X17008500/nw5050sup1.pdf
Acknowledgments
We thank Martin Caffrey and Dianfan Li for advice concerning the LCP crystallization method, Christoph Parthier for assistance in data reduction and Tina Iverson for critically reading the manuscript. Crystallographic data were collected at the Swiss Light Source (SLS), Villigen with support from the European Community’s 7th Framework Programme (FP7/2007-2013) under BioStruct-X (grant agreement No. 283570). The authors declare no conflict of interest.
References
- Alexandrov, A. I., Mileni, M., Chien, E. Y. T., Hanson, M. A. & Stevens, R. C. (2008). Structure, 16, 351–359. [DOI] [PubMed]
- Caffrey, M. (2015). Acta Cryst. F71, 3–18. [DOI] [PMC free article] [PubMed]
- Caffrey, M. & Cherezov, V. (2009). Nature Protoc. 4, 706–731. [DOI] [PMC free article] [PubMed]
- Day, P. W., Rasmussen, S. G. F., Parnot, C., Fung, J. J., Masood, A., Kobilka, T. S., Yao, X.-J., Choi, H.-J., Weis, W. I., Rohrer, D. K. & Kobilka, B. K. (2007). Nature Methods, 4, 927–929. [DOI] [PubMed]
- Drew, D., Newstead, S., Sonoda, Y., Kim, H., von Heijne, G. & Iwata, S. (2008). Nature Protoc. 3, 784–798. [DOI] [PMC free article] [PubMed]
- Drew, D. E., von Heijne, G., Nordlund, P. & de Gier, J. W. (2001). FEBS Lett. 507, 220–224. [DOI] [PubMed]
- Edgar, R. & Bibi, E. (1997). J. Bacteriol. 179, 2274–2280. [DOI] [PMC free article] [PubMed]
- Edgar, R. & Bibi, E. (1999). EMBO J. 18, 822–832. [DOI] [PMC free article] [PubMed]
- Fluman, N., Cohen-Karni, D., Weiss, T. & Bibi, E. (2009). J. Biol. Chem. 284, 32296–32304. [DOI] [PMC free article] [PubMed]
- Fluman, N., Ryan, C. M., Whitelegge, J. P. & Bibi, E. (2012). Mol. Cell, 47, 777–787. [DOI] [PMC free article] [PubMed]
- Heng, J., Zhao, Y., Liu, M., Liu, Y., Fan, J., Wang, X., Zhao, Y. & Zhang, X. C. (2015). Cell Res. 25, 1060–1073. [DOI] [PMC free article] [PubMed]
- Hino, T., Iwata, S. & Murata, T. (2013). Curr. Opin. Struct. Biol. 23, 563–568. [DOI] [PubMed]
- Jaenecke, F., Nakada-Nakura, Y., Nagarathinam, K., Ogasawara, S., Liu, K., Hotta, Y., Iwata, S., Nomura, N. & Tanabe, M. (2017). In the press. [DOI] [PubMed]
- Jeffrey, P. D., Schildbach, J. F., Chang, C. Y., Kussie, P. H., Margolies, M. N. & Sheriff, S. J. (1995). J. Mol. Biol. 248, 344–360. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Karplus, P. A. & Diederichs, K. (2012). Science, 336, 1030–1033. [DOI] [PMC free article] [PubMed]
- Köhler, G. & Milstein, C. (1975). Nature (London), 256, 495–497. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Lewinson, O., Padan, E. & Bibi, E. (2004). Proc. Natl Acad. Sci. USA, 101, 14073–14078. [DOI] [PMC free article] [PubMed]
- Li, D., Shah, S. T. A. & Caffrey, M. (2013). Cryst. Growth Des. 13, 2846–2857. [DOI] [PMC free article] [PubMed]
- Liu, M., Heng, J., Gao, Y. & Wang, X. (2016). Biophys. Rep. 2, 78–85. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Nikaido, H. (2009). Annu. Rev. Biochem. 78, 119–146. [DOI] [PMC free article] [PubMed]
- Pontecorvo, G. (1976). Birth Defects Orig. Artic. Ser. 12, 399–400. [PubMed]
- Putman, M., van Veen, H. W. & Konings, W. N. (2000). Microbiol. Mol. Biol. Rev. 64, 672–693. [DOI] [PMC free article] [PubMed]
- Quistgaard, E. M., Löw, C., Guettou, F. & Nordlund, P. (2016). Nature Rev. Mol. Cell Biol. 17, 123–132. [DOI] [PubMed]
- Sigal, N., Cohen-Karni, D., Siemion, S. & Bibi, E. (2006). J. Mol. Microbiol. Biotechnol. 11, 308–317. [DOI] [PubMed]
- Yan, N. (2013). Trends Biochem. Sci. 38, 151–159. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1 and S2.. DOI: 10.1107/S2053230X17008500/nw5050sup1.pdf