Abstract
Nuclear factor erythroid 2-related factor 2 (Nrf2) is a master regulator that promotes transcription of cytoprotective genes in response to oxidative/electrophilic stress. A large number of natural dietary compounds are thought to protect against oxidative stress and a few have been reported to induce genes involved in antioxidant defense through activating Nrf2. Therefore, a library of 54 natural compounds were collected to determine whether they are Nrf2 activators, and to compare their efficacy and potency to activate Nrf2. The assay utilized AREc32 cells that contain a luciferase gene under the control of antioxidant response element (ARE) promoters. Each natural compound was tested at 13 concentrations between 0.02 and 30 µM. Known Nrf2 activators, tert-butylhydroquinone (tBHQ) and 2-cyano-3,12-dioxooleana-1,9-diene-28-imidazolide (CDDO-Im), were used as positive controls in parallel with the natural compounds. Among the 54 tested natural compounds, andrographolide had the highest efficacy, followed by trans-chalcone, sulforaphane, curcumin, flavone, kahweol, and carnosol, all of which had better efficacy than tBHQ. Among the compounds tested, CDDO-Im was the most potent, having an EC50 of 0.41 µM. Seven of the natural compounds, namely andrographolide, trans-chalcone, sulforaphane, curcumin, flavone, kahweol, and cafestol had lower EC50 values than tBHQ, but higher than CDDO-Im. The present study provides insights into which natural compounds activate the Keap1-Nrf2 pathway and thus might be useful for detoxifying oxidative/electrophilic stress.
Keywords: Nrf2, natural compounds, oxidative stress
Introduction
Nuclear factor erythroid 2-related factor 2 (Nrf2) is a transcription factor that induces a battery of cytoprotective genes in response to oxidative/electrophilic stimuli. Under physiological conditions, Nrf2 is bound to Kelch-like ECH associating protein 1 (Keap1) in the cytosol. Keap1 functions as an adapter protein that retains Nrf2 in the cytoplasm by interacting with the cytoskeleton, and facilitates degradation of Nrf2 by binding with Cullin 3-based E3 ligase, which ubiquitinates Nrf2 protein [1]. With the negative regulatory system of Keap1, Nrf2 is constitutively synthesized and rapidly degraded by the proteasome under unstressed conditions. In response to oxidative/electrophilic stress, Nrf2 is released from Keap1, translocates into the nucleus, and induces its target genes by binding to the antioxidant response element (ARE) in the promoter regions of its target genes [1].
The classical Nrf2 target genes are mainly involved in reducing oxidative stress, including genes encoding enzymes that synthesize glutathione (e.g., glutamate--cysteine ligase and glutathione synthase), genes encoding enzymes that reduce hydrogen peroxide (e.g., glutathione peroxidase and peroxiredoxin), genes encoding proteins and enzymes that reduce oxidized protein (e.g., thioredoxin, thioredoxin reductase, and sulfiredoxin), and genes encoding proteins that sequester free ions (e.g., ferritin) [2,3]. Nrf2 also plays a critical role in induction of genes involved in drug metabolism and distribution, including multiple genes encoding non-cytochrome P450 phase-I drug metabolism enzymes (e.g., aldo-keto reductases, carboxylesterases, and carbonyl reductases), a large number of genes encoding phase-II drug metabolism enzymes (e.g., glutathione-S-transferases), and genes encoding efflux transporters (e.g., multidrug resistance-associated protein and breast cancer resistance protein) in mouse liver [4].
A number of natural compounds are known to be antioxidants. Some of these natural compounds have been shown to exhibit antioxidant effects through activating the Nrf2 pathway. Recently, it was reported that oleanolic acid stimulates the nuclear translocation of Nrf2 and induces Nrf2-dependent genes, which protects from acetaminophen hepatotoxicity [5]. Carnosol and curcumin, purified natural compounds, as well as extracts from rosemary and turmeric, respectively, as well as extracts from coffee, thyme, broccoli, and red onion activates the Keap1-Nrf2 pathway in vitro and in vivo [6]. Other numerous natural compounds, including sulforaphane, allyl sulfides, resveratrol, and a battery of dietary flavonoids also activate the Keap1-Nrf2 pathway and have a chemopreventive effect in various cell lines and tissues [7].
In addition to descriptive studies showing protective effects of natural compounds against oxidative/electrophilic stress through activating Nrf2, studies revealing molecular mechanisms of how natural compounds activate the Keap1-Nrf2 pathway have provided further knowledge to develop Nrf2 activators as potential therapeutic drugs. For example, quercetin, a plant derived compound found in many fruits, vegetables, grains, and leaves, increases Nrf2 mRNA and protein synthesis, and reduces Keap1 protein through post-translational modification in HepG2 cells [8]. Carnosol activates the PI3K-Akt pathway to induce phosphorylation and translocation of Nrf2 into the nucleus of P12 cells [9]. Sulforaphane, a compound found in cruciferous vegetables such as broccoli, causes Nrf2 to escape from Keap1-mediated degradation in NIH3T3 cells [10], as well as ubiquitination and degradation of Keap1 to facilitate Nrf2 activation in COS-1 cells [11].
Collectively, various natural chemicals act as antioxidants and are used for chemoprevention and therapy of oxidative/electrophilic stress-induced tissue damage. However, given the fact that Nrf2 activation also leads to induction of certain phase-I and phase-II drug metabolism genes as well as efflux transporters, Nrf2 activators may be involved in drug-drug interactions and chemotherapeutic resistance. Despite studies that investigated the effect and mechanisms of individual natural compounds on the Keap1-Nrf2 pathway, there are no data comparing the efficacy and potency of natural compounds to activate Nrf2. In the present study, 54 natural compounds were tested as potential Nrf2 activators, and their efficacy and potency to activate Nrf2 was examined using AREc32 cells, which contains a luciferase gene under control of eight copies of rat ARE in the promoter region. Results from this screening provide novel insights into which natural compounds might detoxify oxidative/electrophilic stress.
Material and Methods
Materials
2-Cyano-3,12-dioxooleana-1,9-diene-28-imidazolide (CDDO-Im) was a generous gift from Dr. Michael Sporn (Dartmouth College, Hanover, New Hampshire). tert-Butylhydroquinone (tBHQ) was purchased from Sigma-Aldrich (St. Louis, MO). The chemical names, CAS-numbers, and sources of all the natural chemicals are summarized in Table 1. The purity of CDDO-Im was 95%, and that of tBHQ and other tested compounds were greater than 98%.
Table 1.
Chemical names, CAS-numbers and sources of all the test natural chemicals.
| Compound | CAS | Source |
|---|---|---|
| Allyl disulfide | 2179-57-9 | Sigmaa |
| Apigenin | 520-36-5 | Sigma |
| Andrographolide | 5508-58-7 | Sigma |
| Baicalin | 21967-41-9 | Sigma |
| Berberine | 2086-83-1 | Sigma |
| Cafestol | 469-83-0 | LKTb |
| Caffeic acid | 331-39-5 | Caymanc |
| Caffeic acid phenethyl ester | 104594-70-9 | Sigma |
| Carnosol | 5957-80-2 | Cayman |
| (+)-Catechin | 7295-85-4 | Acrosd |
| CDDO-Im | NA | Sporne |
| Chlorogenic acid | 327-97-9 | LKT |
| Chlorophyllin | 15611-43-5 | Sigma |
| Chrysin | 480-40-0 | Sigma |
| Chrysophanic acid | 481-74-3 | Sigma |
| Crocin | 42553-65-1 | Sigma |
| Curcumin | 458-37-7 | Acros |
| Daidzein | 486-66-8 | Sigma |
| Daidzin | 552-66-9 | Sigma |
| (−)-Epicatechin | 490-46-0 | Sigma |
| Epigallocatechin 3-gallate | 989-51-5 | Sigma |
| Evodiamine | 5956-87-6 | Sigma |
| Flavone | 525-82-6 | Sigma |
| Fisetin | 528-48-3 | Sigma |
| Galangin | 548-83-4 | Sigma |
| Genistein | 446-72-0 | Acros |
| Glycitein | 40957-83-3 | Cayman |
| Glycyrrhetinic acid | 471-53-4 | Quality |
| Hesperetin | 520-33-2 | Acros |
| Kahweol | 6894-43-5 | LKT |
| tBHQ | 1948-33-0 | Sigma |
| Indole-3-carbinol | 700-06-1 | Sigma |
| Icariin | 489-32-7 | Sigma |
| Kaempferol | 520-18-3 | Sigma |
| Lupeol | 545-47-1 | LKT |
| Luteolin | 491-70-3 | Cayman |
| Matrine | 519-02-8 | Qualityf |
| Morin | 480-16-0 | INDOF |
| Myricetin | 529-44-2 | Sigma |
| Naringin | 10236-47-2 | Sigma |
| Naringenin | 480-41-1 | LKT |
| Oleanolic acid | 508-02-1 | Quality |
| Oxymatrine | 16837-52-8 | Sigma |
| Quercetin | 117-39-5 | Sigma |
| Quercitin | 522-12-3 | Sigma |
| Resveratrol | 501-36-0 | Sigma |
| Rutin | 153-18-4 | Sigma |
| Sennoside B | 128-57-4 | Sigma |
| Silibinin | 22888-70-6 | Acros |
| Silymarin | 65666-07-1 | INDOFg |
| Sulforaphane | 4478-93-7 | Sigma |
| Synephrine | 94-07-5 | Sigma |
| (s,s)-(+)-Tetrandrine | 518-34-3 | Sigma |
| Zebularine | 3690-10-6 | LKT |
Sigma-Aldrich (St. Louis, MO)
LKT Laboratories (St. Paul, MN)
Cayman Chemicals (Ann Arbor, MI)
Acros Organics (Geel, Belgium)
generous gift from Dr. Michael Sporn (Dartmouth College, Hanover, New Hampshire)
Quality Phytochemicals, LLC (Edison, NJ)
INDOFINE Chemical Company (Hillsborough, NJ).
Cell growth and maintenance
AREc32 is a stable cell line derived from the human MCF7 breast carcinoma cell line. The generation of the cell line was described previously [12] and the cell line was licensed from CRX Biosciences (Dundee, Scotland). AREc32 cells were maintained in Dulbecco’s modified Eagle’s medium containing glutamax (Invitrogen, Carlsbad, CA), supplemented with 10% fetal calf serum and 500 µg/mL G418. The cell line was grown at 37°C in the presence of 5% CO2. Before screening, AREc32 cells were seeded onto tissue-culture treated 384-well plates (flat-bottom white, opaque, sterile, with lids). Cells were seeded at a density of 3,500 cells/well by a Wellmate bulk dispenser (ThermoFisher Scientific, Waltham, MA) in complete media with 50µL of cell suspension per well. Cell plates were incubated at ambient room temperature for 30 min following seeding, to allow for even cell settling. Cell plates were then incubated at 37°C, 5% CO2 in a 95% humidified incubator for 20 h.
Compound preparation and treatment
All compounds were prepared as 10 mM stock solutions in DMSO, and then diluted into working solutions at various concentrations. The working solutions were added to cells by transferring 175 nL from each compound well into 50 µL of cell culture medium in the receiving well to make final concentrations of 0.02–30 µM. Four controls were used on each plate of cells: (1) cells treated with tBHQ, (2) cells treated with CDDO-Im, (3) cells in media containing 0.35% DMSO (vehicle control), and (4) cells in media containing no DMSO to measure background luminescence.
Quantification of ARE activation by Promega Steady-Glo luciferase assay system
The AREc32 cell line is a stably transfected MCF7 cell line that contains a luciferase gene construct under the control of eight copies of the rat Gsta2 ARE. The luciferase reporter provides a rapid and convenient method for the quantification of ARE activation. The AREc32 cell line was exposed to natural compounds for 24 h and then removed from the incubator and left at ambient room temperature for 20 min to equilibrate the plate and contents to room temperature. The Matrix Wellmate dispensed Steady-Glo (Promega, Madison, WI) luciferase assay reagent to all wells, 10 µL per well, and plates were shaken for 1 min at 1600 rpm. Thirty min later, the luminescence of each well was determined (Tecan Safire2, Tecan, Durham, NC) to quantify induction of ARE. Three independent experiments were performed as biological triplicates.
Quantification of Nqo1 mRNA by qPCR analysis
Hepa-1c1c7 cells were maintained in Dulbecco’s modified Eagle’s medium containing glutamax, supplemented with 10% fetal bovine serum and Penicillin/Streptomycin. The cell line was grown at 37°C in the presence of 5% CO2. Before screening, Hepa-1c1c7 cells were seeded onto 24-well plates at a density of 100000 cells/well. After overnight culture for attachment, cells were treated with phytochemicals at 0.1–30µM. Twenty-four hours after treatment, cells were lysed in Trizol reagent (Life Technologies, Grand Island, NY) for total RNA isolation. Total RNA was reverse-transcribed into first-strand cDNA using multiscript reverse transcriptase from a High Capacity RT kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. Messenger RNA of genes of Nqo1 was determined with quantitative real time-PCR performed on an Applied Biosystems Prism 7900HT sequence detection system. The reaction system contains 2 ng of cDNA, 150 nM of eachprimer, and 5 µ of Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) in a total volume of 10 µL. The specific primers used to quantify Nqo1 expression are 5’-tatccttccgagtcatctctagca-3’ (forward) and 5’-tctgcagcttccagcttcttg-3’ (reverse). The relative mRNA levels were calculated by cycle threshold (Ct) values, which were normalized to the internal control glyceraldehyde 3-phosphate dehydrogenase (Gapdh) mRNA.
MTS cell viability assay
Hepa-1c1c7 cells were seeded onto 96-well plates at a density of 30000 cells/well. After overnight culture for attachment, cells were treated with phytochemicals at 0.1–30µM. Twenty-four hours after treatment, MTS assay were performed to estimate cell viability using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI).
Results
Induction of ARE-regulated luciferase activity by known Nrf2 activators
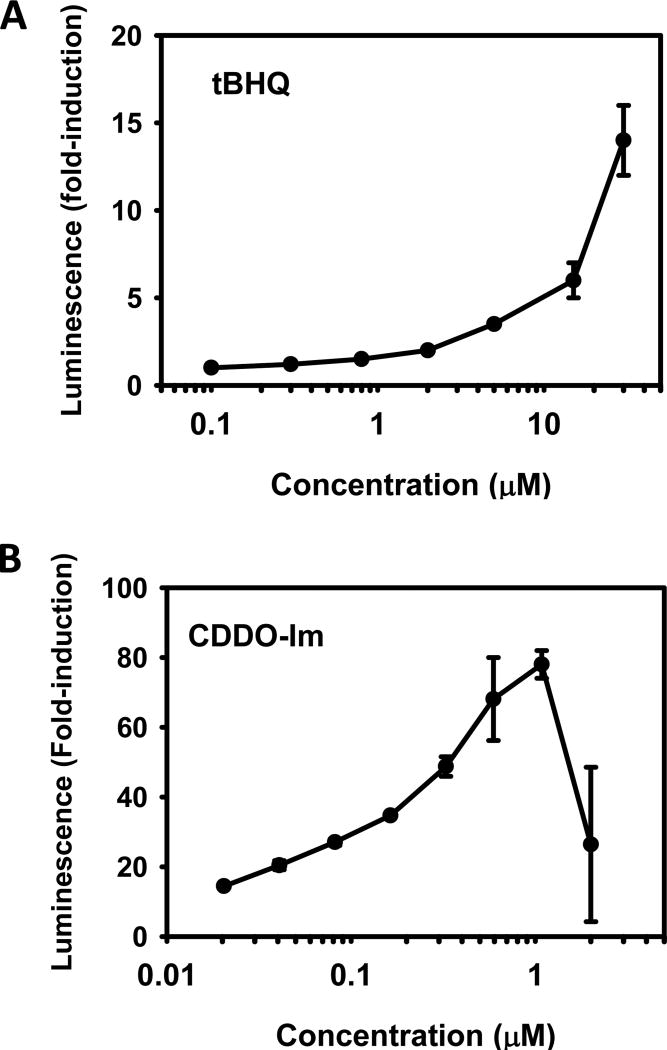
To validate the AREc32 cell line model for screening Nrf2 activators from a natural compound library, tBHQ, a prototypical Nrf2 activator, and CDDO-Im, an extremely potent Nrf2 activator, were tested in the assay. Both tBHQ and CDDO-Im increased the luminescence signal in a concentration-dependent manner. tBHQ increased the luciferase activity at concentrations higher than 2 µM, and the luciferase activity was increased 14-fold at 30 µM (Fig. 1A). In contrast, CDDO-Im increased the luciferase activity 13-fold at 0.02 µM, had maximum induction at 1 µM (77-fold), but the induction of luciferase activity was blunted at higher concentrations (Fig. 1B).
Figure 1.
Dose-response activation of ARE luciferase reporter construct in AREc32 cells by (A) tBHQ and (B) CDDO-Im. AREc32 cells were seeded at 3,500 cells/well in a 384-well plate, 46µL/well. 24 h after seeding, cells were treated with compound (tBHQ or CDDO-Im) or DMSO vehicle. 24 h after treatment, luciferase activity was assessed using the Steady-Glo luciferase assay with a luminescence readout. The luminescence of each well was divided by the median luminescence of the DMSO vehicle control wells to generate the fold Nrf2 activation. Data are presented as the average from three independent experiments.
Induction of ARE-regulated luciferase activity by natural compounds
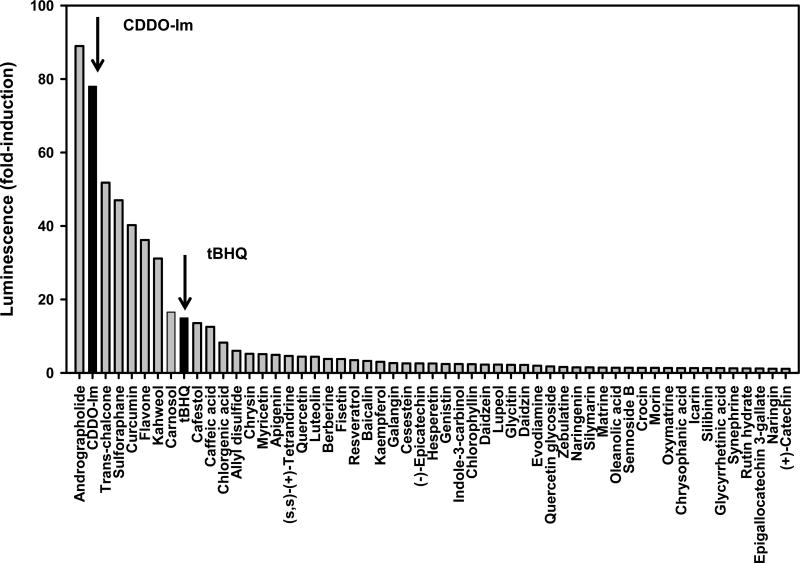
Fifty-four natural compounds were screened using the AREc32 cell line. The mean and S.E. of fold-induction of each compound at each concentration was listed in Supplemental Table 1. The maximum-fold induction and EC50 concentration of each individual compound are summarized in Figure 2 and 3, and the maximum fold-induction value as well as EC50 concentrations for tBHQ and CDDO-Im are listed as positive controls. As shown in Fig. 2, andrographolide produced the highest maximum-fold induction over vehicle controls, with an 88-fold increase in luminescence at 30 µM. Six natural compounds had a higher induction of luminescence signal than tBHQ but lower than CDDO-Im: trans-chalcone (50-fold), sulforaphane (46-fold), curcumin (39-fold), flavone (35-fold), kahweol (30-fold), and carnosol (17-fold). Six additional natural compounds, namely cafestol, caffeic acid, chlorgenic acid, allyl disulfide, chrysin, and myricetin were less effective than tBHQ, but still produced moderate induction of the luminescence signal (5–14 fold).
Figure 2.
Maximum fold-induction of the ARE luciferase reporter by tBHQ, CDDO-Im, and natural compounds. AREc32 cells were seeded at 3,500 cells/well in a 384-well plate, 46 µL/well. 24 h after seeding, cells were treated with compound (0.02–30 µM) or DMSO vehicle. 24 h after treatment, luciferase activity was assessed using the Steady-Glo luciferase assay with a luminescence readout. The luminescence of each well was divided by the median luminescence of the DMSO vehicle control wells to generate the fold Nrf2 activation.
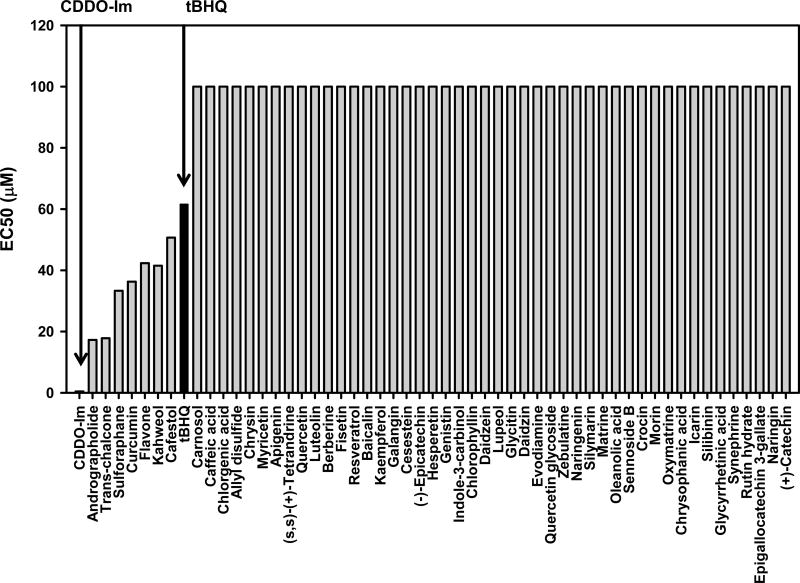
Figure 3.
EC50 of tBHQ, CDDO-Im, and natural compounds to activate Nrf2. AREc32 cells were cultured and treated as described in figure 2. The EC50s were calculated based on the data from concentration-response curves of each individual compound and constrained to 100-fold induction.
EC50 values of tBHQ, CDDO-Im, and the natural compounds to activate the Nrf2 pathway are summarized in Fig. 3. The concentration-response curve of each compound was fitted to a log-linear extrapolation model and EC50 values were calculated as constrained to a 100-fold induction. Among the compounds tested, CDDO-Im was the most potent, with an EC50 of 0.41 µM. There were seven compounds that had higher EC50 values than CDDO-Im, but lower EC50 values than tBHQ: andrographolide (17 µM), trans-chalcone (18 µM), sulforaphane (33 µM), curcumin (36 µM), flavone (42 µM), kahweol (42 µM), and cafestol (51 µM). All other natural compounds had EC50 values higher than 100 µM for activating the Nrf2 pathway.
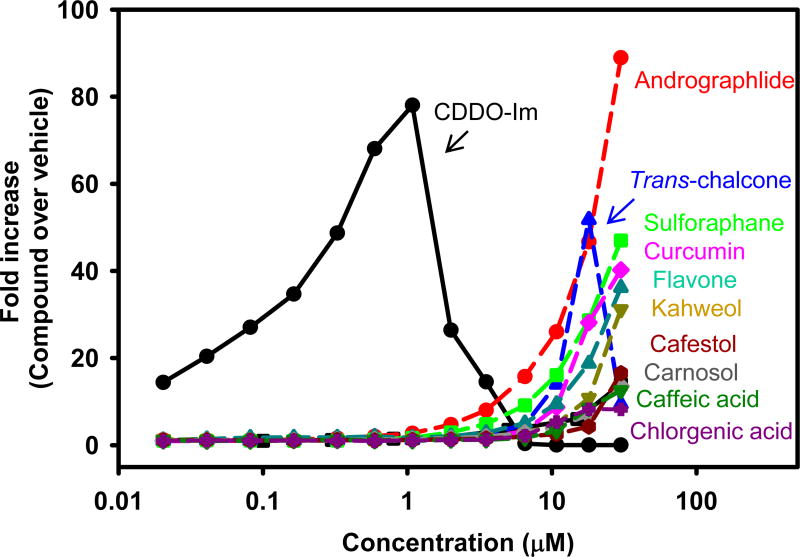
Concentration-response of top 10 hits
Figure 4 depicts concentration-response curves of the top 10 natural compounds as well as tBHQ and CDDO-Im as positive controls. CDDO-Im induced the luminescence signal 13-fold at the lowest concentration tested (20 nM), with a continued increase in luminescence signal as the concentration increased, reaching a maximum fold-induction at 1 µM. The luminescence decreased to basal levels at higher concentrations of CDDO-Im. Trans-chalcone increased the luminescence signal at concentrations higher than 3.5 µM, reached a maximum fold-induction at 18 µM, but the induction decreased to basal levels at 30 µM. In contrast, tBHQ and the other nine natural compounds did not increase the luminescence at concentrations lower than 3.5 µM, and reached the maximum fold-induction at the highest concentration tested (30 µM).
Figure 4.
Concentration-response activation ARE luciferase reporter construct in AREc32 cells by tBHQ, CDDO-Im, and the 10 best natural compounds to activate the Keap1-Nrf2-ARE pathway. AREc32 cells were cultured and treated as described in figure 2. The luminescence of each well was divided by the median luminescence of the DMSO vehicle control wells to generate the fold Nrf2 activation. Data are presented as the average from three independent experiments.
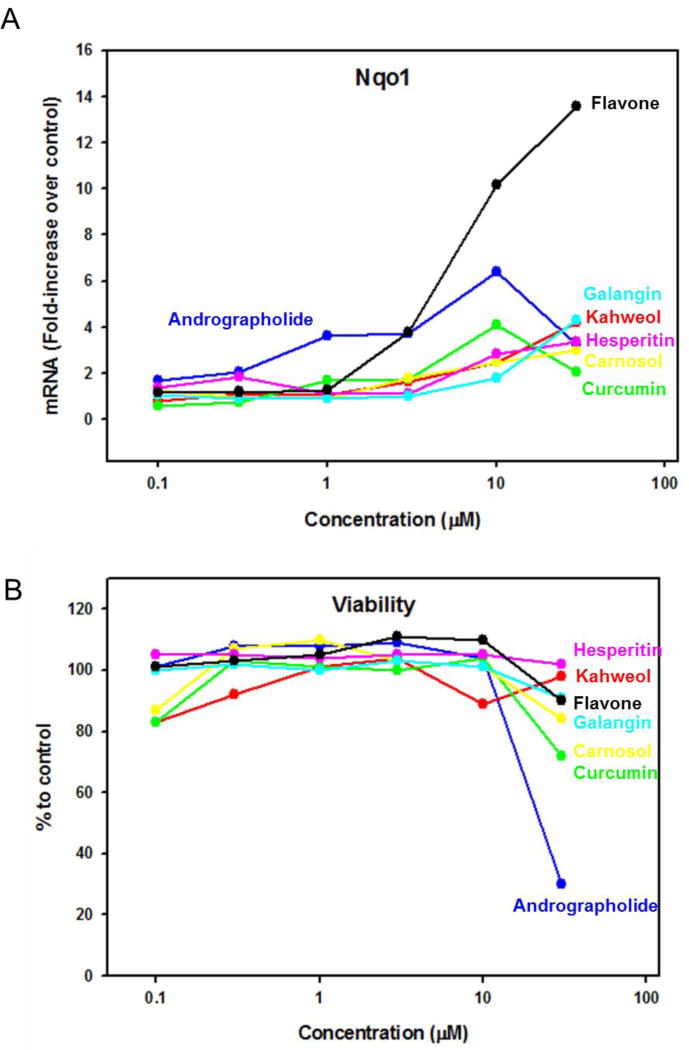
Induction of Nqo1 mRNA by natural compounds in Hepa-1C1c7 cells
OF all the natural compounds tested in Hepa-1C1C7 cells, only seven of them produced a concentration-dependent induction of Nqo1, the prototypical Nrf2 target gene. Figure 5A depicts concentration-response curves of these seven natural compounds. Andrographolide and curcumin resulted in maximum Nqo1 induction at 10µM. Kahweol, carnosol, hesperitin, galangin, and flavoneIn resulted in maximum Nqo1 induction at the highest concentration tested (30µM). Among these top seven compounds, flavone had the best efficacy with 13-fold induction of Nqo1 mRNA. For cytotoxicity, only andrographolide and curcumin produced substantial decreases in cell viability at 30µM, and the other natural compounds did not affect cell viability at any concentration tested (Fig. 5B).
Figure 5.
Concentration-response of (A) Nqo1 induction and (B) cell viability in Hepa- 1C1C7 cells by the 7 natural compounds. Data are presented by fold-induction or percentage of cell viability over DMSO control cells.
Discussion
The present study compared the efficacy and potency of two synthetic Nrf2 activators as well as 54 natural compounds to activate the Keap1-Nrf2 pathway using the AREc32 cell line system. Results showed that seven natural compounds, namely andrographolide, trans-chalcone, sulforaphane, curcumin, flavone, kahweol, and carnosol were more effective than the classic Nrf2 activator tBHQ, but none of the natural compounds were more potent than CDDO-Im, the most potent synthetic Nrf2 activator that has been identified.
tBHQ is a prototypical Nrf2 activator and protects against oxidative stress-induced toxicity in vitro and in vivo. For example, tBHQ has been shown to activate the Keap1-Nrf2 pathway and prevent ethanol-induced apoptosis in cranial neural crest cells in vitro [13]. tBHQ also inhibits LPS-induced microglial activation by Nrf2-dependent mechanism in microglial cells [14]. In addition, tBHQ protects against mitochondrial stress by induction of the Nrf2-driven antioxidant response in mice [15]. CDDO-Im has emerged in recent years as a protective compound against oxidative stress. CDDO-Im activates the Nrf2-Keap1 pathway [16], and leads to a cytoprotective response through the up-regulation of gene expression. CDDO-Im protects against acetaminophen-induced hepatotoxicity in mice [17]. In addition to the known synthetic Nrf2 activators, two well-established natural Nrf2 activators, namely sulforaphane and curcumin, were also included [18]. The present study shows both synthetic and natural compounds can increase the luminescence signal in the AREc32 cell line in a concentration-dependent manner, indicating that this system is excellent for determining the efficacy and potency of potential Nrf2 activators.
Among the 54 natural compounds tested in the AREc32 cell line system, compounds with a diterpene structure, including andrographolide, kahweol, cafestol, and carnosol, were the most active compounds. Andrographolide was even more effective than CDDO-Im, a triterpenoid known to be an extremely strong activator of the Keap1-Nrf2 pathway. Andrographolide is a diterpene lactone that exists in leaves and stems of Andrographis paniculata, which is a plant used for medicinal purposes in India [19]. Andrographolide has strong anti-inflammatory effects through inactivation of NFκB [20], and anti-cancer effects by inhibition of androgen receptor signaling [21]. Recently, andrographolide was shown to induce heme oxygenase-1 (HO1) by activating Nrf2 in human endothelial cells, and the Nrf2-HO1 pathway was suggested to be involved in the andrographolide-induced suppression of inflammation [22]. Kahweol and cafestol are coffee specific diterpenes that produce chemoprevention to a broad range of carcinogens, including 7,12-dimethylbenz[a]anthracene, aflatoxin B1, benzo[a]pyrene, and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, at least partially through induction of phase-II drug metabolism genes, as well as genes involved in cellular antioxidant defense [23]. Recently, it was reported that kahweol and cafestol induce cytoprotective genes primarily by Nrf2 [24]. Similarly, carnosol, a compound found in rosemary, also activates phase-II drug metabolism genes and antioxidant genes through activating Nrf2 in vitro [25] and in vivo [6]. In parallel with the previous observations, results from the present study indicate that these diterpenes (andrographolide, kahweol, cafestol, and carnosol) are highly effective activators of the Nrf2 pathway. The exact mechanism of diterpenes to activate Nrf2-ARE is unknown. However, andrographolide [26] and carnosol [9] are known electrophiles and have potential to modify cysteine reduces on Keap1 protein to activate Keap1-Nrf2 pathway through Michael reaction.
In addition to andrographolide, kahweol, cafestol, and carnosol, trans-chalcone were reported to activate Nrf2 in certain cell lines or animal models, and also activate Nrf2 in the present study. Trans-chalcone was the top hit of 43 potential chemopreventive chemicals to increase glutathione content in rat liver cells [27]. Recently, a series of chalcone derivatives were synthesized, and 2-trifluoromethyl-2'-methoxychalone was shown to be a potent Nrf2 activator in human lung epithelial cells and in mice [28]. It should be noted that trans-chalcone contains an enone structure and can become a Michael Acceptor to react with a Michael Donor. Thus, like diterpenes, trans-chalcone also may activate Nrf2 through modifying cysteine residues on Keap1 protein.
In the present study, both caffeic acid and its derivative caffeic acid phenethyl ester (CAPE) were shown to activate Nrf2, with caffeic acid more effective than CAPE (13-fold versus 8-fold maximum increase of luminescence signal). In previous reports, CAPE activates Nrf2 in multiple cell lines. However, whether caffeic acid activates the Keap1-Nrf2 pathway was unknown. CAPE induces Nrf2 target genes in porcine renal epithelial proximal tubule cells and rat kidney epithelial cells [29], rat liver tissue macrophages [30], and human colon carcinoma cells [31]. The present study indicates that caffeic acid has the potential to be a novel Nrf2 activator, and more in vivo studies are necessary to test the antioxidative activity of caffeic acid in whole animals.
Dietary flavonoids are a family of natural compounds that have cancer preventive effects through inducing phase-II drug metabolism genes [32], and antioxidative effects against ultraviolet radiation [33]. In addition, a large number of dietary flavonoids, including luteolin [34], epigallocatechin-3-gallate [35], and quercetin [36] induce Nrf2 target genes in various cell lines and animal models. Thus, 27 dietary flavonoids were screened in the present study to test whether these dietary flavonoids activate the Nrf2 pathway. Surprisingly, only flavone of these dietary flavonoids was highly effective (33-fold increase in luminescence) in activating Nrf2, and the remaining flavonoids tested exhibited slight or no increase in Nrf2 activation. Similarly, several natural compounds, including resveratrol, xanthohumol, isoliquiritigenin, and oleanolic acid have been reported to activate Nrf2, but were inactive in the present study. For example, resveratrol was shown to protect against oxidative/electrophilic stress via inducing Nrf2 target genes in human lung epithelial cells [37], rat primary hepatocytes [38], coronary arterial endothelial cells, and branches of the femoral artery in mice [39]. Xanthohumol and isoliquiritigenin alkylate cysteine 151 on Keap1 protein, leading to Nrf2 activation in vitro and in vivo [40]. Oleanolic acid has been shown to activate Nrf2 in mouse liver [5] and rat vascular smooth muscle cells [41]. However, resveratrol, xanthohumol, isoliquiritigenin, and oleanolic acid did not activate Nrf2 in the present study, indicating that activation of Nrf2 by those natural chemicals may be cell line specific. In addition, oleanolic acid was recently shown to inhibit estradiol-17β-glucuronide uptake by OATP1B1, an uptake transporter, suggesting that oleanolic acid is an OATP1B1 substrate [42], and uptake transport may be required to increase the bioavailability of oleanolic acid. Collectively, oleanolic acid, together with other natural compounds may require transport and/or bio-transformation to activate the Keap1-Nrf2 pathway.
To investigate whether the validated hits from the primary screen can also induce Nrf2 target genes, a secondary screen was designed to test the ability of the hit compounds to induce Nqo1, the prototypical Nrf2 target gene, in Hepa1c1c7 cells. The results show that most of compounds that induced AREc32 cells also increased Nqo1 mRNA in Hepa-1c1c7 cells in a concentration-dependent manner. Surprisingly, two compounds (hesperitin and galangin) only increased Nqo1 mRNA in Hepa-1c1c7 cells. The difference in activity of these two compounds may result from cell line difference (human breast cancer cells versus mouse hepatoma cells), or hesperitin and galangin induced Nqo1 mRNA through Nrf2-independent mechanisms. Because the Nrf2 pathway also serves as an adaptive signal in response to oxidative stress, it is possible that some of these natural compounds may induce the Nrf2 pathway through cytotoxic effects. Thus, cell viability assays were performed in parallel with validation assays of Nqo1 induction. The results showed that all the positive compounds were either non-toxic, or induced Nqo1 expression at low concentrations when cell viability was not affected.
In conclusion, among the 54 natural compounds tested, andrographolide, trans-chalcone, sulforaphane, curcumin, flavone, kahweol, and carnosol have higher efficacy than the synthetic antioxidant tBHQ, indicating that these natural compound may serve as candidates to protect against oxidative stress-induced pathogenesis. In addition, derivatives can be designed and synthesized according to the chemical structure of these natural compounds to develop Nrf2 activators with better potency and efficacy. However, it should be noted that Nrf2 activation results in enhanced phase-II conjugation and excretion, and thus may alter the pharmacokinetics of certain drugs. The possibility of drug-drug interactions should be noted when using these natural compounds and derivatives as therapeutic drugs or dietary supplements.
Supplementary Material
Acknowledgments
The authors would like to thank all the graduate students and postdoctoral fellows for technical support of the experiments as well as revision of the manuscript. The authors would like to thank Eric Reimer for the help of realtime PCR analysis in the Nqo1 induction assay. Some of the phytochemicals used in the present study were kindly provided by Drs Baoting Zhu and Xiaochao Ma at the University of Kansas Medical Center. This work was supported by NIH grants DK-081461 and ES-009649.
Abbreviations
- ARE
antioxidant response element
- CAPE
Caffeic acid phenethyl ester
- CDDO-Im
2-cyano-3,12-dioxooleana-1,9-diene-28-imidazolide
- Gclc
glutamate-cysteine ligase, catalytic subunit
- GSH
glutathione
- Gsr
glutathione reductase
- Gss
glutathione synthase
- Gpx
glutathione peroxidase
- Ho-1
heme oxygenase-1
- Keap1
Kelch-like ECH associating protein 1
- Nrf2
nuclear factor erythroid 2-related factor 2
- Prdx
peroxiredoxin
- ROS
reactive oxygen species
- Srxn
sulfiredoxin
- tBHQ
tert-butylhydroquinone
- Trxn1
thioredoxin-1
- Txnrd1
thioredoxin reductase-1
Footnotes
Conflict of interest statement
There are no conflicts of interest to disclose for any of the authors.
References
- 1.Cullinan SB, Gordan JD, Jin J, et al. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 3.Wang LJ, Zhou X, Wang W, et al. Andrographolide inhibits oral squamous cell carcinogenesis through NF-kappaB inactivation. J Dent Res. 2011;90:1246–1252. doi: 10.1177/0022034511418341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu KC, Cui JY, Klaassen CD. Effect of Graded Nrf2 Activation on Phase-I and -II Drug Metabolizing Enzymes and Transporters in Mouse Liver. PLoS One. 2012;7:e39006. doi: 10.1371/journal.pone.0039006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reisman SA, Aleksunes LM, Klaassen CD. Oleanolic acid activates Nrf2 and protects from acetaminophen hepatotoxicity via Nrf2-dependent and Nrf2-independent processes. Biochem Pharmacol. 2009;77:1273–1282. doi: 10.1016/j.bcp.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balstad TR, Carlsen H, Myhrstad MC, et al. Coffee, broccoli and spices are strong inducers of electrophile response element-dependent transcription in vitro and in vivo - studies in electrophile response element transgenic mice. Mol Nutr Food Res. 2011;55:185–197. doi: 10.1002/mnfr.201000204. [DOI] [PubMed] [Google Scholar]
- 7.Surh YJ, Kundu JK, Na HK. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta Med. 2008;74:1526–1539. doi: 10.1055/s-0028-1088302. [DOI] [PubMed] [Google Scholar]
- 8.Tanigawa S, Fujii M, Hou DX. Action of Nrf2 and Keap1 in ARE-mediated NQO1 expression by quercetin. Free Radic Biol Med. 2007;42:1690–1703. doi: 10.1016/j.freeradbiomed.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Martin D, Rojo AI, Salinas M, et al. Regulation of heme oxygenase-1 expression through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J Biol Chem. 2004;279:8919–8929. doi: 10.1074/jbc.M309660200. [DOI] [PubMed] [Google Scholar]
- 10.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang DD, Lo SC, Sun Z, et al. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 12.Natsch A, Emter R. Skin sensitizers induce antioxidant response element dependent genes: application to the in vitro testing of the sensitization potential of chemicals. Toxicol Sci. 2008;102:110–119. doi: 10.1093/toxsci/kfm259. [DOI] [PubMed] [Google Scholar]
- 13.Yan D, Dong J, Sulik KK, et al. Induction of the Nrf2-driven antioxidant response by tert-butylhydroquinone prevents ethanol-induced apoptosis in cranial neural crest cells. Biochem Pharmacol. 2010;80:144–149. doi: 10.1016/j.bcp.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh K, Cha Y, Kim S, et al. tBHQ inhibits LPS-induced microglial activation via Nrf2-mediated suppression of p38 phosphorylation. Biochem Biophys Res Commun. 2009;380:449–453. doi: 10.1016/j.bbrc.2009.01.082. [DOI] [PubMed] [Google Scholar]
- 15.Shih AY, Imbeault S, Barakauskas V, et al. Induction of the Nrf2-driven antioxidant response confers neuroprotection during mitochondrial stress in vivo. J Biol Chem. 2005;280:22925–22936. doi: 10.1074/jbc.M414635200. [DOI] [PubMed] [Google Scholar]
- 16.Liby K, Hock T, Yore MM, et al. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65:4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- 17.Reisman SA, Buckley DB, Tanaka Y, et al. CDDO-Im protects from acetaminophen hepatotoxicity through induction of Nrf2-dependent genes. Toxicol Appl Pharmacol. 2009;236:109–114. doi: 10.1016/j.taap.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thimmulappa RK, Mai KH, Srisuma S, et al. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 19.Mishra K, Dash AP, Dey N. Andrographolide: A Novel Antimalarial Diterpene Lactone Compound from Andrographis paniculata and Its Interaction with Curcumin and Artesunate. J Trop Med. 2011;2011:579518. doi: 10.1155/2011/579518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu WJ, Lee JJ, Chou DS, et al. A novel role of andrographolide, an NF-kappa B inhibitor, on inhibition of platelet activation: the pivotal mechanisms of endothelial nitric oxide synthase/cyclic GMP. J Mol Med (Berl) 2011;89:1261–1273. doi: 10.1007/s00109-011-0800-0. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Nadiminty N, Tummala R, et al. Andrographolide targets androgen receptor pathway in castration-resistant prostate cancer. Genes Cancer. 2011;2:151–159. doi: 10.1177/1947601911409744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu AL, Lu CY, Wang TS, et al. Induction of heme oxygenase 1 and inhibition of tumor necrosis factor alpha-induced intercellular adhesion molecule expression by andrographolide in EA.hy926 cells. J Agric Food Chem. 2010;58:7641–7648. doi: 10.1021/jf101353c. [DOI] [PubMed] [Google Scholar]
- 23.Cavin C, Holzhaeuser D, Scharf G, et al. Cafestol and kahweol, two coffee specific diterpenes with anticarcinogenic activity. Food Chem Toxicol. 2002;40:1155–1163. doi: 10.1016/s0278-6915(02)00029-7. [DOI] [PubMed] [Google Scholar]
- 24.Higgins LG, Cavin C, Itoh K, et al. Induction of cancer chemopreventive enzymes by coffee is mediated by transcription factor Nrf2. Evidence that the coffee-specific diterpenes cafestol and kahweol confer protection against acrolein. Toxicol Appl Pharmacol. 2008;226:328–337. doi: 10.1016/j.taap.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi T, Tabuchi T, Tamaki Y, et al. Carnosic acid and carnosol inhibit adipocyte differentiation in mouse 3T3-L1 cells through induction of phase2 enzymes and activation of glutathione metabolism. Biochem Biophys Res Commun. 2009;382:549–554. doi: 10.1016/j.bbrc.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 26.Woo AY, Waye MM, Tsui SK, et al. Andrographolide up-regulates cellular-reduced glutathione level and protects cardiomyocytes against hypoxia/reoxygenation injury. J Pharmacol Exp Ther. 2008;325:226–235. doi: 10.1124/jpet.107.133918. [DOI] [PubMed] [Google Scholar]
- 27.White EL, Ross LJ, Schmid SM, et al. Screening of potential cancer preventing chemicals for induction of glutathione in rat liver cells. Oncol Rep. 1998;5:507–512. [PubMed] [Google Scholar]
- 28.Kumar V, Kumar S, Hassan M, et al. Novel chalcone derivatives as potent Nrf2 activators in mice and human lung epithelial cells. J Med Chem. 2011;54:4147–4159. doi: 10.1021/jm2002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balogun E, Hoque M, Gong P, et al. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem J. 2003;371:887–895. doi: 10.1042/BJ20021619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess A, Wijayanti N, Neuschafer-Rube AP, et al. Phorbol ester-dependent activation of peroxiredoxin I gene expression via a protein kinase C, Ras, p38 mitogen-activated protein kinase signaling pathway. J Biol Chem. 2003;278:45419–45434. doi: 10.1074/jbc.M307871200. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y, Shin DH, Kim JH, et al. Caffeic acid phenethyl ester-mediated Nrf2 activation and IkappaB kinase inhibition are involved in NFkappaB inhibitory effect: structural analysis for NFkappaB inhibition. Eur J Pharmacol. 2010;643:21–28. doi: 10.1016/j.ejphar.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 32.Saracino MR, Lampe JW. Phytochemical regulation of UDP-glucuronosyltransferases: implications for cancer prevention. Nutr Cancer. 2007;59:121–141. doi: 10.1080/01635580701458178. [DOI] [PubMed] [Google Scholar]
- 33.Dinkova-Kostova AT. Phytochemicals as protectors against ultraviolet radiation: versatility of effects and mechanisms. Planta Med. 2008;74:1548–1559. doi: 10.1055/s-2008-1081296. [DOI] [PubMed] [Google Scholar]
- 34.Wruck CJ, Claussen M, Fuhrmann G, et al. Luteolin protects rat PC12 and C6 cells against MPP+ induced toxicity via an ERK dependent Keap1-Nrf2-ARE pathway. J Neural Transm Suppl. 2007:57–67. doi: 10.1007/978-3-211-73574-9_9. [DOI] [PubMed] [Google Scholar]
- 35.Tsai PY, Ka SM, Chang JM, et al. Epigallocatechin-3-gallate prevents lupus nephritis development in mice via enhancing the Nrf2 antioxidant pathway and inhibiting NLRP3 inflammasome activation. Free Radic Biol Med. 2011;51:744–754. doi: 10.1016/j.freeradbiomed.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Miyamoto N, Izumi H, Miyamoto R, et al. Quercetin induces the expression of peroxiredoxins 3 and 5 via the Nrf2/NRF1 transcription pathway. Invest Ophthalmol Vis Sci. 2011;52:1055–1063. doi: 10.1167/iovs.10-5777. [DOI] [PubMed] [Google Scholar]
- 37.Kode A, Rajendrasozhan S, Caito S, et al. Resveratrol induces glutathione synthesis by activation of Nrf2 and protects against cigarette smoke-mediated oxidative stress in human lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2008;294:L478–488. doi: 10.1152/ajplung.00361.2007. [DOI] [PubMed] [Google Scholar]
- 38.Rubiolo JA, Mithieux G, Vega FV. Resveratrol protects primary rat hepatocytes against oxidative stress damage: activation of the Nrf2 transcription factor and augmented activities of antioxidant enzymes. Eur J Pharmacol. 2008;591:66–72. doi: 10.1016/j.ejphar.2008.06.067. [DOI] [PubMed] [Google Scholar]
- 39.Ungvari Z, Bagi Z, Feher A, et al. Resveratrol confers endothelial protection via activation of the antioxidant transcription factor Nrf2. Am J Physiol Heart Circ Physiol. 2010;299:H18–24. doi: 10.1152/ajpheart.00260.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y, Eggler AL, Liu D, et al. Sites of alkylation of human Keap1 by natural chemoprevention agents. J Am Soc Mass Spectrom. 2007;18:2226–2232. doi: 10.1016/j.jasms.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng J, Zhang P, Chen X, et al. PI3K and ERK/Nrf2 pathways are involved in oleanolic acid-induced heme oxygenase-1 expression in rat vascular smooth muscle cells. J Cell Biochem. 2011;112:1524–1531. doi: 10.1002/jcb.23065. [DOI] [PubMed] [Google Scholar]
- 42.Roth M, Araya JJ, Timmermann BN, et al. Isolation of Modulators of the Liver-Specific Organic Anion-Transporting Polypeptides (OATPs) 1B1 and 1B3 from Rollinia emarginata Schlecht (Annonaceae) J Pharmacol Exp Ther. 2011;339:624–632. doi: 10.1124/jpet.111.184564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.