Abstract
PKC-ε concentration at phagocytic cups mediates the membrane fusion necessary for efficient IgG-mediated phagocytosis. The C1B and pseudosubstrate (εPS) domains are necessary and sufficient for this concentration. C1B binds diacylglycerol; the docking partner for εPS is unknown. Liposome assays revealed that the εPS binds phosphatidylinositol 4-phosphate (PI4P) and PI(3,5)P2. Wortmannin, but not LY294002, inhibits PKC-ε concentration at cups and significantly reduces the rate of phagocytosis. As Wortmannin inhibits PI4 kinase, we hypothesized that PI4P mediates the PKC-ε concentration at cups and the rate of phagocytosis. PKC-ε co-localizes with the Trans Golgi network PI4P reporter the P4M, suggesting its' tethering at the TGN. Real time imaging of GFP-PKC-ε expressing macrophages revealed a loss of Golgi-associated PKC-ε during phagocytosis, consistent with a Golgi-to-phagosome translocation. Treatment with PIK93, a PI4K inhibitor, reduces PKC-ε at both the TGN and the cup, decreases phagocytosis, and prevents the increase in capacitance that accompanies membrane fusion. Finally, expression of the Golgi-directed PI4P phosphatase, hSac1-K2A, recapitulates the PIK93 phenotype, confirming that Golgi-associated PI4P is critical for efficient phagocytosis. Together these data are consistent with a model in which PKC-ε is tethered to the TGN via an εPS-PI4P interaction. The TGN-associated pool of PKC-ε concentrates at the phagocytic cup where it mediates the membrane fusion necessary for phagocytosis. The novelty of these data lies in the demonstration that εPS binds PI4P and PI(3,5)P2 and that PI4P is necessary for PKC-ε localization at the TGN, its translocation to the phagocytic cup, and the membrane fusion required for efficient FcγR-mediated phagocytosis.
Introduction
PKC-ε regulates IgG-dependent phagocytosis by mediating the membrane fusion necessary for pseudopod extension (1). Wild type, but not PKC-ε-/-, macrophages spread on IgG surfaces, adding membrane as measured by increased membrane capacitance; re-expression of PKC-ε in null cells restores spreading, confirming a role for PKC-ε in membrane fusion (2). This is consistent with PKC-ε involvement in exocytosis, including release of dense granules from chromaffin cells (3), secretion by lacrimal cells (4), and insulin release (5). How PKC-ε mediates membrane fusion is an open question.
PKC-ε is a member of the Ca2+-independent, diacylglycerol activated subset of PKCs and is unique in that it contains an actin-binding motif. This implicates it in cytoskeletal functions; it is associated with actin filaments and keratin and is involved in integrin-dependent cell motility and spreading (6). Interestingly, its overexpression in NIH3T3 fibroblasts elicits a transformed phenotype (7) and its down-regulation inhibits metastasis (8). As metastatic cells are quite motile, PKC-ε is likely involved in cell motility in cancer models. Indeed, PKC-ε is upregulated in prostate (8) and breast (9) cancers and is considered an oncogene (10). However, its mechanism of action has yet to be determined.
Structurally, all PKCs consist of regulatory and catalytic domains connected by a hinge. Catalytic domains are homologous, containing the ATP binding pocket as well as the substrate- binding site. The regulatory domains vary and the isoforms are categorized by their lipid and calcium binding properties (11). All regulatory regions contain a pseudosubstrate sequence that, when bound in the active site, maintains the enzyme in an inactive state. Interaction of the regulatory domain with lipids and RACK (Receptor for Activated C Kinases) causes conformational changes that release the pseudosubstrate leading to enzyme activation. Although studies with chimeric PKCs suggested that the regulatory domain targets the catalytic domain to its site of action (12, 13), numerous exceptions have been reported. For example, PKC-βII V5 domain (in the catalytic region) interacts with RACK1 for PKC targeting in cardiac myocytes (14), the isolated regulatory domain of PKC-ε supports neurite extension (15), and catalytically inactive PKC-α activates PLD (16). Thus, intact PKC can signal via regulatory domain targeting of the catalytic region, the regulatory domain alone can promote cellular responses, and catalytically inactive PKC can mediate signaling. The studies detailed here establish that the pseudosubstrate of PKC-ε binds PI4P, tethering it to the TGN. This TGN-associated pool of PKC-ε is necessary for its' concentration at the phagocytic cup and FcγR-dependent membrane fusion. This is the first demonstration that a PKC pseudosubstrate-lipid interaction directs PKC concentration and membrane fusion in response to receptor ligation.
Materials and methods
Buffers and Reagents
ACK lysis buffer: 0.15 M NH4Cl, 1 mM KHCO3, and 0.1 mM EDTA, pH 7.4. HBSS: HBSS++: Hanks' Balanced Salt Solution (HBSS) containing 4 mM sodium bicarbonate, 10 mM HEPES, and 1.5 mM each CaCl2 and MgCl2. Bone marrow media: Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum, 20% L-cell conditioned media, and 0.03% NaHCO3, and 50μg/ml gentamicin. PIK93 was from Echelon Biosciences (Salt Lake City, UT). Wortmannin and LY were from Cayman Chemical (Ann Arbor, MI).
Mice and cells
PKC-ε-/+ heterozygotes on the C57/Bl6 background were purchased from The Jackson Laboratory (stock# 004189, Bar Harbor, ME) and bred in the Albany Medical Center Animal Resources Facility. Heterozygotes were crossed to produce the wild type and PKC-ε-/- mice. All animal procedures were approved by the Albany Medical Center Institutional Animal Care and Use Committee. Cells obtained from one mouse represent one independent experiment.
Bone marrow-derived macrophages
PKC-ε+/+ and PKC-ε-/- mice were euthanized and their femurs removed. The bone marrow was removed and the RBC lysed with ACK lysis buffer. Bone marrow stem cells were differentiated by incubation in phenol-red free (high glucose) DMEM containing 10% fetal bovine serum, 20% L-cell conditioned media, and sodium bicarbonate. Bone marrow-derived macrophages (BMDM) were used seven to 14 days after harvesting.
RAW 264.7 mouse macrophages
The RAW cell subclone, LacR/FMLPR.2 (RAW cells) (17) was used. Cells were maintained in (high glucose) DMEM, non-essential amino acids, sodium pyruvate, GlutaMAX® (all from Gibco Life Technologies), and 10% newborn calf serum (HyClone).
Liposome binding assay
Production of GST-peptides
Oligonucleotides were designed to encode the pseudosubstrate domain of PKC-ε (aa147-165) or PKC-δ (aa136-154) flanked by BamHI and EcoRI sites. Complementary oligonucleotides were annealed in 10 mM Tris pH 8.0, 50 mM NaCl, 1 mM EDTA at 50 μM, phosphorylated, and subcloned into pGEX-2T (GE Healthcare, WI) linearized with BamHI and EcoRI. Constructs were transformed into Rosetta 2 cells (EMD Millipore). Isopropyl-β-D-thiogalactopyranoside (IPTG, 1 mM) was added to logarithmically growing bacteria for 2h. Protein was extracted with BPERII (ThermoFisher) supplemented with 10 mM MgCl2, protease inhibitors, 1% lysozyme, and DNaseI. GST fusion proteins were purified from clarified bacterial lysates using glutathione-agarose beads (Sigma-Aldrich) and glutathione elution according to manufacturers' directions.
Liposome binding assay
Biotinylated PolyPIPosomes® were purchased from Echelon Bioscience (Salt Lake City, UT) and the assay done according to manufacturers' instructions. Briefly, incubations containing 1 μl of liposome and 5 nMol GST-peptide in 500 μl TBST were rotated (room temperature) for 60 min. Liposomes were immobilized on Streptavidin magnetic beads (Dynabeads®, Life Technologies) in a magnetic field, washed twice with TBST, and subjected to Western blot analysis for GST. An aliquot of the input was run in parallel to confirm equivalent addition of GST protein.
IgG-coated targets and Surfaces
IgG-opsonized beads (BIgG)
Acid washed 2 μm borosilicate beads (Duke Standards, Thermo Scientific USA) were sequentially coated with poly-L-lysine, dimethylpimelimidate, 2 HCl, and Alexa 568-conjugated (IgG-free) BSA. Free active groups were blocked by a 1 hour incubation in 0.5 M Tris, pH 8.0 before opsonizing with rabbit anti-BSA IgG.
IgG-coated surfaces
15 mm glass coverslips were rinsed with ethanol and distilled H2O and dried with compressed air to prepare for coating. The coverslips were aldehyde silane-coated by sequential incubation with an oxygen (O2) plasma cleaner (5 min) and vaporized silane (11-(Triethoxysilyl) undecanal, 70°C, 1 h, sealed in curing oven). To covalently link proteins to the silane-coated coverslips, the coverslips were washed with PBS and incubated with 25 μg/ml normal rabbit IgG (60 min, room temperature). After protein attachment, the remaining active sites were blocked 0.5 M Tris pH 8 (4°C, overnight). Before use, the coverslips were washed twice in HBSS++.
Phagocytosis
Synchronized phagocytosis was performed as detailed previously detailed (2). Briefly, cells were pre-treated (37°C, 30 min) with inhibitors (100 nM Wt, 50 μM LY, 0.1 or 10 μM PIK93, DMSO served as the no treatment control) and cooled on ice (15 min). IgG-opsonized beads were added (4 beads:cell) and allowed to bind (15 min, 4°C), then transferred to a 37°C water bath. Cells were fixed after 5 min (4% PFA/PBS) and imaged by spinning disk confocal microscopy (Olympus IX81-DSU) with a 100×/1.4 N.A. oil objective; Hamamatsu electron-multiplying charge-coupled device camera, driven by MetaMorph software (Molecular Devices, Sunnyvale, CA, USA). Image analysis for concentration at cups and nascent phagosomes in RAW cells was performed using ImageJ/FIJI and Localization Index calculated as detailed (2) . Briefly, the target associated membrane was selected using the 3 pixel brush and the intensity normalized to an equivalent area of non-involved membrane.
For live imaging, the ice steps were eliminated. Cells were pre-treated with inhibitors, brought into focus on the stage of the spinning disk, and IgG-opsonized beads were added. Images were taken every 5 s for 10 min. The rate of bead internalization was calculated as the number of frames from the first indentation of the plasma membrane through the first frame showing a completely enclosed particles × 5 (sec between frames) (2).
Plasmids
The GFP plasmid was purchased from Clonetech (Mountain View, CA). GFP-P4M, and GFP-hSac1-K2A (K2A) were generous gifts from Tamas Balla (18), and Peter Mayinger (19), respectively. Construction of the εK437W and pBM-GW-eGFP IRES-Puro vector and retroviral plasmid for PKC-ε-GFP have been published (2).
Expression of exogenous proteins
Transfection of RAW cells
RAW cells are transfected with Mirus Trans-IT neural at a 3:1 (Mirus:DNA) ratio. Detailed methodology has been published (2, 20).
Virus production and BMDM transduction
The details of retroviral pseudovirion production and BMDM transduction have been published (2). Briefly, Phoenix ecotropic packaging cells (National Gene Vector Biorepository, Indianapolis, IN) were transfected with the viral plasmids and the supernatants harvested 72 h post-infection. Viral transduction of bone marrow cells was done by spinfection 24 and 48 h after harvesting. Puromycin selection began ∼96 h after harvesting and cells differentiated BMDM were used 8-14 days after isolation; ∼90% of the selected cells expressed the transduced protein.
Patch Clamping
Whole cell patch-clamping was done essentially as described (2). Briefly, BMDM were removed from plates and treated in suspension (HBSS++) with inhibitors (30 min, room temperature), allowed to settle onto IgG-coated coverslips (20 min), then placed at 37°C to promote cell spreading. At 15 min, the coverslips were transferred to a holder and bathed in a balanced salt solution (10 mM HEPES, pH7.4, containing 145 mM NaCl, 5 mM KCl, 1 mM CaCl2, 2 mM MgCl2, and 5 mM glucose) for capacitance measurements. Cells were maintained at a 0 mV holding potential during experiments. Data is reported as mean capacitance ± SEM. Statistical significance was determined by one-way ANOVA with Bonferroni's correction.
Results
Live imaging of PKC-ε-GFP during IgG-mediated phagocytosis in RAW 264.7 cells and primary macrophages (MØ) reveal two temporally distinct waves of concentration, early at the phagocytic cup and later, as a “flash” on nascent phagosomes [(2, 20) and Movie 1]. The (diacylglycerol binding) C1B and pseudosubstrate (εPS) domains of PKC-ε are necessary and sufficient for concentration at both sites (2). C1B binds DAG, released by phospholipase C-γ1 (20); the docking partner for εPS is unknown.
εPS binds PI4P and PI(3,5)P2
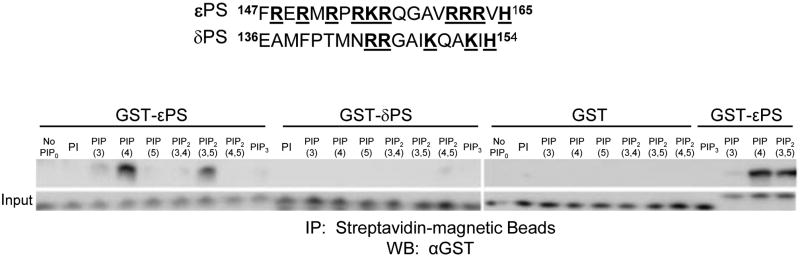
The pseudosubstrate of PKC-ε contains 12 basic amino acids, including two triplets (Fig. 1). Such polybasic clusters are a signature of phosphoinositide phosphate (PIP) binding proteins (21), leading us to test the ability of εPS to bind PIPs. Using GST-conjugated peptides and PolyPIPosomes® (liposomes that mimic the composition and curvature of cellular membranes), we identified PI4P and PI(3,5)P2 as binding partners for εPS (Fig. 1). The homologous region of PKC-δ, which neither contains polybasic triplets nor concentrates during phagocytosis (1), does not bind PIPs.
Figure 1. εPS binds PI4P and PI(3,5)P2.
The εPS sequence with basic amino acids bolded; the corresponding sequence from PKC-δ. These peptides were expressed as GST fusion proteins. Liposome binding assays were done according to manufacturer's directions using biotinylated PolyPIPosomes® containing 5 Mole% of the indicated PIP. Liposomes were recovered using Streptavidin-magnetic beads. The corresponding PS domain from PKC-δ [which does not concentrate during phagocytosis (1)] and unconjugated GST served as controls. Representative of 3 experiments.
PKC-ε concentration in the Golgi and at cups is sensitive to wortmannin, but not LY294002
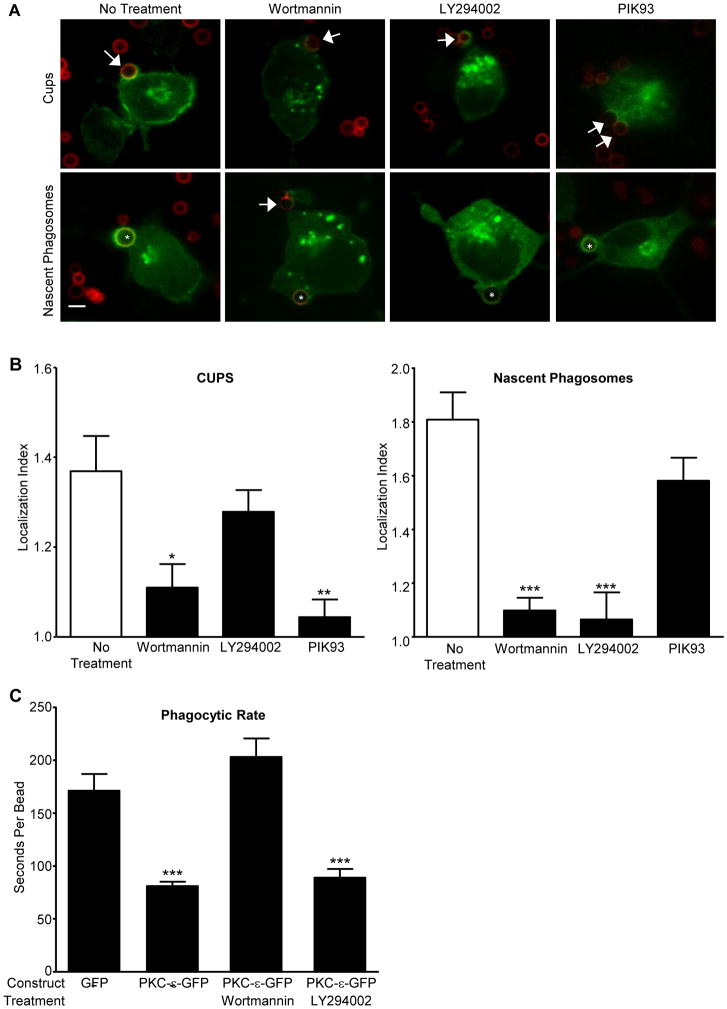
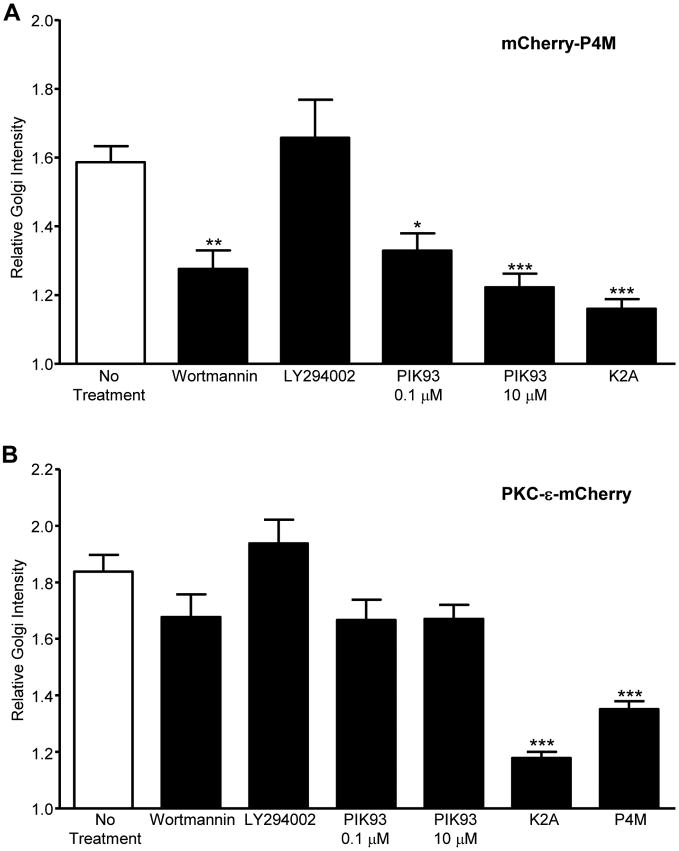
Given that εPS binds PI4P and PI(3,5)P2, we asked if LY294002 (LY) and/or wortmannin (Wt) block PKC-ε concentration at cups. This pair of inhibitors was chosen as both inhibit PI3 Kinase but Wt also blocks the type III PI4 Kinases. While Wt blocks PKC-ε concentration at cups and nascent phagosomes (NP), LY inhibits PKC-ε concentration only at NP (Fig. 2A, B). Additionally, Wt, but not LY, significantly slows the rate of target internalization (Fig. 2C), consistent with a role for PI4P in PKC-ε-dependent phagocytosis. To determine if Wt depletes PI4P under the conditions that inhibit phagocytosis, we expressed the PI4P reporter P4M (18) in RAW cells and quantified the effect of Wt and LY on P4M intensity. Wt, but not LY, significantly reduces perinuclear PI4P levels (Fig 3A) consistent with the hypothesis that a PI4P-PKC-ε interaction is necessary for PKC-ε concentration at cups and regulation of phagocytosis. When the same experiment was done using PKC-ε-GFP, we found a modest, but not statistically significant, decrease in Golgi-associated PKC-ε (Fig 3B). This apparent disparity may reflect the fact that Wt prevents PI4P formation, which would have little effect on that pool of PI4P already bound to PKC-ε.
Figure 2. Wortmannin and PIK93 inhibit PKC-ε localization during phagocytosis; LY294002 blocks PKC-ε at nascent phagosomes, but not cups.
(A) RAW cells expressing PKC-ε-GFP were treated with Wortmannin (Wt), LY294002 (LY), or PIK93 and subjected to synchronized phagocytosis. Arrow: cups, *: nascent phagosomes. (B) Quantitation of localization index at cups and nascent phagosomes (30-50 events, 4 experiments). Data are presented as mean ± SEM. Significance determined by ANOVA with Bonferroni's correction; * p<0.05, ** p<0.01, *** p<0.001 compared to No Treatment. (C) Quantitation of phagocytic rate. RAW cells expressing PKC-ε-GFP were treated with Wt or LY as in (A) and imaged in real time during phagocytosis; unconjugated GFP served as the transfection control. Expression of exogenous PKC-ε decreased the time necessary for target internalization (ie, beads taken up faster, compare GFP and PKC-ε-GFP). Wt significantly slowed internalization while LY had no effect. Data are presented as mean ± SEM. Significance determined by ANOVA with Bonferroni's correction; ***p< 0.001 compared to unconjugated GFP.
Figure 3. PI4P modulating treatments reduce Golgi PI4P/PKC-ε intensity.
RAW cells expressing mCherry-P4M (A) or PKC-ε-mCherry (B) were treated with Wt, LY, or PIK93 (30 min, 37°C). Alternatively, cells were co-transfected with PKC-ε-mCherry and GFP-hSac1-K2A (A,B) or GFP-P4M (B). Cells with were imaged by confocal microscopy and perinuclear P4M (A) or PKC-ε (B) intensity quantified. Results are reported as Relative Golgi Intensity, that is, the pixel intensity at the Golgi normalized to an equivalent area in the cytosol (mean ± SEM). Significance was determined by ANOVA with Bonferroni's correction. * p<0.05, ** p<0.01, *** p<0.001 compared to No Treatment. n=20-25 cells (A) or 40-70 cells (B) compiled from 3 independent experiments). A relative ratio of 1 = no concentration. Representative images of cells co-expressing PKC-ε-mCherry and GFP-hSac1-K2A or GFP-P4M are provided in Fig 5.
Given the body of literature concluding the PI3K is necessary for phagocytosis (22), our results suggesting that the Wt effect at cups is PI3K independent was somewhat surprising. The disparity likely arises from the differences in experimental design. Fig 2B reports the rate of bead internalization, extracted from live imaging; published studies quantify the number of particles internalized after 60 min. While LY does not decrease the initial rate of phagocytosis, it does inhibit phagocytosis at 60 min (data not shown), in agreement with the literature. Notably, our data parallel the findings that the exocytosis phase of phagocytosis is PI3K independent (23). Thus, PI3P acts later in phagocytosis, likely at the level of membrane/receptor recycling.
PI4P mediates PKC-ε concentration at cups, membrane fusion, and phagocytosis
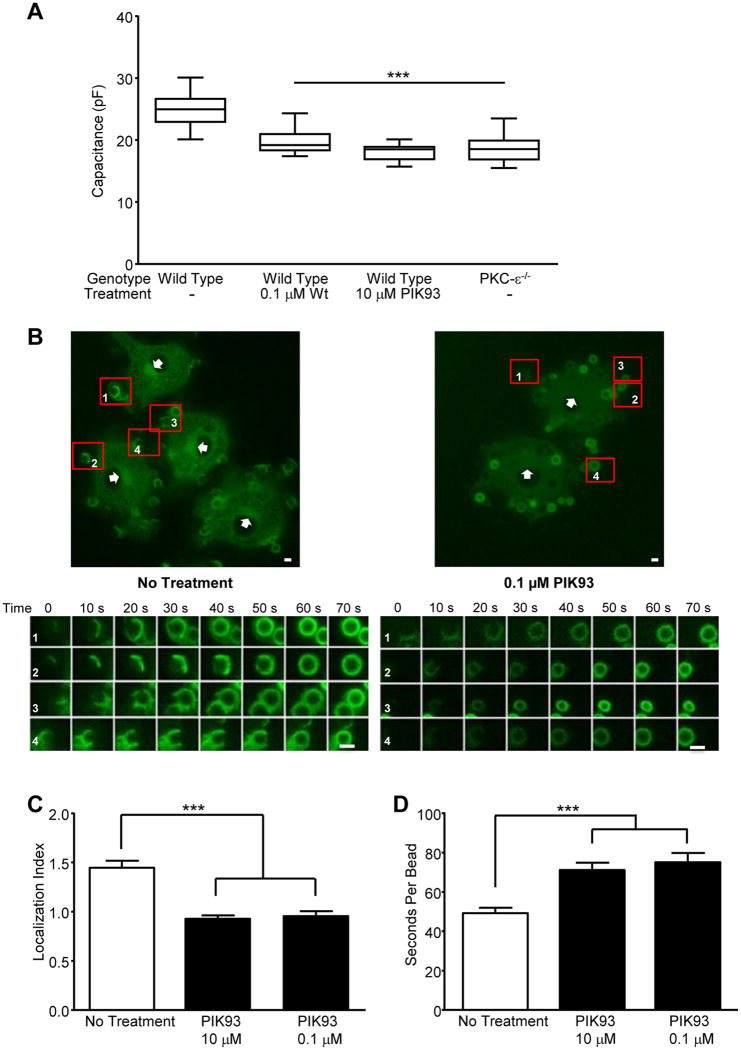
Given that PKC-ε translocation is necessary for membrane fusion for IgG-mediated phagocytosis (2), if PI4P is required for PKC-ε translocation, then inhibiting PI4 kinase (PI4K) should block FcγR-dependent membrane fusion. Thus, we tested the effect of PIK93, an inhibitor of the Wt-sensitive PI4Ks (24) on macrophage spreading on IgG surfaces. Using whole cell patch clamping, we determined that cells treated with Wt or PIK93 have significantly lower capacitance than solvent treated controls (Fig 4A). The fact that the capacitance of WT cells treated with either Wt or PIK93 is equivalent to that of PKC-ε-/- bone marrow derived macrophages (BMDM) supports the hypothesis that PI4P is required for PKC-ε-mediated membrane fusion during phagocytosis.
Figure 4. Inhibition of PI4K blocks membrane fusion, PKC-ε concentration at cups, and slows target uptake.
(A) Wild type BMDM, treated with PIK93 or Wt, and PKC-ε-/- BMDM, were attached to IgG surfaces (20 min, 37°C) and capacitance quantified by whole cell patch-clamping. Data is reported as mean ± SEM, 15 cells/condition from two animals of each genotype. (B) BMDM expressing PKC-ε-GFP were treated with PIK93 and phagocytosis followed in real time (Movies 1 and 2). A selected frame from Movies 1 and 2 is shown with 4 targets identified (1-4) and tracked from attachment to phagosome closure; 0 defines the time of target binding. Bars = 2 μm. From the movies, PKC-ε concentration at cups (C) and the rate of internalization (D) were determined. Results are reported as mean ± SEM (n = 40-49 events compiled from 3 independent experiments). Significance was tested by ANOVA with Bonferroni's correction. *** p< 0.001 compared to No treatment
Patch clamping utilized 10 μM PIK93 as it inhibits both Wt-sensitive PI4KIIIα and IIIβ. As PIK93 has an isoform selectivity, we used 0.1 and 10 μM to test the dose dependency going forward: 0.1 μM preferentially blocks PI4K IIIβ (Tamas Balla, unpublished data) with10 μM needed to substantively inhibit PI4KIIIα (24). Both concentrations reduce PI4P Golgi concentration in RAW cells (Fig 3A). When tested for their effects in primary BMDM, both concentrations significantly reduce PKC-ε concentration at cups (Fig 4B,C) and slow IgG-mediated phagocytosis (Fig 4D).
PI4P tethers PKC-ε at the TGN
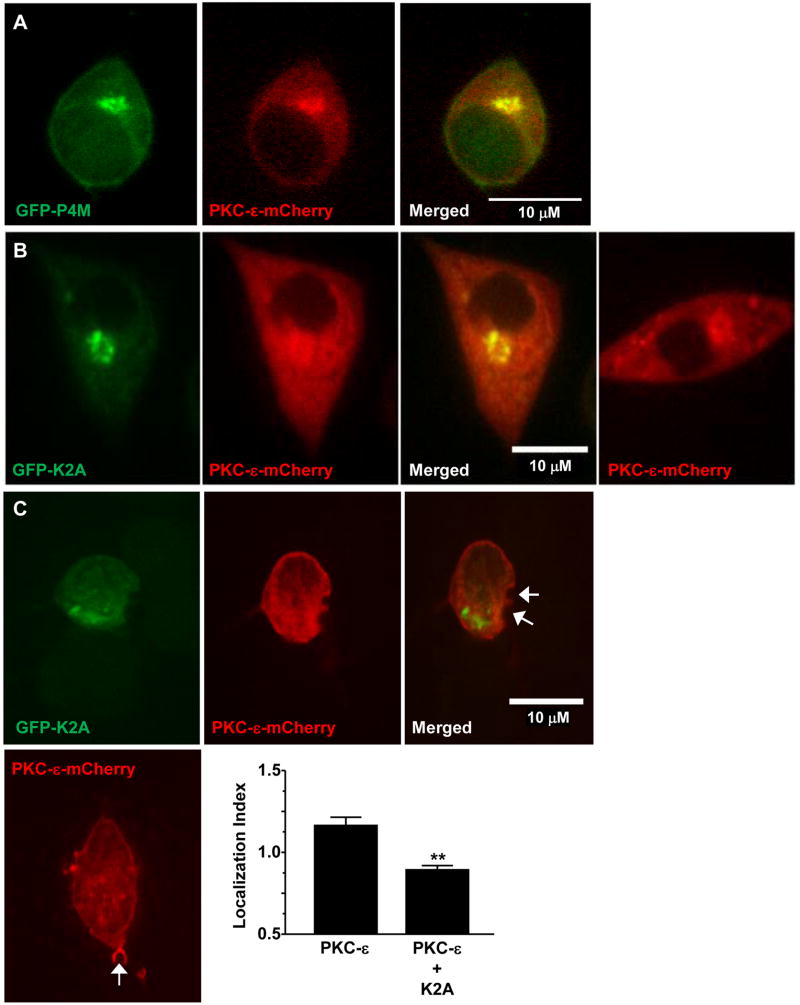
Live imaging of BMDM expressing PKC-ε-GFP reveals a decrease in perinuclear PKC-ε intensity during active phagocytosis (Movie 1). 0.1 μM PIK93 dramatically reduces PKC-ε concentration at the Golgi (Fig 4B, Movie 2, and Supplemental Fig 1), consistent with its' selective inhibition of PI4KIIIβ, the isoform producing PI4P in the Golgi (25). The hypothesis that PKC-ε associates with PI4P in the TGN is supported by several lines of evidence: ∼20% of PKC-ε in macrophages is membrane associated (26), PKC-ε concentrates in the perinuclear region in macrophages ((2), Movie 1 and arrowheads, Fig 4A), and PI4P is enriched in the Trans Golgi Network (25). If PKC-ε binds PI4P in the Golgi, then PI4P and perinuclear PKC-ε should co-localize. This is the case, as evidenced by a Pearson's correlation of 0.86 (40 cells from 3 independent experiments) in macrophages co-expressing the PI4P reporter, P4M and PKC-ε (Fig 5A); co-localization of PKC-ε with the Golgi marker GM130 (Supplemental Fig 2) with a Pearson's coefficient of 0.64 confirms that perinuclear PKC-ε is indeed Golgi-associated. Notably, the perinuclear intensity of PKC-ε is significantly lower in cells co-expressing P4M (Fig 3B), which would be expected if P4M and PKC-ε compete for Golgi PI4P. Secondly, if PKC-ε is tethered to the Golgi via PI4P, then selective removal of Golgi-associated PI4P should reduce perinuclear PKC-ε. This was tested by co-expressing the Golgi directed PI4 phosphatase, hSac1-K2A (K2A) (27) with either P4M or PKC-ε. K2A expression substantively reduces both perinuclear PI4P (Fig 3A) and PKC-ε (Representative image, Fig 5B, quantitation Fig 3B), paralleling the effects of 0.1 μM PIK93 on Golgi-associated PI4KIIIβ (24) and perinuclear PKC-ε in RAW cells (Fig 3) and BMDM (Note lack of perinuclear PKC-ε in Movie 2 compared to Movie 1, arrowheads in Fig 4B, and Suppl Fig. 1). Additionally, if the pool of PKC-ε that concentrates at cups originates in the Golgi, we would predict that expression of K2A would prevent/reduce that concentration. Indeed, co-expression of K2A and PKC-ε revealed a statistically significant decrease in PKC-ε at cups in cells expressing K2A compared to those expressing PKC-ε alone (Fig 5C). Together, these data support the conclusion that PKC-ε binds PI4P in Golgi and this interaction is necessary for the translocation of PKC-ε to the phagocytic cup and the membrane fusion necessary for efficient phagocytosis.
Figure 5. PKC-ε is tethered to the Golgi via PI4P.
RAW264.7 cells were co-transfected with PKC-ε-mCherry and either the PI4P reporter GFP-P4M (A) or the Golgi-directed PI4 phosphatase, GFP-K2A (B,C). B,C Representative images showing co-localization of PKC-ε with P4M and loss of perinuclear PKC-ε upon co-expression of K2A. Quantitation and statistical analysis is included in Figure 3. (C) Cells expressing the indicated constructs were subjected to synchronized phagocytosis and localization index at the cup (arrows) quantified. Co-expression of K2A and PKC-ε (upper panels) significantly reduced cup localization compared to PKC-ε alone (lower image). Data is reported as mean ± SEM (n= 28 events/condition). Statistical significant was determined by unpaired t-test. ** p< 0.01
Discussion
PKC-ε concentrates in two waves during phagocytosis, at the forming phagosome (the cup) and after phagosome closure, appearing as a “flash” ((2, 20) and Movie 1). The C1B and εPS domains were shown to be necessary and sufficient for both waves of concentration (2). C1B binds diacyglycerol but the binding partner for εPS was unknown. We narrowed the required εPS sequence to 19 amino acids (diagrammed in Fig 1, identified in (2)) and noted the predominance of basic amino acids, a signature of PIP binding (21). While the polybasic nature of εPS might suggest electrostatic binding, this is clearly not the case as εPS binds PI4P but not the isomers PI3P and PI5P, compounds with the same net charge. Similarly, there is a specificity of PI(3,5)P2 over isomers PI(3,4)P2 and PI(4,5)P2 and no apparent interaction with the most negatively charged PIP, PI(3,4,5)P3. Precedence for such selectivity comes from the PH domain literature. Indeed, this selectivity has been exploited in generating a toolbox of lipid reporters (28), including the P4M utilized herein.
Based on the liposome binding assays, and the acknowledged role for PI3K in IgG-mediated phagocytosis (22), we hypothesized that the εPS-PI(3,5)P2 interaction was responsible for PKC-ε concentration. Thus, the differential effects of Wt and LY on PKC-ε concentration at the cup were surprising and focused our attention on the ability of Wt to inhibit the Type III PI4 kinases. PI4P modulators, combined with the P4M lipid reporter provided us with tools with which to modulate PI4P (PIK93, K2A), monitor its presence (P4M), and reproduce the Wt findings. Notably, all treatments that decreased Golgi-associated PI4P reduced perinuclear PKC-ε, its' concentration at cups, and slowed phagocytosis. Conversely, LY did not substantively reduce perinuclear PKC-ε (Fig 2) nor did it reduce cup-associated PKC-ε or alter phagocytic rate, providing a “control” lipid inhibitor that did not alter PI4P. Our conclusion, based on the totality of the results, is that PKC-ε associates with PI4P in the TGN and it is this pool that concentrates at the cup. One argument against this model is that brefeldin A, a drug that disrupts the Golgi, does not alter IgG-mediated phagocytosis (29). When tested in our system, brefeldin A had no effect on the PKC-ε perinuclear localization (data not shown), consistent with reports that it neither disrupts TGN trafficking (30) nor inhibits IgG-mediated phagocytosis (29). This provides additional evidence that PKC-ε is localized in the TGN. While not directly tested, several pieces of evidence argue that PI4KIIIβ is producing the PI4P for PKC-ε tethering. These include: localization of PI4KIIIβ to the trans Golgi (31) and loss of Golgi-associated PI4P/disruption of phagocytosis at a concentration of PIK93 that targets PI4KIIIβ (24) (Fig 2, 3).
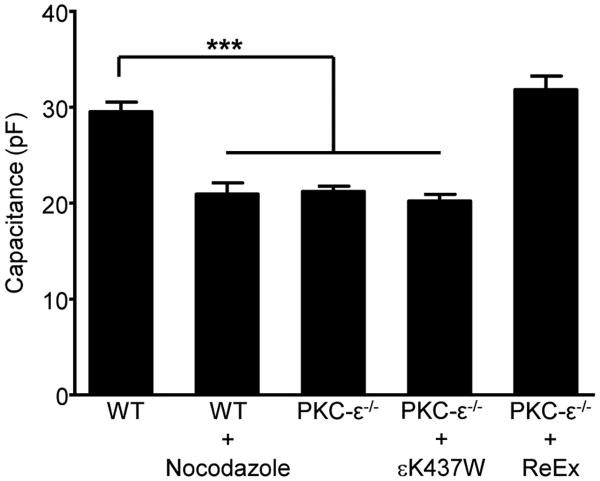
The simplest model that incorporates our data and the literature is that the PKC-ε that concentrates at the cup originates in the Golgi and travels on vesicles in a PI4P dependent manner; PI4KIIIβ facilitates the Golgi to PM trafficking (25, 31). PKC-ε-positive vesicles would fuse into the cup, providing membrane for pseudopod extension, explaining the loss of membrane fusion in response to PI4K inhibition (Fig 4A). As Golgi-to-plasma membrane vesicles traffic on microtubules, one test of the model would be to determine the effect of nocodazole on membrane fusion. Whole cell patch clamping revealed that nocodazole prevents FcγR-mediated membrane fusion, with nocodazole-treated wild type cells having significantly lower capacitance than their untreated counterparts, capacitance equivalent to that of PKC-ε-/- BMDM (Fig 6). These data support a vesicular trafficking model. That re-expression of full length, but not catalytically inactive, PKC-ε in PKC-ε-/- cells restored membrane fusion bridges these studies to our previous findings that catalytically inactive PKC-ε translocates, but does not support phagocytosis (2). Thus, the regulatory domain directs vesicle targeting, with catalytic activity needed for membrane fusion.
Figure 6. Nocodazole blocks membrane fusion in response to FcγR ligation.
Wild type BMDM (WT) expressing exogenous PKC-ε-GFP were treated with nocodazole (10 μM, 20 min). Intact (ReEx) or catalytically inactive (εK437W) PKC-ε was re-expressed in PKC-ε-/- BMDM. Cells were attached to IgG surfaces (20 min, 37°C) and capacitance quantified by whole cell patch-clamping; untreated WT and PKC-ε-/- BMDM served as controls. Data is reported as mean ± SEM, 15-20 cells/condition from three independent experiments. Statistical significance was determined by ANOVA with Bonferroni's correction. ***p < 0.005.
In conclusion, while these studies focused on PKC-ε at the cup, it of interest to consider that treatments that block PKC-ε association with the Golgi, block its' concentration at the cup, and retard phagocytosis but have little effect on PKC-ε concentration at nascent phagosomes (Fig 2 and Supplemental Fig 3). This suggests there may be two pools of PKC-ε: one at the Golgi regulated by εPS-PI4P and a second pool, potentially regulated by εPS-PI(3,5)P2 binding (Fig 1). The differential regulation of these two putative pools is intriguing and currently under investigation.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Lynn Cassimeris for her insightful analysis of our real time imaging. Additionally, KateTubbesing for her help with Imaris imaging, Dr. Elaine Raines and lab for guidance on viral constructs and production of pseudovirions, Tiffany Wood-Cammock for generation of preliminary data, and Drs. Daniel J. Loegering and James Drake for helpful discussions and reading the manuscript. Finally, many thanks to Deborah Moran for all her work on the preparing the manuscript and the Johnathan R. Vasiliou Foundation for their generous support of this work.
Abbreviations used in this paper
- BMDM
Bone marrow-derived macrophages
- εPS (Pseudosubstrate domain of PKC-ε) FcγR
Fc [gamma] Receptor
- PKC-ε
Protein Kinase C-epsilon, K2A: Golgi-directed PI4 phosphatase, hSac1-K2A(27)
- NP
nascent phagosomes, Wt, Wortmannin, LY, LY29004
Footnotes
Author contributions: M.R. Lennartz conceived the project. C.M. Hanes, A.E. D'Amico, A. C. Wong, T. Ueyama, X. Zhang, performed experiments, collected and analyzed data; W.F. Hynes and N.C. Cady developed the methodology and produced the activated glass surfaces; N.C. Cady, M Trebak, M.M. Barroso, N. Saito advised on experimental design and analysis. All authors read, edited, and approved the final manuscript.
The authors declare no competing financial interests.
The study was supported by National Institutes of Health Grants AI50821 and GM090325 (MRL), The Johnathan R. Vasiliou Foundation (MRL), and an Albany Medical College Bridge Grant (MRL).
The authors declare no commercial affiliations or conflicts of interest.
References
- 1.Larsen EC, Ueyama T, Brannock PM, Shirai Y, Saito N, Larsson C, Loegering D, Weber PB, Lennartz MR. A role for PKC-varepsilon in FcgammaR-mediated phagocytosis by RAW 264.7 cells. J Cell Biol. 2002;159:939–944. doi: 10.1083/jcb.200205140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood TR, Chow RY, Hanes CM, Zhang X, Kashiwagi K, Shirai Y, Trebak M, Loegering DJ, Saito N, Lennartz MR. PKC-epsilon pseudosubstrate and catalytic activity are necessary for membrane delivery during IgG-mediated phagocytosis. J Leukoc Biol. 2013;94:109–122. doi: 10.1189/jlb.1212634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park YS, Hur EM, Choi BH, Kwak E, Jun DJ, Park SJ, Kim KT. Involvement of protein kinase C-epsilon in activity-dependent potentiation of large dense-core vesicle exocytosis in chromaffin cells. J Neurosci. 2006;26:8999–9005. doi: 10.1523/JNEUROSCI.2828-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jerdeva GV, Yarber FA, Trousdale MD, Rhodes CJ, Okamoto CT, Dartt DA, Hamm-Alvarez SF. Dominant-negative PKC-epsilon impairs apical actin remodeling in parallel with inhibition of carbachol-stimulated secretion in rabbit lacrimal acini. Am J Physiol Cell Physiol. 2005;289:C1052–1068. doi: 10.1152/ajpcell.00546.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendez CF, Leibiger IB, Leibiger B, Hoy M, Gromada J, Berggren PO, Bertorello AM. Rapid association of protein kinase C-epsilon with insulin granules is essential for insulin exocytosis. J Biol Chem. 2003;278:44753–44757. doi: 10.1074/jbc.M308664200. [DOI] [PubMed] [Google Scholar]
- 6.Akita Y. Protein kinase Cε: multiple roles in the function of, and signaling mediated by, the cytoskeleton. FEBS Journal. 2008;275:3995–4004. doi: 10.1111/j.1742-4658.2008.06557.x. [DOI] [PubMed] [Google Scholar]
- 7.Tachado SD, Mayhew MW, Wescott GG, Foreman TL, Goodwin CD, McJilton MA, Terrian DM. Regulation of tumor invasion and metastasis in protein kinase C epsilon-transformed NIH3T3 fibroblasts. J Cell Biochem. 2002;85:785–797. doi: 10.1002/jcb.10164. [DOI] [PubMed] [Google Scholar]
- 8.Hafeez BB, Zhong W, Weichert J, Dreckschmidt NE, Jamal MS, Verma AK. Genetic ablation of PKC epsilon inhibits prostate cancer development and metastasis in transgenic mouse model of prostate adenocarcinoma. Cancer Res. 2011;71:2318–2327. doi: 10.1158/0008-5472.CAN-10-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan Q, Bao LW, Kleer CG, Sabel MS, Griffith KA, Teknos TN, Merajver SD. Protein Kinase Cε Is a Predictive Biomarker of Aggressive Breast Cancer and a Validated Target for RNA Interference Anticancer Therapy. Cancer Research. 2005;65:8366–8371. doi: 10.1158/0008-5472.CAN-05-0553. [DOI] [PubMed] [Google Scholar]
- 10.Gorin MA, Pan Q. Protein kinase C epsilon: an oncogene and emerging tumor biomarker. Mol Cancer. 2009;8:9. doi: 10.1186/1476-4598-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newton AC. Protein kinase C: poised to signal. Am J Physiol Endocrinol Metab. 2010;298:E395–402. doi: 10.1152/ajpendo.00477.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang QJ, Acs P, Goodnight J, Giese T, Blumberg PM, Mischak H, Mushinski JF. The catalytic domain of protein kinase C-d in reciprocal de chimeras mediates phorbol ester-induced macrophage differentiation of mouse promyelocytes. J Biol Chem. 1997;272:76–82. doi: 10.1074/jbc.272.1.76. [DOI] [PubMed] [Google Scholar]
- 13.Acs P, Wang QJ, Bogi K, Marquez AM, Lorenzo PS, Biro T, Szallasi Z, Mushinski JF, Blumberg PM. Both the catalytic and regulatory domains of protein kinase C chimeras modulate the proliferative properties of NIH 3T3 cells. J Biol Chem. 1997;272:28793–28799. doi: 10.1074/jbc.272.45.28793. [DOI] [PubMed] [Google Scholar]
- 14.Stebbins EG, Mochly-Rosen D. Binding specificity for RACK1 resides in the V5 region of beta II protein kinase C. J Biol Chem. 2001;276:29644–29650. doi: 10.1074/jbc.M101044200. [DOI] [PubMed] [Google Scholar]
- 15.Zeidman R, Lofgren B, Pahlman S, Larsson C. PKCepsilon, via its regulatory domain and independently of its catalytic domain, induces neurite-like processes in neuroblastoma cells. J Cell Biol. 1999;145:713–726. doi: 10.1083/jcb.145.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JS, Exton JH. Regulation of phospholipase D2 activity by protein kinase C alpha. J Biol Chem. 2004;279:22076–22083. doi: 10.1074/jbc.M311033200. [DOI] [PubMed] [Google Scholar]
- 17.Cox D, Chang P, Zhang Q, Reddy PG, Bokoch GM, Greenberg S. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J Exp Med. 1997;186:1287–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond GRV, Machner MP, Balla T. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. The Journal of Cell Biology. 2014;205:113–126. doi: 10.1083/jcb.201312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayinger P. Regulation of Golgi function via phosphoinositide lipids. Semin Cell Dev Biol. 2009;20:793–800. doi: 10.1016/j.semcdb.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheeseman KL, Ueyama T, Michaud TM, Kashiwagi K, Wang D, Flax LA, Shirai Y, Loegering DJ, Saito N, Lennartz MR. Targeting of PKC-ε during FcγR-dependent Phagocytosis Requires the εC1B Domain and Phospholipase C-γ1. Mol Biol Cell. 2006;17:799. doi: 10.1091/mbc.E04-12-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox D, Tseng CC, Bjekic G, Greenberg S. A requirement for phosphatidylinositol 3-kinase in pseudopod extension. J Biol Chem. 1999;274:1240–1247. doi: 10.1074/jbc.274.3.1240. [DOI] [PubMed] [Google Scholar]
- 23.Di A, Nelson DJ, Bindokas V, Brown ME, Libunao F, Palfrey HC. Dynamin regulates focal exocytosis in phagocytosing macrophages. Mol Biol Cell. 2003;14:2016–2028. doi: 10.1091/mbc.E02-09-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tóth B, Balla A, Ma H, Knight ZA, Shokat KM, Balla T. Phosphatidylinositol 4-Kinase IIIβ Regulates the Transport of Ceramide between the Endoplasmic Reticulum and Golgi. Journal of Biological Chemistry. 2006;281:36369–36377. doi: 10.1074/jbc.M604935200. [DOI] [PubMed] [Google Scholar]
- 25.Clayton EL, Minogue S, Waugh MG. Mammalian phosphatidylinositol 4-kinases as modulators of membrane trafficking and lipid signaling networks. Prog Lipid Res. 2013;52:294–304. doi: 10.1016/j.plipres.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen EC, DiGennaro JA, Saito N, Matha S, Loegering DJ, Mazurkiewicz JM, Lennartz MR. Differential requirement for classic and novel PKC isoforms in respiratory burst and phagocytosis in RAW 264.7 cells. J Immunol. 2000;165:2809–2817. doi: 10.4049/jimmunol.165.5.2809. [DOI] [PubMed] [Google Scholar]
- 27.Bishe B, Syed GH, Field SJ, Siddiqui A. Role of Phosphatidylinositol 4-Phosphate (PI4P) and Its Binding Protein GOLPH3 in Hepatitis C Virus Secretion. J Biol Chem. 2012;287:27637–27647. doi: 10.1074/jbc.M112.346569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platre MP, Jaillais Y. Guidelines for the Use of Protein Domains in Acidic Phospholipid Imaging. Methods Mol Biol. 2016;1376:175–194. doi: 10.1007/978-1-4939-3170-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Cox D, Tseng CC, Donaldson JG, Greenberg S. A requirement for ARF6 in fcgamma receptor-mediated phagocytosis in macrophages. J Biol Chem. 1998;273:19977–19981. doi: 10.1074/jbc.273.32.19977. [DOI] [PubMed] [Google Scholar]
- 30.Wood SA, Brown WJ. The morphology but not the function of endosomes and lysosomes is altered by brefeldin A. J Cell Biol. 1992;119:273–285. doi: 10.1083/jcb.119.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hausser A, Storz P, Martens S, Link G, Toker A, Pfizenmaier K. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat Cell Biol. 2005;7:880–886. doi: 10.1038/ncb1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.