Abstract
Aspiration of gastrointestinal contents has been linked to worse outcomes following lung transplantation but uncertainty exists about underlying mechanisms. We applied high-resolution metabolomics of bronchoalveolar lavage fluid (BALF) in patients with episodic aspiration (defined by bile acids in the BALF) to identify potential metabolic changes associated with aspiration. Paired samples, one with bile acids and another without, from 29 stable lung transplant patients were studied. Liquid chromatography coupled to high-resolution mass spectroscopy was used to interrogate metabolomic contents of these samples. Data were obtained for 7068 ions representing intermediary metabolites, environmental agents and chemicals associated with microbial colonization. A substantial number (2302) differed between bile acid positive and negative samples when analyzed by false discovery rate at q = 0.01. These included pathways associated with microbial metabolism. Hierarchical cluster analysis defined clusters of chemicals associated with bile acid aspiration that were correlated to previously reported biomarkers of lung injury including T cell granzyme B level and the chemoattractants CXCL9 and CXCL10. These data specifically link bile acids presence in lung allografts to inflammatory pathways known to segregate with worsening allograft outcome, and provide additional mechanistic insight into the association between reflux and lung allograft injury.
Keywords: Aspiration, bile acid, lung transplantation, metabolomics
Introduction
Gastroesophageal reflux disease (GERD) has been linked to worsening lung allograft function. One of the hypotheses underlying this association is that patients with GERD are predisposed to aspiration of stomach and duodenal contents that are injurious to the pulmonary epithelium, either directly or indirectly through activation of pathways that stimulate innate inflammation and subsequent acquired immune system activation. Supporting this idea are reports that patients who have detectable bile acids or pepsin in their bronchoalveolar lavage fluid (BALF) have increased rates of development of bronchiolitis obliterans (1–3). In addition, a rodent model of chronic aspiration using gastric fluid as the inciting substance has shown that such aspiration induced a strong immune inflammatory response ultimately leading to rejection (4,5). Based on these findings, there is a growing interest in the lung transplant community to consider surgical measures such as fundoplication to limit aspiration. Yet a program of universal preemptive fundoplication may be premature at the present time as many unanswered questions remain regarding the mechanistic associations between GERD, aspiration and lung dysfunction. A central need in this field is understanding precisely how to detect and quantify such risk. To better inform what the indices of risk from GERD and aspiration are, we undertook a project in BALF metabolomic profiling (MP). We prospectively identified patients who at various time points in the posttransplant period had detectable bile acids in the BALF using a simple, validated biochemical test. We then performed BALF MP on these patients in the setting of bile acid aspiration (BAA) and at a time when BAA was absent (each patient was their own control). We show here that BAA is associated with a distinct BALF MP, which itself is correlated to other biomarker measures of immune activation known to be associated with lung allograft injury. Our results both relate GERD to acute immune augmentation in a highly controlled setting, and introduce BALF MP as a robust platform for discovery of metabolic patterns correlated to injury in lung transplantation.
Methods
BALF collection
Lung lavage fluid was obtained during fiberoptic bronchoscopy performed for routine surveillance for rejection. Our clinical protocols call for bronchoscopy to be performed at eight predetermined intervals in the posttransplant setting. The samples utilized here were taken from patients presenting for these protocol bronchoscopies. In all patients, the bronchoscope was wedged in either the right middle lobe bronchus or the lingular bronchus. Five aliquots of 30 mL saline were instilled into the lungs followed by serial aspiration. The first 30 mL of fluid returned was sent for routine clinical microbiology. The remainder of the lung lavage fluid was filtered through a 40-μm filter, then subjected to centrifugation to remove the cellular component. All supernatants were aliquoted to avoid repeated freezing and thawing for subsequent studies and all samples were stored at −80°C until analysis.
Patient selection
Patients were screened at the Emory Transplant Center and had provided informed consent for BALF samples to be used under an Institutional Review Board approved tissue acquisition protocol. A total of 384 samples from 51 patients were selected with the criteria that each patient had at least four BALF samples obtained from the first posttransplant year. Samples were obtained through the first posttransplant year. All of the samples utilized from this study were obtained from patients who were free from bronchiolitis obliterans syndrome (BOS) at the time of sample collection. From this group, 29 pairs of samples were chosen for MP. All subjects included in this analysis had undergone screening to test for esophageal reflux. This testing included pretransplant barium esophagram, ambulatory esophageal monitoring of reflux by pH and impedance and esophageal manometry. Patient characteristics of these 29 subjects are shown in Table 1. Of note three patients underwent fundoplication during the period of sample collection, and two patients had undergone fundoplication more than 1 year prior to transplantation.
Table 1.
Baseline characteristics and status of GERD testing of subjects assessed by BALF MP
| Testing for reflux | 8 Patients with reflux on barium swallow |
| 24 h ambulatory pH monitoring: 26 patients tested, median DeMeester score 11.2, range 0.8–58 | |
| Antireflux therapy | 26 Patients (90%), PPI 25 patients, H2 blocker 1 patient |
| Patient age | Mean 55, range 16–67 |
| Patient gender | 13 Female, 16 male |
| Fundoplication | 3 Patients treated with gastrofundoplication between samples, 2 patients with pretransplant gastrofundoplication |
| Esophageal function | Normal 10 patients; decreased peristalsis 10 patients; hypertensive lower esophageal sphincter 7 patients; untested 2 patients |
| Patients at lowest risk for aspiration | DeMeester <14, no reflux on barium esophagram, normal esophageal motility: 5 patients (17%) |
BALF MP, bronchoalveolar lavage fluid metabolomic profiling; COPD, chronic obstructive pulmonary disease; GERD, gastroesophageal reflux disease; IPF, idiopathic pulmonary fibrosis; PPI, proton pump inhibitor.
Disease indication for lung transplant: COPD 12, IPF 13, sarcoid 2, obliterative bronchiolitis 1, cystic fibrosis 1.
Screening for bile acids
All samples were screened for bile acids using a BioQuant (San Diego, CA) colormetric kit. All samples were run in triplicate, with bile acid concentration calculated using bile acid standards included in the kit. As a positive control, a duodenal aspirate obtained on a nonlung transplant patient was utilized. This fluid was diluted 10:1 in normal saline, the same fluid used to obtain BALF. Serial 1:10 dilutions of this stock duodenal aspirate were used in each experiment. The limit of detection of bile acids in this positive control was a 1:10 000 dilution, corresponding to a bile acid standard concentration of 0.195 μM. Samples were considered positive if they were greater than 0.39 μM, and considered negative if they were less than 0.195 μM. Normal saline as well as two plasma samples from nonlung transplant patients were utilized for every experiment and constituted the negative control. Both normal saline and the control plasma had absorbance values below that of the 0.195 μM standard. Of the 51 patients screened, 11 patients had no bile acids detected on any of their BALF samples. Three BALF samples obtained from normal humans were screened as well. Two of these samples were negative, while the third sample demonstrated detectability at the threshold of the test. Of the 40 lung transplant patients who had at least one sample with detectable bile acids, 29 patients pairs (58 samples) were chosen for BALF MP analysis. The criteria for analysis were that both the positive and negative samples were negative for infection at the time of sample collection and also no higher than minimal grade (A1) rejection by International Society of Heart and Lung Transplant criteria. Therefore, of the 40 subjects who had at least one episode of aspiration, 11 were excluded due to the significant rejection or infection detected in the bile positive bronchoscopy.
BALF MP
The metabolomic profiling was performed using liquid chromatography (LC) coupled to high-resolution mass spectrometry that we developed using an Orbitrap FTQ-Velos mass spectrometer (Thermo Scientific, Waltham MA) (6). Each sample was run in duplicate. This platform allows for differential assessment of chemical compounds with a mass to charge ratio (m/z) between 85 and 2000, which includes constituents ranging from small hydrocarbons to oligopolypeptides. Peak extraction and quantification of ion intensities were performed by an adaptive processing of liquid chromatography mass spectroscopy (apLCMS) software package (7) that provided tables containing m/z values, retention time and integrated ion intensity for each m/z feature. The total number of features with apLCMS was 7068 features from C18 reverse phase chromatography coupled with mass spectrometry.
Metabolite annotation and identification
The m/z features detected by high-resolution metabolomics include intermediary metabolites, chemicals derived from food and dietary supplements, products of the microbiome, drugs and their metabolites and environmental agents (8). Identities of common intermediates, such as amino acids and related metabolites, have been previously established (9–11), and LC retention time and accurate mass m/z provide unambiguous reference information for those without established chemical identities. Discriminatory features from statistical and bioinformatic analyses (below) were mapped into known endogenous metabolic pathways published in Kyoto Encyclopedia for Genes and Genomes (KEGG) (www.genome.jp/kegg) and annotated using the Metlin Database (www.metlin.scrips.edu). For selected features, ion dissociation (tandem mass spectroscopy [MS/MS]) was used to confirm chemical identity relative to reference spectra (Metlin) or authentic standards.
Biostatistics and bioinformatics
False discovery rate (FDR) analysis of log-transformed data with q = 0.01 was used to test for metabolites that differed between conditions, bile acids present or absent. Principal component analysis (PCA) with autoscaling was used to visualize differences between the two conditions, and the top 5% of metabolites were identified that contributed to 95% separation of bile acid present and bile acid absent. Similarly, principal component regression analysis (PCRA) was used to identify top metabolites associated with variation of specific biomarkers.
Hierarchical clustering analysis (HCA) was performed to identify patterns of metabolites associated with and without BAA. Common chemical features included into the HCA were those found by all three approaches: FDR, PCA and PCRA. Before running HCA, the features were log2 transformed and normalized by performing quantile normalization across samples. Then LIMMA package in R (Linear Models for Microarray Data) was applied to identify significant features at a significance threshold of 0.05 after FDR adjustment. The heatmap.2 function in the R package gplots (http://cran.r-project.org/web/packages/gplots) was used for HCA.
Correlation of HCA families to known biomarkers of risk. Our previous work from this cohort of lung transplant recipients has shown associations between the transcription factor Forkhead box P3 (FoxP3), the serine protease granzyme B and the soluble chemokines CXCL9, CXCL10 and monocyte chemoattractant protein 1 (MCP-1) with relevant transplant outcomes (12–14). We used Spearman correlation to test for associations between the 11 metabolite clusters present or absent in the setting of BAA, to the value of CD8, granzyme B, CD4, FoxP3 and soluble CXCL9, CXCL10 and MCP-1 from the same sample for the 42 samples where these data were available.
Results
High-resolution metabolomics of BALF reveal bias toward increased small chemicals in association with BAA
We extracted information obtained from LC–MS from the BALF of the 58 samples using the R package apLCMS. apLCMS analysis resolved 7608 metabolic features seen as individual peaks defined by m/z and retention time on the chromatography column. In order to better define the metabolic pathways associated with aspiration, we mapped the LC–MS data onto the KEGG database of human metabolism. The 7608 metabolomic features included 1128 chemicals present in this database. Because of the shear magnitude of chemical features seen, we used a rigorous FDR of q = 0.01 to adjust for multiple comparisons. With this approach we found 2302 features that were significantly different. In order to give a high-order view of the metabolomic differences associate with aspiration, we assessed the strength of the difference (p-value) as a function of m/z. The Manhattan plot of the −log p for metabolites as a function of m/z showed that the majority of the variance between the two conditions occurred with smaller chemicals (m/z in the 80–500 range) (Figure 1). The relevance of this finding is that in contrast to other systems in which metabolomic differences appear to be normally distributed as a function of m/z, the differences are decidedly skewed toward small molecules in association with BAA.
Figure 1. Manhattan plot on 7608 features from C18 showed that 2302 features were significantly different at FDR q = 0.01.
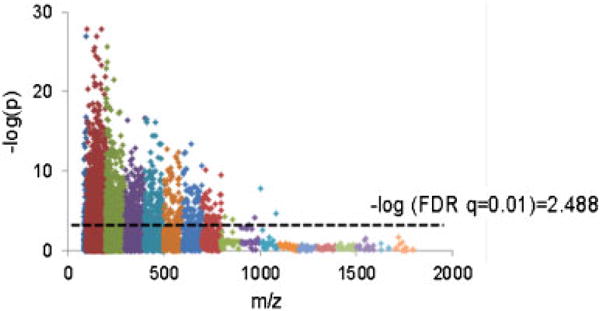
X-axis is m/z ranging 85–2000. Y-axis shows −log p-value for each metabolite detected between the two conditions. The higher the y-axis value, the more significant the difference was between bile positive and negative samples. Dot line is −log of FDR 0.01. FDR, false discovery rate; m/z, mass to charge ratio.
PCA discriminates bile acid positive from bile acid negative samples
PCA is an unsupervised approach to reduce complexity in large data sets, where the differences between two groups are reduced into a set of eigenvectors termed principal components. Hence, the value of PCA as a qualitative tool is that it can define whether two populations are different and give a qualitative assessment of that difference based on how completely the populations are separated and how many components are required to observe these differences. Color coding of the no bile acid (blue) and bile acid (red) samples in the PCA score plot of the 7068 features showed complete separation (Figure 2A) with only two principal components, corroborating the findings from apLCMS that BALF in the setting of bile acids is vastly different from BALF when no bile acids are present. We selected the top 5% of features associated with 95% group identity as bile acid present or absent, and found these 472 metabolites to extensively overlap with the metabolites detected by FDR (Figure 2B).
Figure 2.
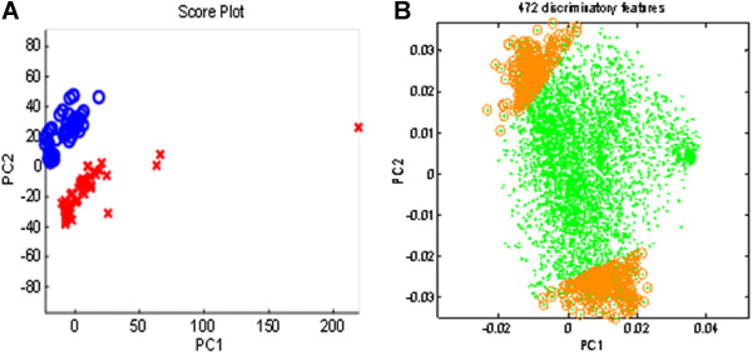
(A) PCA on total 7068 metabolites from 85 to 2000 m/z on C18 reverse phase coupled with Orbitrap Velos. Red = X bile present, Blue 0 = bile absent. (B) Principal component loading statistics (PCLS) determined 472 discriminatory metabolites. Green dots represent the metabolites with 95% correlation to the first two principal components. Gold circles denote the top 5% of these metabolites that most closely associated with groups. m/z, mass to charge ratio; PCA, principal component analysis.
Integrated use of bioinformatics reveals candidate features associated with BAA
As a complement to the PCA and FDR approaches, we employed PCRA on the BALF from the two conditions. PCRA is a regression analysis that uses PCA when estimating regression coefficients. PCRA was applied to determine significantly correlated metabolites to bile acid concentration with permutation and Bonferroni test. The PCRA included the 2411 features that were present in all runs for all samples. Hence, this approach was utilized on a subset of the total chemical features present in the entire 58 sample set. PCRA revealed 461 features that were significantly related to bile acid presence (Figure 3). We then assessed the interaction of PCA, PCRA and FDR to define the chemical species differentially present in all approached. As shown in the Venn diagram (Figure 4), there were 292 chemical features that were different between bile acid positive and negative samples by all three approaches.
Figure 3. Principal component regression analysis (PCRA) on 2411 features which appears in all runs of all samples.
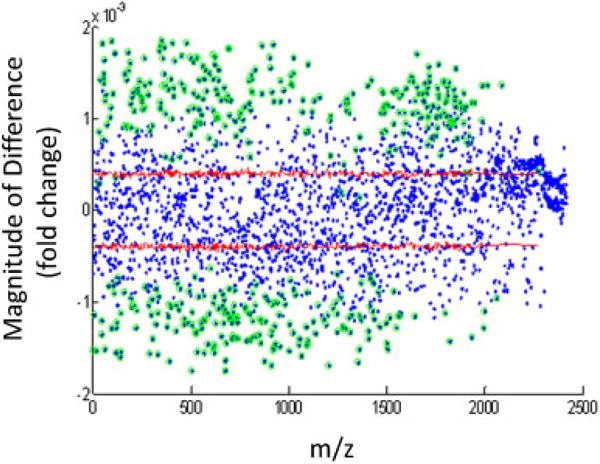
Four hundred sixty-one features were significantly related to bile acids. Green circles represent significant variables by the permutation and Bonferroni testing of PCRA loadings (regression vector). The red line represents two-sided 95% confidence intervals of the variables coefficient (i.e. effect) by permutation tests. The magnitude of the value on the y-axis represents the fold difference between bile positive samples compared with bile negative samples. Metabolites with a positive value were increased with BAA, while metabolites with a negative value were decreased with BAA. BAA, bile acid aspiration.
Figure 4. Venn diagram was generated using unique metabolites in each set.
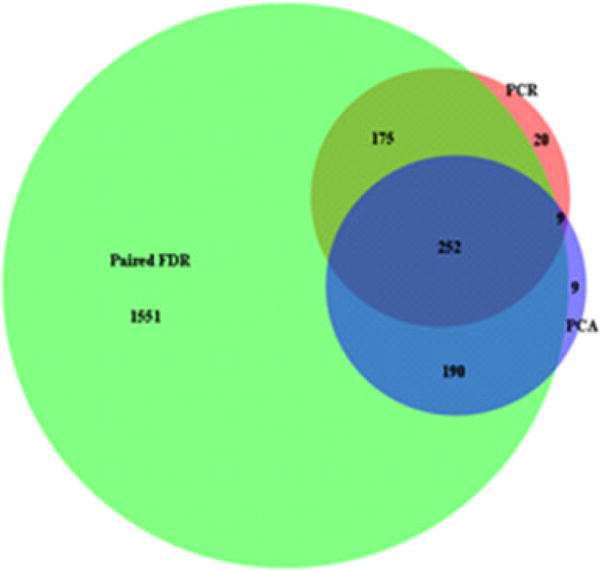
Green region represented paired FDR at q = 0.01 significant features. Blue region represented discriminatory features from PCA. The region of dark green contained the significant metabolites from PCRA with permutation Bonferroni test (p = 0.05). FDR, false discovery rate; PCA, principal component analysis; PCRA, principal component regression analysis.
HCA of bile acids and the association of these clusters with known markers of lung injury
After the common significant features were selected with Venn diagram using three different analyses, HCA was performed on the 292 common metabolomic features to see the association between clusters and significant metabolites. HCA sequentially links metabolomics features that are most commonly observed together. In our analysis, we resolved six clusters of metabolites (three associated with bile and three associated without bile) (Figure 5). Using the six metabolomic clusters from samples with and without bile acids present from our cohort, we performed correlational analysis between the differentially present BALF MP clusters and the other biomarkers that we had previously collected. Included were the concentration in μg/mL of CXCL9, CXCL10 and MCP-1 as determined by ELISA (12,14), the percentage of CD8 cells expressing granzyme B as determined by flow cytometry (14) and the percentage of CD4 cells expressing the transcription factor FoxP3 as determined by flow cytometry (13) (Figure 6). Statistically, a correlation coefficient of at least ±0.248 corresponded to a p-value of 0.05. Based on this, CD4 FoxP3 frequency had the strongest cross-omics correlation with bile acid associated metabolomic clusters, followed by BAL CXCL10 concentration. The presence of bile itself in a sample showed only modest correlation with the other biomarkers of danger (r-values 0.17–0.26, p-values 0.09–0.3). Finally, given that BAA has been associated with increased airway neutrophilia (15), we assessed each of our 29 sample pairs for the frequency of neutrophils and overall cellularity. There was a nonstatistically significant trend toward increased neutrophils and overall BALF cellularity in samples testing positive for bile (Figure S1).
Figure 5. Heatmap of hierarchical clustering on common metabolites (filtered by false discovery rate, PCLS and PCRA Bonferroni and permutation on 58 samples on duplicated runs.
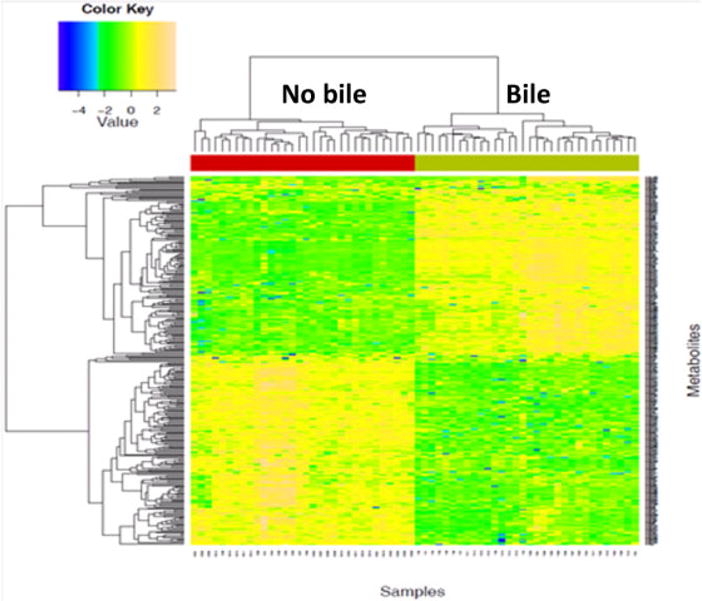
Green bar represents bile presence, and red bar signifies the absence of bile. Each column represents a single patient. PCLS, principal component loading statistics; PCRA, principal component regression analysis.
Figure 6. Chemokine-metabolite Spearman correlations.
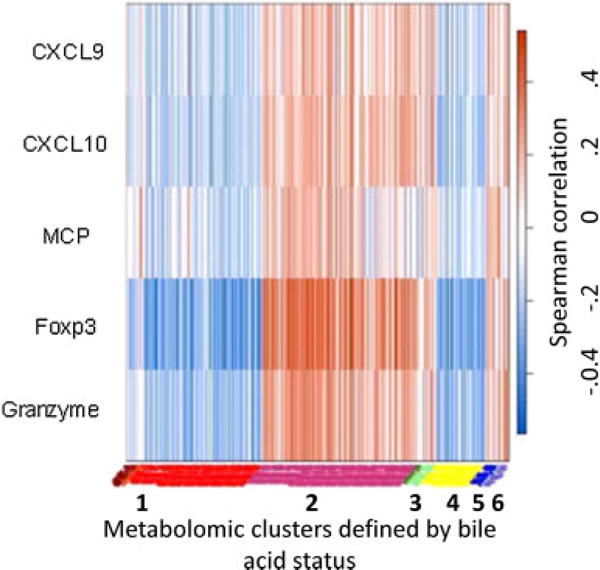
Two hundred sixty-five metabolites associated with biomarkers of risk for graft failure (left y-axis) were assessed for correlation with BALF common significantly regulated metabolites clustered around bile acid positive tests (x-axis). Spearman correlation is shown on the right y-axis with red values showing positive correlation and blue values negative correlation. A correlation coefficient of ±0.248 was the threshold for a p-value of 0.05. Cluster 1 is the major clusters associated with bile acid absence, and Cluster 2 is the major cluster associated with bile acid presence. Clusters 3 and 6 associate with bile, while Clusters 4 and 5 associate with no bile. BALF, bronchoalveolar lavage fluid.
Metabolomic differences from BAA are associated with a diverse set of endogenous and exogenous pathways
The common and differentially present MP features were subjected to database queries. Three hundred forty-one of the features matched known endogenous human metabolic pathways (KEGG Mapper). Additional database queries found identities for the many of the additional metabolites (Table 2). Included in the endogenous metabolites found at higher values in the setting of bile aspiration was the bile acid, 3-Oxocholic acid, validating the results of the biochemical test used to screen the samples. Differentially expressed metabolites that were not part of the KEGG database included p-cresol, a phenol derivative found in cleaning products. P-cresol levels were decreased in the bile acid positive samples, which is noteworthy as metabolism of this exogenous chemical is primarily a function of bacterial species such as Pseudomonas. Fosfomycin was increased in samples containing bile acids. Fosfomycin is sometimes used as a pharmaceutical agent, but exists in nature as an anti-microbial agent produced endogenously by several species of bacteria. None of our patients have been treated with fosfomycin, and neither p-cresol nor fosfomycin are constituents of the cleaning solutions used to sterilize bronchoscopes. Finally, we noted decreased levels of oxalureate in samples with bile acids. This purine metabolite is formed from the oxidation of ureidoglycolate, a process that is efficiently driven through bacterial enzymes. Based on these associations, we assessed the microbiologic data of the BAL samples used in these studies. None of the samples tested were associated with bacterial growth greater than 10 000 colonies, which is the general standard we utilize to differentiate clinical infection. Selected differentially expressed metabolites are shown in Table 2.
Table 2.
BALF metabolites that are differentially found in the setting of BAA and are associated with microbial metabolism
| Metabolite | Association with BAA |
|---|---|
| Glycocholic acid | Fourfold increase |
| Dehydrocholic acid | Twofold increase |
| Oxalureate | Twofold increase |
| P-cresol | Threefold decrease |
| C4-HSL | Twofold decrease |
| Fosfomycin | Threefold increase |
| Glutathionylspermine | 3.5-fold increase |
| Cystathionine | 5.8-fold decrease |
BAA, bile acid aspiration; BALF, bronchoalveolar lavage fluid.
Discussion
Lung allografts are inherently connected to the gastrointestinal tract and GERD is common after lung transplantation (16–18). GERD may predispose to graft loss by increasing the risk of aspiration. This is supported by studies showing that lung transplant patients are more prone to proximal reflux (19), and that surgical correction of GERD decreases many indices of inflammation within the lung (20). Further, patients with gastroparesis, another risk factor for aspiration, are also at a heightened risk for BOS (21). While it has been established that the presence of bile in the lung is a risk factor for subsequent chronic graft failure, significant work remains in establishing the pathways that lead to graft loss in the setting of aspiration. Here, we have employed a robust analytical tool in a discovery phase study to show that the chemical milieu of BALF is affected by reflux and aspiration of gastrointestinal contents. Assessing the data at the most basic level shows that the difference between samples with aspiration and no aspiration is most pronounced for small molecules (low m/z). We speculate that aspiration of bile or bile-associated constituents weakens epithelial barrier function. The concept that aspiration could affect the epithelial barrier is supported by studies linking aspiration to lowered levels of the surfactant protein A (SP-A) (22). This may have the combined effect of increasing shear stress across the alveolar membrane and at the same time lowering resistance to microbial infection as SP-A is felt to have intrinsic anti-microbial properties (23), as well as limiting inflammation induced by lipopolysaccharide (24). Further, recent in vitro work has shown that bile salts decrease barrier function in esophageal and respiratory cells (25,26).
It is important to note that our targeted analysis of differentially represented features indicated a bias toward features associated with microbial colonization including cresols, homoserine lactone (HSL), novel purine analogs and antibiotics endogenously produced by bacteria such as fosfomycin. These are substances that exogenous to humans. Also, it is important to note that some metabolites such as C4-HSL and p-cresol are conspicuously decreased in the setting of aspiration, indicating that the effects seen here cannot be ascribed to permeability changes alone. The association between reflux and pseudomonas colonization has been noted clinically (27). In our study, samples were excluded if they were positive by standard microbiologic techniques for bacterial or viral infection. We acknowledge that there are limitations to current microbiologic testing of fluids such as BAL. Therefore, it is plausible that the bile positive samples came from times in which subclinical infection was occurring. We plan to test this prospectively by integrating 16S rRNA and pyrosequencing with BALF MP in a longitudinal manner.
Our findings, derived from a cohort of patients who serve as their own controls, show modest correlation between BAA-associated metabolomic clusters and biomarkers of risk such as CXCL10, a chemokine secreted by pulmonary epithelial cells that is chemoattractive for CXCR3 bearing T lymphocytes, as well as granzyme B, one of the hallmark proteases found predominantly in effector memory CD8 lymphocytes.
Our group and others have found strong correlations between these biomarkers and subsequent graft loss across a range of transplant types in both clinical and experimental transplantation (12,28–31). Hence, these associations serve as validation that the metabolomic differences we see between samples from the same patient are related to other inflammatory changes that are felt to have strong implications on the organ. We did not detect a robust difference in BAL neutrophilia in the setting of bile, but do not believe that this finding exonerates neutrophils from the pathogenesis of aspiration-induced lung injury. It is possible that removal of the first aliquot of BALF for microbiology removed neutrophils present in the terminal airways.
There are likely multiple applications of our findings, which would pertain to lung transplantation as well as lung disease in general. For example, while there is growing consensus that aspiration of gastric contents can participate in the injury leading to BOS, it is still unclear clinically which patients are at greatest risk from such aspiration. By linking MP patterns to graft injury, prospective studies are likely to identify which patients are most likely to benefit from surgical procedures such as fundoplication, and which patients may require anti-microbial therapy for smoldering infection.
Our findings highlight the robust nature of MP in defining patterns of metabolites, which may portend greatest risk. Future studies will be needed to assess these MP patterns in a longitudinal manner to determine the extent to which they predict subsequent graft failure. In this analysis, we used bile as a biomarker for aspiration. The biochemical test used has limitations including the fact that there may be false positives from constituents, which are chemically similar to bile salts, and the cutoff for a clinically important bile test is simply not known. LC–MS validation of the accuracy of the colorimetric assay utilized to assign bile acid positivity of the paired BAL specimens was not performed for this study. Further, it is unclear the extent to which the damage caused by aspiration is directly linked to the bile itself, or to other constituents of the gastrointestinal tract. Thinking mechanistically, one potential benefit of using a tool as robust as MP in a longitudinal manner is that it can reveal the strength of association between various metabolites and subsequent graft failure, ultimately shining a spotlight on the most proximate cause of epithelial injury.
Supplementary Material
Left panel shows 28 pairs of patients in both the bile and no bile sample with y-axis measuring the frequency of the BALF cells which are neutrophils. Median no bile 2% (95% CI 1–18), median bile 6% (95% CI 6.9–27); p = 0.09. Right panel shows 28 pairs of patients in both the bile and no bile sample with y-axis measuring the BALF total cell count. Median no bile 169 cell/mL (95% CI 139–306) median bile 274 cell/mL (95% CI 248–519), p = 0.08. One of the 29 patients was excluded due to missing data.
Acknowledgments
DCN is supported by an NIH career development grant: 5K08AI079166.
Abbreviations
- apLCMS
adaptive processing of liquid chromatography mass spectroscopy
- BAA
bile acid aspiration
- BALF
bronchoalveolar lavage fluid
- BALF MP
bronchoalveolar lavage fluid metabolomic profiling
- BOS
bronchiolitis obliterans syndrome
- FDR
false discovery rate
- FoxP3
Forkhead box P3
- GERD
gastroesophageal reflux disease
- GF
gastrofundoplication
- HCA
hierarchical cluster analysis
- HSL
homoserine lactone
- KEGG
Kyoto Encyclopedia for Genes and Genomes
- LC
liquid chromatography
- MCP-1
monocyte chemoattractant protein 1
- MS/MS
tandem mass spectroscopy
- m/z
mass to charge ratio
- PCA
principal component analysis
- PCRA
principal component regression analysis
- SP-A
surfactant protein A
Footnotes
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
References
- 1.D’Ovidio F, Mura M, Tsang M, et al. Bile acid aspiration and the development of bronchiolitis obliterans after lung transplantation. J Thorac Cardiovasc Surg. 2005;129:1144–1152. doi: 10.1016/j.jtcvs.2004.10.035. [DOI] [PubMed] [Google Scholar]
- 2.Mertens V, Blondeau K, Van Oudenhove L, et al. Bile acids aspiration reduces survival in lung transplant recipients with BOS despite azithromycin. Am J Transplant. 2011;11:329–335. doi: 10.1111/j.1600-6143.2010.03380.x. [DOI] [PubMed] [Google Scholar]
- 3.Stovold R, Forrest IA, Corris PA, et al. Pepsin, a biomarker of gastric aspiration in lung allografts: A putative association with rejection. Am J Respir Crit Care Med. 2007;175:1298–1303. doi: 10.1164/rccm.200610-1485OC. [DOI] [PubMed] [Google Scholar]
- 4.Hartwig MG, Appel JZ, Li B, et al. Chronic aspiration of gastric fluid accelerates pulmonary allograft dysfunction in a rat model of lung transplantation. J Thorac Cardiovasc Surg. 2006;131:209–217. doi: 10.1016/j.jtcvs.2005.06.054. [DOI] [PubMed] [Google Scholar]
- 5.Li B, Hartwig MG, Appel JZ, et al. Chronic aspiration of gastric fluid induces the development of obliterative bronchiolitis in rat lung transplants. Am J Transplant. 2008;8:1614–1621. doi: 10.1111/j.1600-6143.2008.02298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park YH, Lee K, Soltow QA, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology. 2012;295:47–55. doi: 10.1016/j.tox.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu T, Park Y, Johnson JM, Jones DP. apLCMS-adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones DP, Park Y, Ziegler TR. Nutritional metabolomics: Progress in addressing complexity in diet and health. Annu Rev Nutr. 2012;32:183–202. doi: 10.1146/annurev-nutr-072610-145159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson JM, Yu T, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst. 2010;135:2864–2870. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roede JR, Park Y, Li S, Strobel FH, Jones DP. Detailed mitochondrial phenotyping by high resolution metabolomics. PLoS ONE. 2012;7:e33020. doi: 10.1371/journal.pone.0033020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2011;9(1 Suppl):132–143. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neujahr DC, Perez SD, Mohammed A, et al. Cumulative exposure to gamma interferon-dependent chemokines CXCL9 and CXCL10 correlates with worse outcome after lung transplant. Am J Transplant. 2012;12:438–446. doi: 10.1111/j.1600-6143.2011.03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neujahr DC, Cardona AC, Ulukpo O, et al. Dynamics of human regulatory T cells in lung lavages of lung transplant recipients. Transplantation. 2009;88:521–527. doi: 10.1097/TP.0b013e3181b0e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohammed A, Ulukpo O, Lawrence EC, et al. Cumulative exposure to CD8+ granzyme Bhi T cells is associated with reduced lung function early after lung transplantation. Transplant Proc. 2011;43:3892–3898. doi: 10.1016/j.transproceed.2011.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verleden GM, Vanaudenaerde BM, Dupont LJ, Van Raemdonck DE. Azithromycin reduces airway neutrophilia and interleukin-8 in patients with bronchiolitis obliterans syndrome. Am J Respir Crit Care Med. 2006;174:566–570. doi: 10.1164/rccm.200601-071OC. [DOI] [PubMed] [Google Scholar]
- 16.D’Ovidio F, Keshavjee S. Gastroesophageal reflux and lung transplantation. Dis Esophagus. 2006;19:315–320. doi: 10.1111/j.1442-2050.2006.00603.x. [DOI] [PubMed] [Google Scholar]
- 17.Hadjiliadis D, Duane Davis R, Steele MP, et al. Gastroesophageal reflux disease in lung transplant recipients. Clin Transplant. 2003;17:363–368. doi: 10.1034/j.1399-0012.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 18.Young LR, Hadjiliadis D, Davis RD, Palmer SM. Lung transplantation exacerbates gastroesophageal reflux disease. Chest. 2003;124:1689–1693. doi: 10.1378/chest.124.5.1689. [DOI] [PubMed] [Google Scholar]
- 19.Fisichella PM, Davis CS, Gagermeier J, et al. Laparoscopic antireflux surgery for gastroesophageal reflux disease after lung transplantation. J Surg Res. 2011;170:e279–e286. doi: 10.1016/j.jss.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisichella PM, Davis CS, Lowery E, et al. Pulmonary immune changes early after laparoscopic antireflux surgery in lung transplant patients with gastroesophageal reflux disease. J Surg Res. 2012;177:e65–e73. doi: 10.1016/j.jss.2012.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raviv Y, D’Ovidio F, Pierre A, et al. Prevalence of gastroparesis before and after lung transplantation and its association with lung allograft outcomes. Clin Transplant. 2012;26:133–142. doi: 10.1111/j.1399-0012.2011.01434.x. [DOI] [PubMed] [Google Scholar]
- 22.D’Ovidio F, Mura M, Ridsdale R, et al. The effect of reflux and bile acid aspiration on the lung allograft and its surfactant and innate immunity molecules SP-A and SP-D. Am J Transplant. 2006;6:1930–1938. doi: 10.1111/j.1600-6143.2006.01357.x. [DOI] [PubMed] [Google Scholar]
- 23.Wright JR. Immunoregulatory functions of surfactant proteins. Nat Rev Immunol. 2005;5:58–68. doi: 10.1038/nri1528. [DOI] [PubMed] [Google Scholar]
- 24.Stamme C, Wright JR. Surfactant protein A enhances the binding and deacylation of E coli LPS by alveolar macrophages. Am J Physiol. 1999;276:L540–L547. doi: 10.1152/ajplung.1999.276.3.L540. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Oshima T, Shan J, Fukui H, Watari J, Miwa H. Bile salts disrupt human esophageal squamous epithelial barrier function by modulating tight junction proteins. Am J Physiol Gastrointest Liver Physiol. 2012;303:G199–G208. doi: 10.1152/ajpgi.00454.2011. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Oshima T, Tomita T, et al. Acidic bile salts modulate the squamous epithelial barrier function by modulating tight junction proteins. Am J Physiol Gastrointest Liver Physiol. 2011;301:G203–G209. doi: 10.1152/ajpgi.00096.2011. [DOI] [PubMed] [Google Scholar]
- 27.Vos R, Blondeau K, Vanaudenaerde BM, et al. Airway colonization and gastric aspiration after lung transplantation: Do birds of a feather flock together? J Heart Lung Transplant. 2008;27:843–849. doi: 10.1016/j.healun.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Belperio JA, Keane MP, Burdick MD, et al. Critical role for CXCR3 chemokine biology in the pathogenesis of bronchiolitis obliterans syndrome. J Immunol. 2002;169:1037–1049. doi: 10.4049/jimmunol.169.2.1037. [DOI] [PubMed] [Google Scholar]
- 29.Belperio JA, Keane MP, Burdick MD, et al. Role of CXCL9/CXCR3 chemokine biology during pathogenesis of acute lung allograft rejection. J Immunol. 2003;171:4844–4852. doi: 10.4049/jimmunol.171.9.4844. [DOI] [PubMed] [Google Scholar]
- 30.Medoff BD, Wain JC, Seung E, et al. CXCR3 and its ligands in a murine model of obliterative bronchiolitis: Regulation and function. J Immunol. 2006;176:7087–7095. doi: 10.4049/jimmunol.176.11.7087. [DOI] [PubMed] [Google Scholar]
- 31.Jackson JA, Kim EJ, Begley B, et al. Urinary chemokines CXCL9 and CXCL10 are noninvasive markers of renal allograft rejection and BK viral infection. Am J Transplant. 2011;11:2228–2234. doi: 10.1111/j.1600-6143.2011.03680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Left panel shows 28 pairs of patients in both the bile and no bile sample with y-axis measuring the frequency of the BALF cells which are neutrophils. Median no bile 2% (95% CI 1–18), median bile 6% (95% CI 6.9–27); p = 0.09. Right panel shows 28 pairs of patients in both the bile and no bile sample with y-axis measuring the BALF total cell count. Median no bile 169 cell/mL (95% CI 139–306) median bile 274 cell/mL (95% CI 248–519), p = 0.08. One of the 29 patients was excluded due to missing data.


