Abstract
The MALAT1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1) gene encodes a non-coding RNA that is processed into a long nuclear retained transcript (MALAT1) and a small cytoplasmic tRNA-like transcript (mascRNA). Using a RNA sequence- and structure-based covariance model, we identified more than 130 genomic loci in vertebrate genomes containing the MALAT1 3′-end triple helix structure and its immediate downstream tRNA-like structure, including 44 in the green lizard Anolis carolinensis. Structural and computational analyses revealed a coevolution of the 3′-end module. MALAT1-like genes in Anolis carolinensis are highly expressed in adult testis, thus we named them testis-abundant long noncoding RNAs (tancRNAs). MALAT1-like loci also produce multiple small RNA species, including piRNAs, from the antisense strand. The coevolved 3′-ends of tancRNAs serve as potential targets for the PIWI-piRNA complex. Thus, we have identified an evolutionarily conserved class of lncRNAs with similar structural constraints, post-transcriptional processing, subcellular localization and a distinct function in spermatocytes.
Introduction
Long noncoding RNAs (lncRNAs), range in size from 200 nucleotides to greater than 100kb in length, and are typically transcribed by RNA polymerase II (Rinn and Chang, 2012). A number of studies over the past decade including the ENCODE project have identified thousands of lncRNAs, many which exhibit cell-type and tissue- specific expression, diverse subcellular localization, and disease association (Bertone et al., 2004; Carninci et al., 2005; Derrien et al., 2012; Djebali et al., 2012; Harrow et al., 2012; Kapranov et al., 2007; Katayama et al., 2005; Okazaki et al., 2002). Although some lncRNAs have been shown to act as scaffolds for the assembly of nuclear bodies and cytoplasmic complexes, to play an important role in co-transcriptional gene regulation and chromatin remodeling, or to serve as a molecular decoy to titrate miRNAs or RNA-binding factors (Chodroff et al., 2010; Guttman et al., 2009; Lee et al., 2016; Mao et al., 2011; Orom et al., 2010; Rinn et al., 2007; Shevtsov and Dundr, 2011; Ulitsky and Bartel, 2013), the function of the majority of lncRNAs has yet to be determined. In addition, their evolution on both the structural and functional levels and the correlation between structure and function remains largely unclear. As there is a lack of structure-function relationships and sequence conservation among lncRNAs de novo prediction of lncRNA functional domains is quite challenging (Batista and Chang, 2013).
MALAT1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1) lncRNA, first identified as being upregulated in lung tumors that have the propensity to metastasize (Ji et al., 2003), has subsequently been found to be highly expressed in a large number of tumor types (reviewed in (Gutschner et al., 2013a)). Unlike most lncRNAs that are expressed at very low levels, MALAT1 is also highly expressed in many normal tissues (Zhang et al., 2012) and its expression is regulated during development (Bernard et al., 2010; Hutchinson et al., 2007; Ji et al., 2003). Several Malat1 knockout mouse models have been established and they exhibit no major phenotype in regard to pre- or post-natal development, they exhibit normal growth and fertility, and Malat1 deletion does not affect global gene expression or pre-mRNA splicing (Eissmann et al., 2012; Nakagawa et al., 2012; Zhang et al., 2012). The lack of phenotype upon the in vivo loss of Malat1 lncRNA transcripts could be attributed to functional redundancy with other RNA transcripts or to its subtle cellular and developmental function, which may be compensated or manifest phenotypes only under certain physiological conditions, as occurs with respect to many protein-coding genes (Zhang et al., 2012). Interestingly, knockdown of MALAT1 in several cultured cell lines results in altered pre-mRNA splicing (Tripathi et al., 2010) or E2F1-regulated cell cycle progression (Yang et al., 2011). In addition, two genome-wide studies have indicated that MALAT1 binds to Transcription Start Sites (TSS) and to gene bodies of actively transcribing genes together with lncRNA NEAT1 (West et al., 2014) and to nascent pre-mRNAs indirectly via protein partners (Engreitz et al., 2014). Interestingly, human lung tumor cells lacking MALAT1 are impaired in cell migration and are unable to efficiently develop tumor nodules in a mouse xenograft model (Gutschner et al., 2013b). Furthermore, genetic knockout or antisense oligonucleotide (ASO)-mediated knockdown of Malat1 in vivo in a mouse mammary tumor model results in the differentiation of mammary tumors and significant reduction of metastasis (Arun et al., 2016) supporting a context-dependent function of Malat1.
MALAT1 nascent transcripts are processed by RNases P and Z to produce a long nuclear-retained ncRNA with a 3′-end genetically encoded short polyA-like tail and a 58-nucleotide small tRNA-like cytoplasmic RNA (mascRNA) (Wilusz et al., 2008). Secondary and tertiary structures of lncRNAs have been shown to regulate RNA stability and function through different molecular mechanisms. For example, the stability of polyadenylated nuclear (PAN) RNA produced by Karposi's sarcoma-associated herpesvirus is regulated by a nuclear retention element (ENE) near its 3′-end that forms a triple-helix structure to prevent 3′ nuclease cleavage (Brown et al., 2012). Recently, the 3′-ends of MALAT1 and Menβ (Neat1) lncRNAs have also been shown to form a triple-helical structure which functions to stabilize the RNA (Brown et al., 2014; Brown et al., 2012; Wilusz et al., 2012). Evolution of the MALAT1 gene has been examined, but its origin and function remain largely elusive (Pauli et al., 2012; Stadler, 2010; Ulitsky et al., 2011).
Here, we analyzed the secondary and tertiary structures of the 3′-end region of MALAT1 lncRNA from multiple species using a chemical probing approach. Our results are consistent with a triplex helix complex upstream of the RNase P cleavage site. Using computational modeling based on this identified secondary RNA structure, 132 genomic loci resembling the MALAT1 3′ end processing module have been identified among several vertebrate genomes, including 44 in the lizard Anolis carolinensis. Molecular genomic analyses show that MALAT1 is highly conserved during evolution in terms of its length, expression, and subcellular localization. Despite a lack of significant sequence similarity among the 5′ regions of MALAT1 in vertebrates, a high degree of RNA structural conservation is observed in its 3′-end between human, zebrafish and lizard, implicating critical molecular and/or cellular functions of the MALAT1 3′-end. MALAT1-like genes in Anolis carolinensis do not appear to encode proteins and are highly expressed in testis and as such we have named them testis-abundant long non-coding RNAs (tancRNAs). Surprisingly, molecular characterization has identified multiple small RNA species derived from tancRNA loci, including piRNAs from the antisense strand. Interactions between lizard PIWI proteins and tancRNAs, along with sequence complementation between tancRNAs and their associated piRNAs, implicates the coevolved 3′ structure complex of tancRNAs to act as potential targets for the PIWI-piRNA complex. In summary, we define a class of lncRNAs with a conserved structural complex, post-transcriptional processing, and subcellular localization, and demonstrate a function for these RNAs and/or genomic loci in green lizard spermatocytes.
Results
The 3′-end of MALAT1 forms a triple helix structure and a tRNA-like structure
Although present from fish to human, MALAT1's primary sequence conservation is predominantly constrained to its 3′-end, which comprises two highly conserved U-rich regions followed by a conserved A-rich tract upstream of the RNase P cleavage site. A tRNA-like mascRNA is encoded downstream of the A-rich tract (Figure 1a, and (Wilusz et al., 2008)). To experimentally probe the structure of the 3′ end of MALAT1, we performed SHAPE (Selective 2′-hydroxyl acylation analyzed by primer extension) and DMS (Dimethyl sulfate) analysis on the human, lizard and zebrafish MALAT1 3′ ends. Low SHAPE reactivities are consistent with relatively low mobility of the backbone of a nucleotide (specifically the 2′-hydroxyl group). This occurs when the nucleotide participates in a base pair or a highly stable tertiary structure such as a triple helix configuration. Low DMS reactivities occur when a nucleotide is protected by interactions such as base pairing interactions or participation in a triple helix conformation. Raw probing traces were obtained via capillary electrophoresis for all RNAs studied (Figure 1b, Supplementary Figures S1 and S2). In all three RNAs, we observe significant protection of the U-rich and A-rich sequences against SHAPE and DMS reagents, consistent with the formation of a conserved triplex architecture, where U-A-U base triples represent the major organizing motif, intervened by a C-G-C base triplet (Figure 1c, Figure S1 and S2). The human and zebrafish triplexes contain 10 base triples. The lizard MALAT1 structures only contain 9 Watson-Crick base triples; however, this structure contains a G-A-C motif on the end of the triplex adjacent to the loop. The G-A-C motif can also form a base triple, as observed in the NMR structure of the aptamer domain of the preQ riboswitch (Kang et al., 2014). Each of the three structures also contains a conserved stem-loop. This helix protrudes from the triplex and is capped by a small tetraloop or pentaloop, depending on the RNA. Using previously developed RNA homology techniques (Budkevich et al., 2014; Tung et al., 2002), we constructed 3D models of the triplex tertiary structures (Figure 1d). The triplex structures contain base triples consisting of various combinations of A and U or various combinations of G and C along with a stem loop containing a large number of purine bases. These are all features observed in the triple helix revealed by crystallographic studies of the SAM-II riboswitch aptamer domain (Gilbert et al., 2008). The long stem-loop easily stacks onto the triple helix while maintaining reasonable stereochemistry.
Figure 1. The 3′ end of MALAT1 forms a triple helix structure.
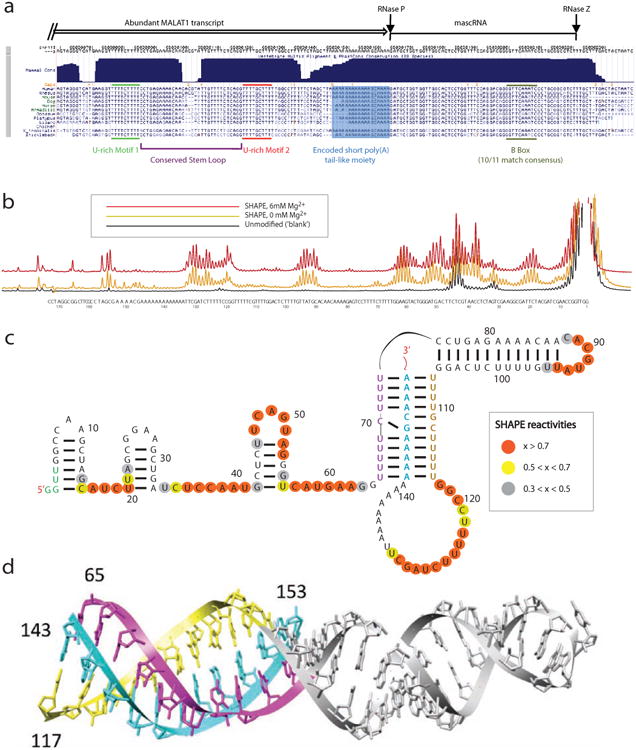
(a) The sequence conservation of the 3′ end of MALAT1. (b) SHAPE chemical probing of the 3′ end of human MALAT1 is consistent with a triple helix. Capillary electrophoresis traces display nucleotides with low and high SHAPE reactivity, corresponding to low and high flexibility. Black, unmodified control; gold, SHAPE reactivity for 0 mM Mg2+; red, SHAPE reactivity for 6 mM Mg2+. (c) Secondary structure of the 3′ end of human MALAT1, as inferred by SHAPE and DMS probing experiments (see supplementary data for DMS). Orange, high reactivity; yellow, medium reactivity; grey, low reactivity; un-circled nucleotides, near-zero reactivity. (d) Tertiary structure model of the 3′ end of human MALAT1 demonstrates stereochemical feasibility of a triple helix. Purple, yellow and cyan strand correspond to similarly colored strands in (c).
In contrast to the highly conserved triplexes, the regions of RNA preceding the triplexes (5′ ends) are highly reactive and vary significantly with respect to each other (data not shown). Human MALAT1 3′-end contains three consecutive stem-loops connected by highly reactive, and likely flexible stretches of RNA. Zebrafish contains a single stem-loop surrounded entirely by flexible bases, as does the lizard MALAT1 3′-end (Supplementary Figure S2).
We also performed SHAPE and DMS analysis on the human mascRNA and observed significant protection of the predicted stem structures against SHAPE and DMS reagents, consistent with the formation of a conserved cloverleaf architecture (Supplementary Figure S3).
A class of genomic loci with structures similar to the MALAT1 3′-end are present in vertebrate genomes
Given the presence of an unusual 3′-end structure (triplex helix and tRNA-like RNA) associated with a unique processing mechanism of the MALAT1 lncRNA that is conserved from fish to human, we hypothesized that this structure and mechanism may exist in other lncRNAs or mRNAs. To search for RNA molecules containing these domains, we built a covariance model (CM) profile (MALAT1-3h-tRNAlike.cm in Supplementary Data SF4), using INFERNAL (Nawrocki and Eddy, 2013) based on structures determined by chemical probing analysis described above (Figure 2a). We identified 132 hits in 35 vertebrate genomes (genome version information in Supplementary Data SF8) with an E-value less than 10-11 (Figure 2a, 2b and 2c, Supplementary Data SF7, SF9, SF10 and SF12). Based on the low E-values, we are confident that all or nearly all of these hits are homologous to the MALAT1 3′-end. However, some hits, particularly in the Medaka genome, have multiple mutations that disrupt the proposed secondary structure indicating that the structure is no longer being conserved in those cases.
Figure 2. A class of genomic loci with structures similar to the MALAT1 3′ end was identified in vertebrate genomes.
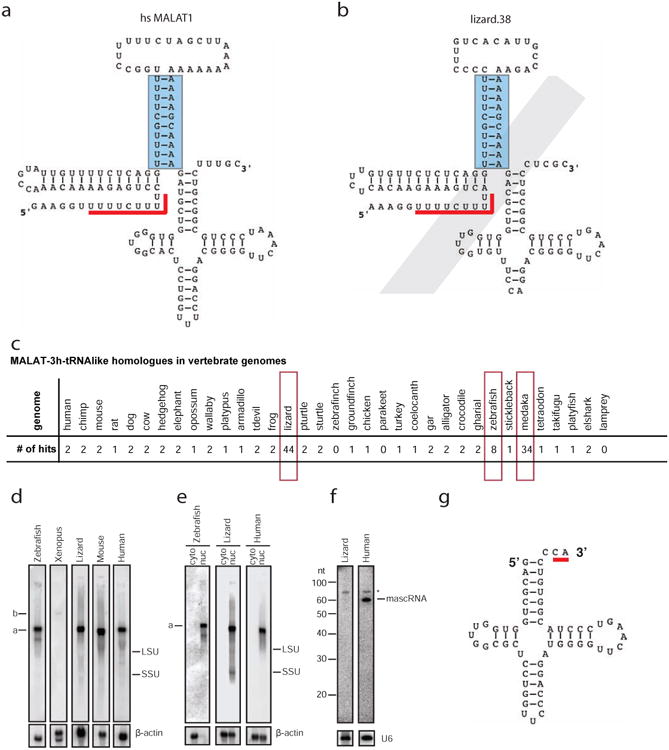
(a) The primary sequence and predicted secondary structure of human MALAT1 3′ module as identified by a homology search using Infernal 1.1rc1 against the human genome using CM file MALAT1-3h-tRNAlike.cm (Supplementary Data SF4). Only basepairs modeled by the CM are indicated with lines connecting two nucleotides. The triple helix is formed between the stem structure indicated by a blue rectangle and its upstream U-rich motif indicated by a red line. (b) One MALAT1 3′ end homologue (lizard.38) with a degenerative tRNA-like structure that was identified in the lizard genome using the MALAT1-3h-tRNAlike.cm CM profile. (c) Summary of MALAT1-3h-tRNAlike hits with an E value less than 1×10-11 in 35 genomes (see Supplementary Data SF7 and SF12). Note that most mammals have one or two hits, while lizard, zebrafish, and medaka have more than two MALAT1-3h-tRNAlike hits (highlighted in red open rectangles). (d) Northern blot analysis shows that MALAT1 orthologues were expressed from cultured cells of zebrafish (ZFL), Xenopus (A6), lizard (IgH-2), mouse (C2C12) and human (U2OS). (e) Northern blot analysis shows that MALAT1 is enriched in nuclei of cultured cells of zebrafish (ZFL), lizard (IgH-2), and human (U2OS). (f) Small RNA Northern blot analysis shows that lizard mascRNA is not detectable in the IgH-2 cell line. (g) Secondary structure of lizard mascRNA in the IgH-2 cell line with noncanonical CA tail modification indicated by a red line. LSU, large subunit of ribosomal RNA; SSU, small subunit of ribosomal RNA; cyto, cytoplasmic fraction; nuc, nuclear fraction; *, non-specific band. β-actin and U6 are loading controls.
Although MALAT1 3′-end homologues are not found in jawless vertebrates, or in any non-vertebrates examined, one or two MALAT1-like genomic loci were identified in most genomes except for zebrafinch (Figure 2c). Searches with BLAST as well as with INFERNAL using a model built only from the turkey hit found no homologs in the zebrafinch genome. We suspect that this may be due to incomplete coverage of that genome sequence. The elephant shark is the most distant organism from human in which we have identified a potential homolog of the MALAT1 triple helix and tRNA-like structure.
These two genes identified in most vertebrate genomes are homologues of the human MALAT1 and NEAT1 genes. These two adjacent lncRNAs with similar 3′-end structures likely represent a genomic duplication during evolution. In Xenopus, these two lncRNAs are oriented in a transcriptionally convergent fashion, which could represent another independent gene duplication event. Interestingly, a significantly greater number of hits were identified in the genomes of lizard (44 hits), zebrafish (8 hits), and medaka (34 hits), suggesting potential expansions of this element in these organisms. Taken together our results suggest that it is likely that the MALAT1 3′-end triplex and tRNA-like structure complex existed in the common ancestor of cartilaginous fish and mammals, and has been maintained through evolution with gene duplication and lineage-specific expansions.
To test whether the triple-helix structure and tRNA-like structure ever occur separately in genomes, we built CM profiles for the triple-helix (MALAT1-3h.cm in Supplementary Data SF5) and tRNA-like regions (MALAT1-tRNAlike.cm in Supplementary Data SF6), separately, and searched the genomes for homologues. In general, whenever there is a high-scoring hit to either the triple helix or tRNA-like region, these hits are adjacent and so a high scoring hit to the combined model (MALAT1-3h-tRNAlike.cm in Supplementary Data SF4) also exists. The main exceptions are in medaka, zebrafish, and one case in lizard. The consistency of the hits between the three models (MALAT1-3h-tRNAlike.cm, MALAT1-3h.cm and MALAT1-tRNAlike.cm, summarized in Supplementary Data SF7) suggests that the MALAT1 3′-end triple-helix and tRNA-like structures co-occurred.
MALAT1 is highly expressed in vertebrates
To determine if MALAT1 orthologues in lizard, Xenopus, and zebrafish are expressed, we performed Northern blot analysis using RNAs extracted from cell lines of each species (Figure 2d). Lizard and zebrafish MALAT1 were expressed at a high level in IgH-2 (lizard) and ZFL (zebrafish) cells, respectively, and exhibit a similar size (∼7 knt) to human and mouse MALAT1 in U2OS and NIH3T3 cells, respectively. While Xenopus MALAT1 is ∼11 knt in length and is expressed at a relatively lower level in Xenopus A6 cells. Similar to human and mouse, lizard and zebrafish MALAT1 are also enriched in the cell nucleus (Figure 2e). Human and mouse MALAT1 are specifically localized to nuclear speckles, where nuclear splicing factors are enriched (Bernard et al., 2010; Hutchinson et al., 2007). RNA FISH analysis of lizard MALAT1 in kidney and IgH-2 cells shows a punctate labeling pattern in nuclei (Supplementary Figure S4, and data not shown for IgH-2 cells), however, whether these domains directly correspond to nuclear speckles in lizard cells has not been directly tested.
Surprisingly, we did not detect mascRNA in lizard IgH-2 cells by Northern blot analysis (Figure 2f), despite high expression of MALAT1 lncRNA. We hypothesize that lizard mascRNA may be unstable as previously suggested for menRNA in some cell types (Wilusz et al., 2011). However, using a more sensitive PCR-based method, we were able to clone and sequence mascRNA from IgH-2 cells and found that lizard mascRNA has a noncanonical 3′ tail modification (Figure 2g and Supplementary Figure S5). Eight clones contained a classical CCA tail addition, while 21 clones contained a truncated CA tail addition. Atypical 3′ CCACCA addition to menRNA has been reported to impact menRNA stability (Kuhn et al., 2015; Wilusz et al., 2011) but it remains to be determined if these noncanonical 3′ modifications are a cause or a consequence of RNA stability and their functional significance. It is noted that the 3′ first non-paired discriminator base of lizard mascRNA is cytosine (C), which may impact its stability, as the discriminator base contributes significantly to the stem structure and stability of the duplex and cytosine is the least frequent discriminator nucleotide observed at position 73 of tRNA sequences (Limmer et al., 1993). In addition, how lizard CCA adding enzyme is able to only add CA to a tRNA-like structure remains to be investigated. Taken together, MALAT1 lncRNA is highly expressed and enriched in the nucleus in lower organisms, while the tRNA-like mascRNA is not stable when it contains a noncanonical 3′ tail modification in lizard.
A class of long non-coding RNAs are produced from MALAT1-like genomic loci in Anolis carolinensis in a tissue-specific manner
To determine if MALAT1-like genomic loci are transcriptionally active, we performed Northern blot analysis using a MALAT1 cDNA probe and consensus oligonucleotide probe (COP) upstream of the RNAse P cleavage site to probe RNAs isolated from IgH-2 cells, and we detected a single RNA species with the expected size of MALAT1 (Figure 3b). Northern blot analysis on different tissues of Anolis carolinensis detected expression of MALAT1 in all tissues examined, while expression of a family of RNA species with different sizes were predominantly present in testis (Figure 3c).
Figure 3. Long noncoding RNAs are produced from MALAT1-like genomic loci in Anolis carolinesis in a tissue-specific manner.
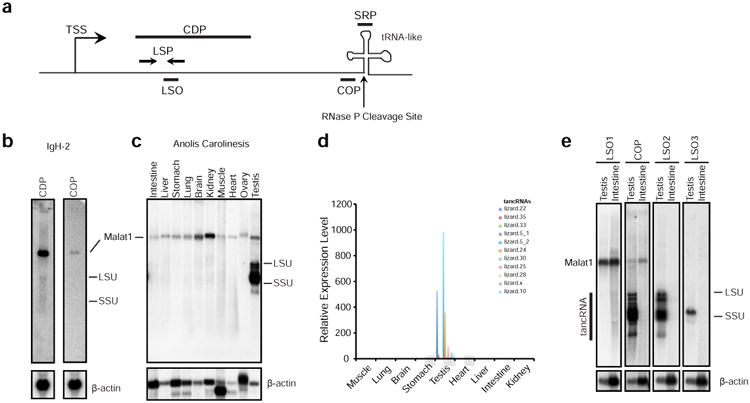
(a) Schematic of the transcriptional unit of the Malat1 3′ module homologous loci. TSS, transcriptional start site; CDP, IgH-2 cDNA Probe (used in panel b); COP, Consensus Oligonucleotide Probe that is derived from the multi-sequence alignment of 44 homologues (used in panels b, c, and e); LSP, locus-specific primers (used in panel d); LSO, locus-specific oligonucleotide (used in panel e); SRP, small RNA probe; DSO, downstream oligonucleotide; the vertical arrow, the predicted RNase P cleavage site just before the tRNA-like structure. (b) Northern blot analysis shows that MALAT1 RNA is highly expressed in lizard IgH2 cells detected by both cDNA probe and COP, but no other RNA species are detected by the COP. (c) Northern blot analysis shows that COP labels MALAT1 in all tissues of Anolis carolinensis and multiple RNA species in testis (named as tancRNAs). (d) qRT-PCR analysis using locus-specific primer (LSP) sets shows that tancRNAs from different genomic loci are specifically expressed in lizard testis. Y axis, relative expression level; X axis, different tissues. (e) Northern blot analysis shows that tancRNAs are expressed from different genomic loci. Note that LSO1 recognizes MALAT1, LSO2 recognizes multiple tancRNA species (lizard.20, lizard. 12, lizard.8), and LSO3 detects one major tancRNA species (lizard.30). β-actin is the loading control.
In order to distinguish if these different sized RNA species are transcripts from different MALAT1-like loci or are processed products of abundant MALAT1, we performed loci-specific qRT-PCR and detected a significant amount of transcripts from upstream regions of multiple MALAT1-like loci (Figure 3d). Using probes recognizing specific MALAT1-like loci, we detected specific RNA species (Figure 3e).
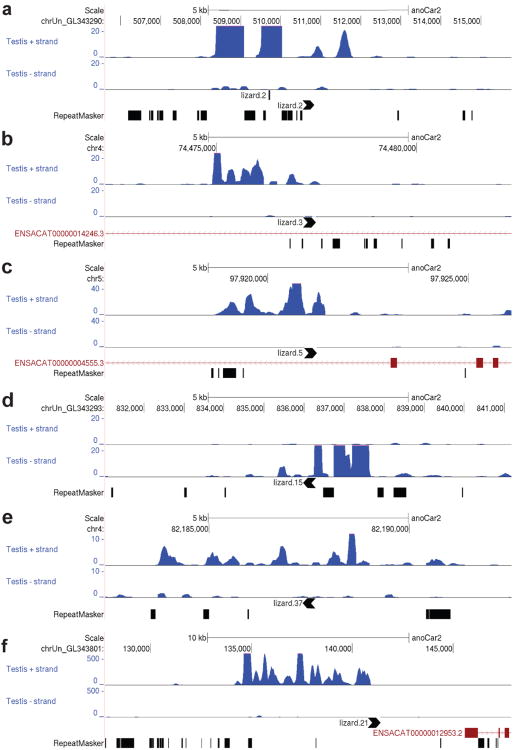
As these RNA species may not have canonical poly-adenosine tails, NSR-seq using “not-so-random” primer sets was performed on both lizard testis and brain mRNAs to obtain full-length, strand-specific profiles of non-ribosomal RNA transcripts independent of their 3′ tail status ((Armour et al., 2009) and methods). 13.6 million reads from testis and 15.6 million reads from brain were mapped to the lizard genome. Consistent with the results from Northern blot analysis, NSR-seq analysis revealed a high expression of MALAT1 in both brain (data not shown) and testis (Figure 4f) and in contrast, transcripts from different MALAT1-3′ end like genomic loci are specifically expressed in testis (Figure 4a-e). Most transcripts, including MALAT1, are derived from the same strand as the orientation of MALAT1-3′ end like elements, which typically sit at their 3′ ends indicated by an apparent drop of downstream read coverage (lizard.2, lizard.3, lizard.5, lizard.15, and lizard.21 in Figure 4a-d and Figure 4f). This supports the idea that MALAT1-3′ end like elements serve as a 3′ end RNA module for processing, stabilization, and/or other functions.
Figure 4. Different examples of transcripts from MALAT1-like genomic loci.
(a) – (f) Normalized RNA-Seq reads from testis (upper track: reads on sense strand, lower track: reads on antisense strand). Black arrowheads denote MALAT1-3h-tRNAlike loci and their direction of transcription. Chromosome location and annotated Ensembl transcripts are indicated. Read depth scale is displayed on the y-axis. Interspersed repeats and low complexity DNA Sequences are indicated in the RepatMasker track. 6 examples are shown; nomenclature according to Supplementary Data SF7.
These transcripts do not contain potential ORFs longer than 100 amino acids. Since multiple MALAT1-like loci are highly transcribed in lizard testis we have named these transcripts testis-abundant non-coding RNAs (tancRNAs). Size heterogeneity of tancRNA species likely comes from divergent sequences, different promoters, and unique transcription initiation sites upstream of each MALAT1 3′-end-like module. Future investigation will shed insights on how this novel class of RNA species is regulated and evolved at a molecular level.
Interestingly, some transcripts, such as that from the lizard.37 locus, appear to be expressed from the opposite strand with a relatively low read coverage and have the MALAT1-3′ end like element in the middle (Figure 4e). These may represent a different RNA species from that described above, and may have a role in tancRNA regulation.
Multiple small RNA species are produced from tancRNA loci
To study the post-transcriptional processing of these RNAs, Northern blot analysis was performed using oligonucleotide probes that hybridized to the triple-helix (COP), the tRNA-like structure (small RNA probe, SRP) (Figure 5b), and a region immediately downstream of the tRNA-like structure (data not shown). Both MALAT1 and tancRNAs in lizard testis, end just upstream of the predicted RNAse P cleavage site, indicating that nascent tancRNA transcripts are processed by the tRNA processing machinery similar to processing of human MALAT1 (Figure 5b). A very light smear was noted in testis, suggestive of an incomplete cleavage. This is also supported by noticeable NSR-seq read coverage downstream of the tRNA-like structure (Figure 4a-d). The structural analysis of the 3′ end of tancRNAs revealed a similar structural complex with a triplex structure associated with a stem-loop as defined in human MALAT1 (Supplementary Figure S6).
Figure 5. tancRNAs are processed and enriched in nuclei and multiple small RNA species are produced from tancRNA loci.
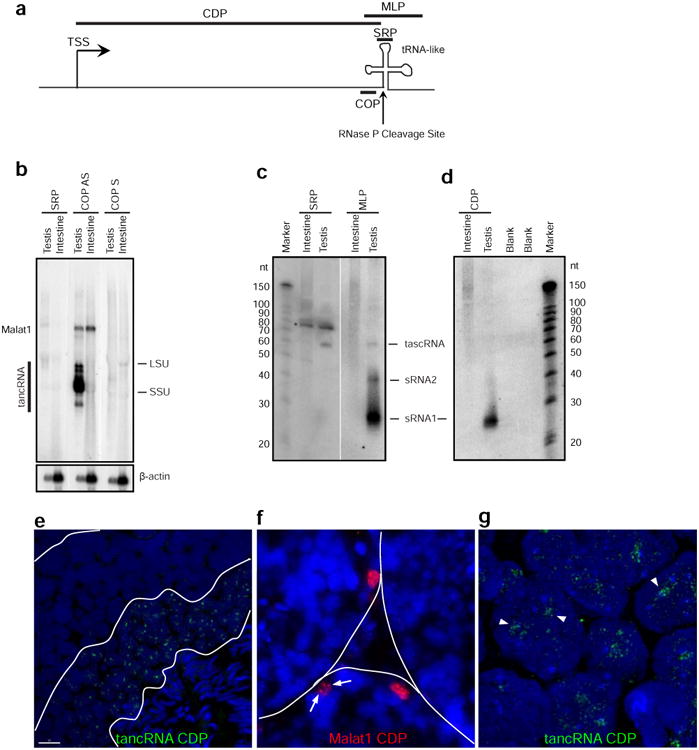
(a) Schematic of the transcriptional unit at the Malat1 3′ module homologous loci. TSS, transcriptional start site; CDP, cDNA Probe (used in panels d, e, f, and g); MLP, MALAT1 3′-like probe (used in panel c); the vertical arrow, the predicted RNase P cleavage site just before the tRNA-like structure. (b) tancRNAs were processed as human MALAT1. Large RNA Northern blot analysis shows that SRP (antisense of mascRNA/tascRNA) does not recognize any significant bands (lane 1). COP AS (tancRNA consensus antisense probe) detects both MALAT1 and tancRNAs (lane 2), while COP S (tancRNA consensus sense probe) does not recognize any significant bands (lane 3). (c) Multiple small RNA species were produced from tancRNA loci. Small RNA Northern blot analysis shows that SRP (mascRNA/tascRNA antisense oligonucleotide probe) detects mascRNA/tascRNA in lizard testis (left), while MLP (consensus PCR pooled probe) detects three small RNA species with sizes of 26, 40, and 60 nt (right). (d) Small RNAs with the size of piRNAs were produced from tancRNA loci. Small RNA Northern blot analysis detects a strong RNA band with the size of 26 nt using CDP (a cDNA probe from the lizard.11 locus, see Supplementary Data SF7 and SF12). (e) RNA FISH shows that tancRNAs are localized to nuclei of pachytene and/or round spermatocytes in adult lizard testis. (f) RNA FISH shows that MALAT1 is enriched in nuclei of the periphery of seminiferous tubules. Arrows indicate nuclear punctuate signal pattern. (g) High-resolution imaging of RNA FISH (in e) shows that tancRNAs occupies distinctive nuclear domains (arrowheads). LSU, large subunit of ribosomal RNA; SSU, small subunit of ribosomal RNA. Equal amount of total RNAs were loaded for each lane. β-actin is the loading control. *, non-specific band.
We refer to the small RNAs associated with the processing of MALAT1-like transcripts as tancRNA-associated small cytoplasmic RNAs (tascRNAs). Northern blots for small RNAs using a consensus probe detected a band with a size of ∼60 nt, corresponding to mascRNA and/or tascRNAs (Figure 4c) enriched in testis. mascRNA was not detectable in intestine, which is consistent with the finding that mascRNA is unstable in the lizard cell line (Figure 2f). Northern analysis using ∼200 bp DNA fragments from five tancRNA loci containing the triple helix and tRNA-like structures also detected the tascRNA, but surprisingly identified two additional RNA species (sRNA1 and sRNA2) with sizes around 45 nt and 28 nt, respectively (Figure 5c). The ∼28 nt small RNAs are the most abundant among the three RNA species. To further test if the region upstream of the RNAse P cleaveage site contributes to the production of the abundant sRNA1, we performed 5′ RACE and determined the 5′ end of one tancRNA and cloned a full-length cDNA, named tancRNA.11 (chr5:105,700,153-105,702,202) from the lizard.11 locus (Supplementary Figure S7). This transcriptional unit is also supported by NSR-Seq analysis (data not shown). Northern analysis using a tancRNA.11 cDNA probe detected the abundant sRNA1, while sRNA2 and tascRNAs were not detected (Figure 5d). These results demonstrated that the processing of the 3′ end triple-helix and tRNA-like structure complex is retained in lizard tancRNAs, and that multiple small RNA species are generated from tancRNA loci.
TancRNAs are enriched in nuclei of round spermatocytes
To determine what cell types express tancRNAs, we performed RNA-FISH using cDNA probes for Malat1 and tancRNA on adult lizard testicular frozen sections. The developmental stage of adult lizard testis was assessed by SYCP3 staining, showing the presence of pachytene nuclei (data not shown). Interestingly, Malat1 is highly expressed in cells localized at the periphery of seminiferous tubules and enriched in a punctuate nuclear distribution (Figure 5f), while tancRNAs (detected by the lizard.11 cDNA probe which recognizes the 3′-end module, which is shared by all tancRNA loci and the upstream sequence of five tancRNA loci) were highly expressed in differentiating germ cells located in the middle portion of seminiferous tubules, consistent with the location of pachytene and round spermatocytes and also enriched in nuclei with a punctuate staining pattern (Figure 5e). While the tancRNA cDNA probe could theoretically also detect small RNAs produced from the same locus, the majority of signals are likely derived from lncRNAs as a similar RNA-FISH pattern was observed using an oligonucleotide probe that recognizes tancRNAs (data not shown). Super-resolution imaging using the OMX microscope demonstrated that tancRNA molecules were dispersed throughout the entire nuclei with an enrichment in interchromatin spaces (Figure 5g). These data indicate that tancRNAs are nuclear lncRNAs in pachytene spermatocytes.
tancRNA loci produce piRNAs from their antisense strand
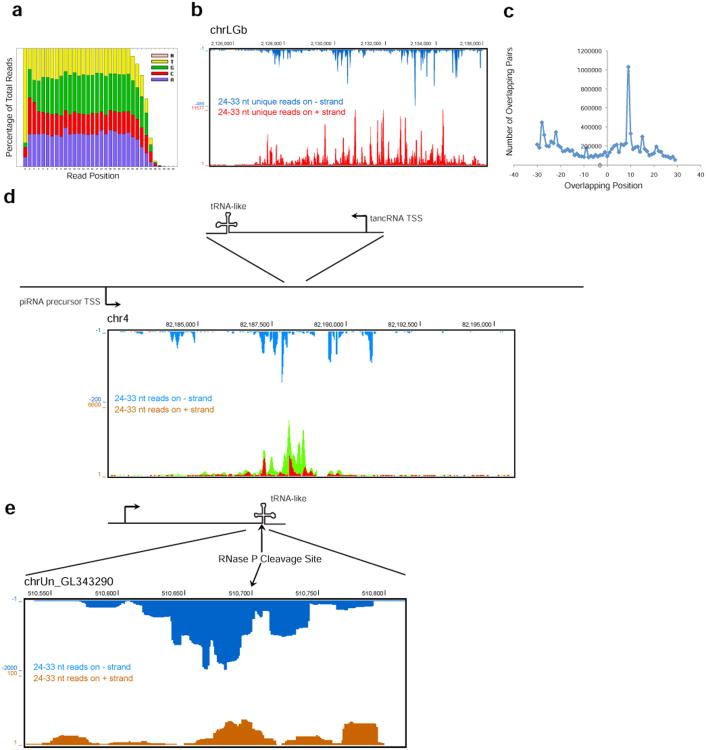
The sRNA1 produced from tancRNA loci has a similar size to piRNAs, PIWI-interactingsmall RNAs specifically expressed in germ cells and associated somatic cells (Aravin and Hannon, 2008). Given the expression pattern of sRNA1 and related tancRNAs, we hypothesized that sRNA1 could represent a type of lizard piRNA, and sRNA2 the intermediate product. To test this possibility, we performed RNA-seq for small RNAs with the size ranges from 19 to 33 nt (19.0 million reads) and 52 to 68 nt (24.6 million reads). RNA molecules with sizes from 24 to 33 nt exhibited classical features of piRNAs, including 5′ U bias (Figure 6a). Interestingly, we noticed a significant 10A bias in the whole population of sRNA1 (Figure 6a). When filtering out 1U RNA molecules, the 10A bias becomes more prominent (data not shown). This suggests that there may be a ping-pong amplification signature in lizard pachytene piRNAs.
Figure 6. piRNAs are produced from the opposite strand of tancRNAs.
(a) Positional distribution of nucleotide composition of small RNAs sequenced from adult lizard testis. A strong 1U bias and a weak 10A enrichment were noticed. (b) Small RNAs with the size of 24-33 nt were uniquely mapped to a dual-strand piRNA cluster on chrLGb. (c) A strong ping-pong signature of piRNAs from the dual-strand cluster on chrLGb (see b). X axis, overlapping index; Y axis, number of overlapping pairs. (d) piRNAs mapped to a tancRNA locus (lizard.37, see Supplementary Data SF7 and SF12) on chr4. Orange, unique mapper on – strand; blue, multiple mapper on – strand; red, unique mapper on + strand; green, multiple mapper on + strand. (e) piRNAs mapped to the 3′ end of a tancRNA locus (lizard.2, see Supplemental File SF7 and SF12) on chrUn_GL343290 with multiple mappers (reporting = 50).
More than 94% of small RNAs with sizes ranging from 24 nt to 33 nt were successfully mapped to the lizard genome (version anoCar2.0). Small RNAs with sizes ranging from 52 to 68 nt had a similar distribution along the lizard genome, indicating that these molecules are likely the intermediate products of piRNA biogenesis. A total of 763 piRNA clusters were identified with 30 unique small RNA species per kb. The average size of these clusters is 9.4 kb. A number of dual strand overlapping piRNA clusters were identified. For example, the piRNA cluster at chrLGb:2,125,001-2,136,000 is ∼11 kb in length with a strong piRNA production from the (+) strand (336,088 unique reads clapped into 4950 unique RNA species) and a relative weaker production from the (–) strand (7522 unique reads clapped into 1460 unique RNA species) (Figure 6b). Analysis of overlapping pairs demonstrated a strong peak for 10 nt overlap, indicating ping-pong amplification for this cluster (Figure 6c). This is a rather unexpected finding since a ping-pong signature has not been reported in non-transposon derived pachytene piRNAs (Beyret et al., 2012), suggesting that lizard pachytene piRNAs biogenesis and action may be different from those in mammalian species or that lizard pachytene piRNAs have a stronger ping-pong signature than their mammalian counterparts.
Eleven tancRNA loci overlap with piRNA clusters (p < 0.001, P(E)=0.20% and P(O)=22.45%). Three prominent piRNA clusters (size: > 15 kb; piRNA density: >80 unique small RNA species per kb), Chr2:120,609,001-120,674,000, chr4:82,182,473-82,194,005, and chrUn_GL343674:2,001-17,000, were identified and overlapping with tancRNA loci, lizard.40, lizard.37, and lizard.33, respectively. piRNAs were only produced from the antisense strand relative to the orientation of tancRNAs or the triple-helix and tRNA-like complex (Figure 6d). This is not a random event and is statistically significant that three of 44 tancRNA loci produce piRNAs from the antisense strand (p < 0.001), and is consistent with the strand orientation of long transcripts from these loci (Figure 4e). Because of primary sequence similarity, many piRNAs produced from three clusters can recognize tancRNAs with the triple helix and tRNA-like structures from different genomic loci. In contrast, the remaining eight tancRNA-associated piRNA clusters are shorter in size (average 8kb) with a much lower piRNA density (average 51piRNA/kb), among which three are from the same strand while five are from the opposite strand of tancRNAs. In addition, there are tancRNA loci which do not overlap with piRNA clusters but can be recognized by piRNAs likely produced from other tancRNA loci in the genome. For example, there are 5,137 small RNA-seq reads mapped to the locus at chrUn_GL343290:510,550-510,800 which actively produces tancRNA (lizard.2, Figure 6e). These results indicate that piRNAs recognize tancRNAs 3′-ends, but do not appear to mediate targeted RNA cleavage (Figure 6e), suggesting a unique mechanism(s) of action for these piRNAs and tancRNAs.
One possibility is that tancRNAs may serve as nuclear targets of the PIWI-piRNA complex. To investigate this, we performed computational analysis of the PIWI protein family in lizard, and found that there are four PIWI homologues, named LIWI1a and LIWI1b, LIWI2, and LILI in lizard (Supplementary Figure S8). All four PIWI homologues are expressed at a relatively high level in testis and ovary at the RNA level (Supplementary Figure S8), while LIWI2 protein is only detected in testis (Supplementary Figure S8). Interestingly, LIWI2 carries a variant residue S at the third residue of the conserved catalytic triad DDH (Song et al., 2004), and is predicted to be enzymatically inactive (Figure 7b). Chromatin RNA immunoprecipitation (ChRIP) analysis showed that LIWI2 protein is associated with tancRNAs (Figure 7c), and also demonstrated that tancRNAs are associated with chromatin (Figure 7a). Taken together, piRNAs produced from the antisense strand are able to recognize tancRNAs through the 3′-end triple-helix/tRNA-like structure complex, and potentially direct the enzymatically inactive LIWI2 protein complexes to chromatin in cis via nascent tancRNA transcription and/or in trans mediated by 5′ tancRNA-associated RNA-chromatin interaction (Figure 7d). The proposed in cis mechanism is analogous to the PIWI-piRNA-pit-RNA mediated sequence-specific methylation of a mouse imprinted locus (Watanabe et al., 2011). The in trans mechanism likely has a profound impact on chromatin remodeling and genome-wide expression during the development of pachytene spermatocytes in lizard, representing a potential mode of action of non-transposon-derived pachytene piRNAs in human and mouse as well, and thus warrants further investigation.
Figure 7. 3′-end of tancRNAs act as targets of antisense piRNAs.
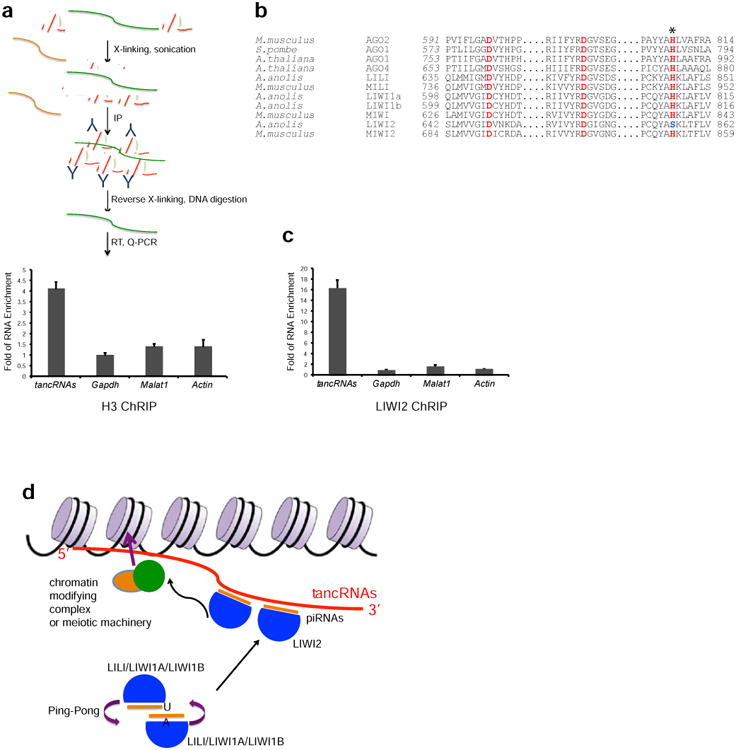
(a) A schematic diagram of Chromatin RNA Immunoprecipitation (ChRIP) analysis. ChRIP using antibody against Histone H3 shows that tancRNAs (primer sequences in Supplementary Data SF14) is associated with chromatin. (b) Sequence alignment showing the catalytic triad (DDH motif) of known Slicers and MIWI. LIWI2 has a variant of the third His to Ser (H856S; marked with an asterisk). (c) ChRIP analysis using antibody against LIWI2 shows that tancRNAs is associated with LIWI2. X axis, RNA examined; Y axis, Fold of RNA enrichment. Sample size n = 3. (d) A proposed working model of tancRNA-piRNA-Liwi2 complex. TancRNAs act as targets of PIWI-piRNA complexes through their 3′ end triplex structure. 5′ end tancRNAs serve as domains to interact with DNA and/or chromatin, therefore tethering PIWI-piRNA complexes to specific genomic loci to regulate chromatin activity.
Discussion
The recent discovery of lncRNAs in different species and their implicated potential roles in a variety of cellular and molecular processes demands a systematic classification of these species of RNA molecules based on their intrinsic properties. However, weak primary sequence conservation makes it a rather challenging task. We analyzed the secondary and tertiary RNA structures of the 3′-end of MALAT1 lncRNA and identified a family of vertebrate lncRNAs with this unique 3′-end structural complex. This analysis suggests a role for these RNAs in lizard, and provides an example to study structure, evolution, and function of lncRNAs.
Chemical probing analysis supports prior studies that demonstrated that the MALAT1 3′-end module harbors a triple helix with an adjacent hairpin loop, followed by a tRNA-like structure (Brown et al., 2014; Brown et al., 2012; Wilusz et al., 2012). All identified MALAT1 3′-end homologues in vertebrate genomes encompass both structural moieties, suggesting that they co-occurred. These structures first appeared in jaw-containing vertebrates, produced NEAT1 incRNA presumably by gene duplication, and expanded in lizard by an unknown mechanism. However, it is still not clear where this highly structured 3′-end module originated. The closest examples are tRNA-like structures (TLS) in the 3′ UTR of the positive-strand of plant RNA viruses, which have roles in viral genome replication and translational enhancement. These TLSs are proceeded by an upstream pseudoknot domain (UPSK), which also provides translational enhancement (Dreher, 2009). It will be interesting to test if the UPSKs can form a triple-helix similar to MALAT1 and tancRNAs. The discovery of a triple helix structure in the 3′-end of MALAT1 RNA, the ENE element of PAN RNA, and in a pseudoknot structure of Telomerase reverse transcriptase RNA component (TERC) demonstrates that both viral and eukaryotic organisms utilize the triplex structure for RNA stability and function, respectively (Brown et al., 2014; Brown et al., 2012; Mitton-Fry et al., 2010; Qiao and Cech, 2008; Wilusz et al., 2012). Taken together, the MALAT1 3′-end represents a unique complex including a co-evolved triple helix and tRNA-like structure for RNA stability and processing, respectively. However, several questions remain to be answered: for example, does the triple helix structure have additional functions besides regulating RNA stability; what is the function of the tRNA-like structure; does it only provide an RNase P cleavage site to produce lncRNAs with a triple helix 3′-tail. The recent identification of one MALAT1 triplex-interacting protein, RNA methyltransferase-like protein 16 (METTL16), suggests the existence of a class of triple-stranded RNA binding proteins (Brown et al., 2016). Future identification of additional proteins associated with the triple-helix and tRNA-like structures will provide further functional insights.
A recent genome-wide bioinformatics search identified ∼200 distinct types of ENE-like structures in the intronless transposable element RNAs mostly in plant and fungal genomes (Tycowski et al., 2016). It was postulated that the acquisition of an ENE compensates for the RNA-destabilizing effects of intron loss during transposable element (TE) horizontal transfer (Tycowski et al., 2016). The presence of multiple copies of MALAT1 3′-end homologues (tancRNAs) in the lizard genome suggests that these genes may behave as transposons and/or their RNA product may have a previously undescribed function. Surprisingly, multiple small RNA species were produced from these loci, including piRNAs from the antisense strand. Despite the high abundance of piRNAs complementary to their 3′ ends, tancRNAs are relatively stable. It is unlikely that these tancRNAs serve as cleavage targets for PIWI-piRNA complexes as they are not degraded. The triplex-complex may be resistant to PIWI-piRNA cleavage. A second possibility is that the PIWI-piRNA complex recognizing tancRNAs may not possess enzymatic activity. The latter is indeed supported by the finding that LIWI2 (lizard homologue of MIWI2) is potentially an enzymatically inactive PIWI protein based on in silico protein sequence analysis. Altogether, we propose that tancRNAs act as targets of PIWI-piRNA complexes through their 3′-end triplex structure and that 5′-end tancRNAs serve as domains to interact with chromatin factors, therefore enabling the tethering of PIWI-piRNA complexes to specifically regulate chromatin activity genome-wide (Figure 7d). Consistent with this hypothesis human MALAT1 and NEAT1 have been shown to associate with the TSS and gene bodies of active genes in somatic cells (Engreitz et al., 2014; West et al., 2014). We hypothesize that germline-specific lncRNAs with similar function exist in other vertebrate genomes. It will be important to identify and characterize them in order to understand the function of the pachytene piRNA-PIWI complex. For example, a lncRNA has been identified to regulate DNA methylation in cis at the imprinted mouse Rasgrf1 locus (Davidovich et al., 2013; Watanabe et al., 2011).
piRNAs are a class of small silencing RNAs in animals that are generated from piRNA clusters, protect the genome, and may carry out additional yet to be defined functions (Aravin et al., 2006; Aravin and Hannon, 2008; Girard et al., 2006; Grivna et al., 2006; Lau et al., 2006). As new transposons migrate in the genome, they eventually land in piRNA clusters, which will generate antisense piRNAs, therefore mediating an “immune” response against this type of transposons. The unique expansion of tancRNAs in Anolis carolinensis led us to speculate that tancRNAs could behave as transposons. Although they are apparently under the control of piRNAs, they are immune to degradation presumably due to the enzymatically inactive form of LIWI2 or the secondary structure of the 3′-end of tancRNAs. Therefore, we propose that tancRNAs not only escaped piRNA-PIWI complex-mediated cleavage, but likely hijacked the complex for chromatin remodeling and/or DNA modification during spermatogenesis.
In mammals, the transposon-silencing response occurs mainly at the embryonic stage, when piRNA clusters are enriched in transposon sequences. However, abundant and transposon-depleted piRNAs produced at pachytene (pachytene piRNAs) are still a mystery (Aravin and Hannon, 2008). RNA-seq analysis of small RNAs from adult Anolis carolinensis testis demonstrated that lizard pachytene piRNA clusters have a low content of transposons (Zhang B and Spector DL, unpublished data), but all dual-strand clusters show a strong ping-pong amplification signature. Moreover, LIWI2 is highly expressed in adult lizard testis, while MIWI2 is specifically expressed in embryonic gonads, suggesting unique mechanisms of piRNA biogenesis and action during lizard spermatogenesis. Therefore, adult lizard spermatogenesis may serve as a model to study biogenesis and function of pachytene piRNAs and PIWIs.
In summary, we have defined a class of lncRNAs with co-evolved 3′-end structures, and have characterized an acquired function of these RNAs as potential targets of the PIWI-piRNA complex in Anolis carolinensis.
Materials and Methods
All animal protocols have been approved by the CSHL Animal Care and Use Committee.
INFERNAL and Covariance Model
We created a covariance model (CM) using an iterative procedure as follows. Using Infernal v1.1rc1, we built a CM based on the human MALAT1 3′ end using the cmbuild program, calibrated it for E-value reporting with the cmcalibrate program, and searched several vertebrate genomes for high scoring hits with the cmsearch program. We then aligned those hits to the CM to create a new alignment and filtered that alignment to 92% sequence identity and built a new CM from the filtered alignment. We repeated this process of building, calibrating, searching, aligning and filtering for two additional iterations with some manual refinement of the alignment after each iteration to produce our final CM in the file MALAT1-3h-tRNAlike.cm (Supplementary data SF4) from our final alignment in the file MALAT1-3h-tRNAlike.stk (Supplementary Data SF1). Default Infernal v1.1rc1 parameters were used for all steps (cmbuild, cmcalibrate, cmsearch, and cmalign programs).
RNA-seq Library Preparation
Libraries for deep sequencing were prepared from RNAs extracted from lizard testis and/or brain using TRIzol (Invitrogen, NY) following the long RNA-Seq (called NSR-Seq) protocol (Armour et al., 2009) and the standard protocol for small RNAs (He et al., 2012). For NSR-seq, briefly, first strand cDNA synthesis was performed using SuperScript III (Invitrogene, CA) and the “not-so-random” forward primer set (5′ TCCGATCTCTNNNNNNN 3′), followed by second strand synthesis using exo- Klenow fragment (New England Biolabs, MA) and the “not-so-random” reverse primer set (5′ TCCGATCTGANNNNNNN 3′). In order to deplete ribosomal RNAs from the libraries, “not-so-random” primer sets were designed by pooling 749 unique hexamers with no perfect match to human cytoplasmic 18S and 28S rRNA and mitochondrial12S and 16S rRNA transcripts. Libraries were subsequently amplified by PCR using primers (5′AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCT 3′ and 5′ CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC TGA 3′) and paired-end sequenced for 50×2 cycles on Illumina Genome Analyzer II according to manufacturers' instructions. For small RNA-seq, briefly, RNA was successively ligated to 3′ and 5′ adaptors, gel purified after each ligation, reverse transcribed, and PCR amplified using Solexa sequencing primers. PCR product was gel purified, quantified, and sequenced for 36 cycles or 76 cycles on Illumina Genome Analyzer II according to manufacturers' instructions.
Sequence Processing, Mapping, and Annotation
For NSR-Seq data, the quality of the raw data was evaluated using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and reads were mapped to anoCar2.0 using STAR v2.4.1(Dobin et al., 2013), resulting in an overall mapping efficiency of 80%. Mapped reads were normalized for read depth as well length, and UCSC tracks were created using HOMER (Heinz et al., 2010), allowing for both unique hits and multi-mappers. Parameters were adjusted for strand-specific paired end sequencing.
For small RNA-seq data, raw reads obtained from the Illumina pipeline were trimmed from 3′ linker and filtered for low-quality reads. Unique sequences 24 nt or longer in length were mapped to the anoCar2.0 assembly of the lizard genome using bowtie (http://bowtie-bio.sourceforge.net/index.shtml) allowing 2 mismatches (Langmead et al., 2009).
Northern Blotting Analysis
Northern blots were performed as described previously (Zhang et al., 2012) with minor modifications. Briefly, 10-20 μg RNAs longer than 200 nt were resolved by 1% denaturing agarose gel electrophoresis and transferred to Hybond-N membrane (GE Healthcare, WI) by capillary transfer, followed by UV cross-linking. Small RNAs were separated by 15% denaturing polyacrylamide gel electrophoresis and electroblotted to Hybond NX membrane (GE Healthcare, WI), followed by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide cross-linking at 55°C for 1 hr. cDNA probes (sequence information in Supplementary Data SF14) were labeled with [α-32P]dCTP using the Prime-It RmT Random Primer Labeling Kit (Stratagene, CA). Oligonucleotide probes (sequence information in Supplementary Data SF14) were labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs, MA). Hybridization was carried out in NorthernMax Prehyb/Hyb Buffer (Ambion, NY) at 42°C for cDNA probes. Hybridization was carried out in ULTRAhyb-Oligo Hybridization Buffer (Ambion, NY) for oligonucleotide probes. Blots were visualized using the Fujifilm Life Science FLA-5100 imaging system (Fujifilm Life Science USA, CT).
Other experimental procedures are included in Supplementary Information (S1).
Supplementary Material
Document S1: Supplemental Experimental Procedures and Figures S1-S8.
Supplemental Dataset: Information of alignment, cm model, cmsearch results, and secondary structure of MALAT1-3h-tRNAlike (SF1-SF13, Related to Figure 2) and sequence information of oligonucleotides and primers (SF14, Related to Figures 2, 3, 5, 7).
Fourteen supplementary datasets are compressed in this file (SF1): The alignment used to build the covariance model MALAT1-3h-tRNAlike.cm (SF4) in Stockholm format. (SF2): The alignment used to build the covariance model MALAT1-3h.cm (SF5) in Stockholm format. (SF3): The alignment used to build the covariance model MALAT1-tRNAlike.cm (SF6) in Stockholm format. (SF4): The covariance model of the combined MALAT1 triple helix complex and the tRNA-like structure. (SF5): The covariance model of the MALAT1 triple helix complex. (SF6): The covariance model of the MALAT1 tRNA-like structure. (SF7): A text file with cmsearch information on 132 MALAT1-3h-tRNAlike elements using three models of MALAT1-3h-tRNAlike.cm, MALAT1-3h.cm, and MALAT1-tRNAlike.cm. (SF8): List of the genomes searched and the versions of each of those genomes. (SF9): A FASTA file containing 132 MALAT1-3h-tRNAlike elements. (SF10): Stockholm-formatted alignment of 132 MALAT1-3h-tRNAlike elements. (SF11): Consensus model of secondary structure of MALAT1-3h-tRNAlike elements. (SF12): Secondary structure of 132 MALAT1-3h-tRNAlike elements. (SF13): Secondary structure template of MALAT1-3h-tRNAlike element. (SF14): Sequence information of oligonucleotides and primers.
Acknowledgments
We thank Carmen Berasain, Jan Bergmann, Megan Bodnar, Melanie Eckersley-Maslin, Stephen Hearn, Ileng Kumaran, Jingjing Li, Cinthya Zepeda-Mendoza, Junwei Shi, and Rui Zhao of the Spector laboratory and Gregory J. Hannon for helpful discussions. We thank Vasily V. Vagin, Yang Yu, Miao He, and Wanhe Li for technical assistance. We acknowledge Los Alamos National Laboratory Directed Research and Development (LDRD). RNA-seq data was deposited to GEO (GSE97451). This work was supported by grants from NCI 5P01CA013106-Project 3 and NIGMS 42694 (to D.L.S.). Portions of this study was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine (E.P.N.). B.Z. was supported by a Department of Defense Prostate Cancer Research Program postdoctoral fellowship (W81XWH-10-1-0190). Y.S.M. was supported by a National Cancer Center postdoctoral fellowship. The CSHL Microscopy and DNA Sequencing Shared Resources are supported by NCI 2P30CA45508. References of photos used in the graphical abstract are included in Supplementary Information (S1).
Footnotes
Accession Numbers: The accession number for the sequencing data reported in this work is GEO: GSE97451.
Author contributions. BZ, YTM, and DLS conceived and designed all experiments. BZ and YTM performed the experiments. IVN and KYS planned chemical probing experiments. IVN performed and analyzed chemical probing experiments. CST and KYS planned tertiary structure model. CST constructed the model. EPN, TAJ, and SE performed and analyzed INFERNAL data. S.D.D. performed RNA-seq analysis. BZ, YTM, and WJL analyzed the data. BZ, YTM, and DLS wrote the paper with inputs from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–290. doi: 10.1101/sqb.2008.73.058. [DOI] [PubMed] [Google Scholar]
- Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, et al. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nature methods. 2009;6:647–649. doi: 10.1038/nmeth.1360. [DOI] [PubMed] [Google Scholar]
- Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. Embo J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Beyret E, Liu N, Lin H. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res. 2012;22:1429–1439. doi: 10.1038/cr.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:14013–14018. doi: 10.1073/pnas.1614759113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budkevich TV, Giesebrecht J, Behrmann E, Loerke J, Ramrath DJ, Mielke T, Ismer J, Hildebrand PW, Tung CS, Nierhaus KH, et al. Regulation of the mammalian elongation cycle by subunit rolling: a eukaryotic-specific ribosome rearrangement. Cell. 2014;158:121–131. doi: 10.1016/j.cell.2014.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnar Z, Ponting CP. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome biology. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW. Role of tRNA-like structures in controlling plant virus replication. Virus Res. 2009;139:217–229. doi: 10.1016/j.virusres.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013a;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013b;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Kang M, Eichhorn CD, Feigon J. Structural determinants for ligand capture by a class II preQ1 riboswitch. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E663–671. doi: 10.1073/pnas.1400126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kuhn CD, Wilusz JE, Zheng Y, Beal PA, Joshua-Tor L. On-enzyme refolding permits small RNA and tRNA surveillance by the CCA-adding enzyme. Cell. 2015;160:644–658. doi: 10.1016/j.cell.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmer S, Hofmann HP, Ott G, Sprinzl M. The 3′-terminal end (NCCA) of tRNA determines the structure and stability of the aminoacyl acceptor stem. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6199–6202. doi: 10.1073/pnas.90.13.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome research. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol. 2008;15:634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Stadler P. Evolution of the Long Non-coding RNAs MALAT1 and MENβ/ε. Advances in Bioinformatics and Computational Biology. 2010;6268:1–12. [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung CS, Joseph S, Sanbonmatsu KY. All-atom homology model of the Escherichia coli 30S ribosomal subunit. Nat Struct Biol. 2002;9:750–755. doi: 10.1038/nsb841. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Steitz JA. Myriad Triple-Helix-Forming Structures in the Transposable Element RNAs of Plants and Fungi. Cell reports. 2016;15:1266–1276. doi: 10.1016/j.celrep.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Molecular cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. tRNAs marked with CCACCA are targeted for degradation. Science. 2011;334:817–821. doi: 10.1126/science.1213671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell reports. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1: Supplemental Experimental Procedures and Figures S1-S8.
Supplemental Dataset: Information of alignment, cm model, cmsearch results, and secondary structure of MALAT1-3h-tRNAlike (SF1-SF13, Related to Figure 2) and sequence information of oligonucleotides and primers (SF14, Related to Figures 2, 3, 5, 7).
Fourteen supplementary datasets are compressed in this file (SF1): The alignment used to build the covariance model MALAT1-3h-tRNAlike.cm (SF4) in Stockholm format. (SF2): The alignment used to build the covariance model MALAT1-3h.cm (SF5) in Stockholm format. (SF3): The alignment used to build the covariance model MALAT1-tRNAlike.cm (SF6) in Stockholm format. (SF4): The covariance model of the combined MALAT1 triple helix complex and the tRNA-like structure. (SF5): The covariance model of the MALAT1 triple helix complex. (SF6): The covariance model of the MALAT1 tRNA-like structure. (SF7): A text file with cmsearch information on 132 MALAT1-3h-tRNAlike elements using three models of MALAT1-3h-tRNAlike.cm, MALAT1-3h.cm, and MALAT1-tRNAlike.cm. (SF8): List of the genomes searched and the versions of each of those genomes. (SF9): A FASTA file containing 132 MALAT1-3h-tRNAlike elements. (SF10): Stockholm-formatted alignment of 132 MALAT1-3h-tRNAlike elements. (SF11): Consensus model of secondary structure of MALAT1-3h-tRNAlike elements. (SF12): Secondary structure of 132 MALAT1-3h-tRNAlike elements. (SF13): Secondary structure template of MALAT1-3h-tRNAlike element. (SF14): Sequence information of oligonucleotides and primers.