Abstract
Structural studies of chromatin complexes composed of chromatin factors or enzymes bound to the nucleosome have been constrained by the ability to produce high quality complexes in the amounts appropriate for biophysical studies and by the difficulty of crystallizing these complexes. We describe here procedures and approaches to prepare chromatin complexes, to crystallize chromatin complexes and to improve diffraction properties through post-crystallization soaks. Special attention is paid to evaluating the quality of the purified chromatin complexes as well as assessing the presence of the chromatin protein or enzyme in crystals. The methods described for preparing and purifying chromatin complexes should be applicable to biochemical, biophysical and other structural approaches including cryo-electron microscopy.
1. Introduction
As the fundamental repeating unit of chromatin, the nucleosome is a hub of genetic activity in the eukaryotic nucleus. This activity involves interactions of chromatin factors and enzymes with not just the individual histone and DNA components of the nucleosome, but with the nucleosome as a unit (McGinty, Makde, & Tan, 2016). Despite the fundamental importance of such interactions to eukaryotic biology, we currently possess only a limited view of the molecular details of nucleosome interactions due to the relatively few three dimensional structures available (Armache, Garlick, Canzio, Narlikar, & Kingston, 2011; Barbera et al., 2006; Kato et al., 2013; Makde, England, Yennawar, & Tan, 2010; Maskell et al., 2015; McGinty, Henrici, & Tan, 2014; Zhou et al., 2015). Remedying this deficiency will require determining atomic structures of different classes of chromatin factors and enzymes with the nucleosome so that paradigms for chromatin interactions can be elucidated. We present here procedures for the crystal structure determination of chromatin complexes with special attention to preparing such complexes, applicable to other structural methods including cryoelectron microscopy, and post-crystallization soaks to improve crystal diffraction quality.
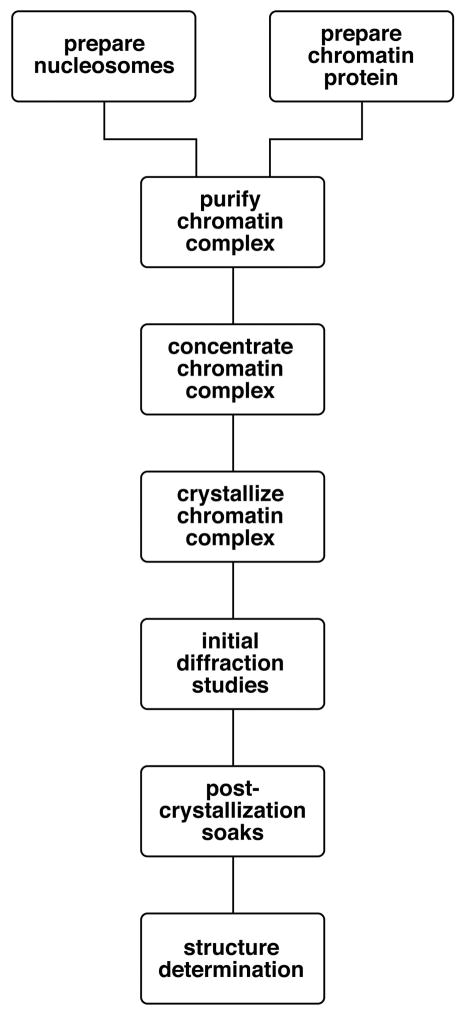
The basic workflow involved in the crystal structure determination of a chromatin complex is shown in Figure 1, and each item except for structure determination is discussed here (there are few aspects of crystal structure determination which are specific to chromatin complexes). The linear form of this ideal workflow does not, of course, reflect the reality of most projects where practical issues force one to proceed in a decidedly non-linear fashion, for example to redesign chromatin protein or nucleosome constructs. Crystallizing chromatin complexes remains a challenging endeavor, and it is the unusual chromatin complex project that progresses smoothly through purification, concentration, crystallization, diffraction studies and structure determination. However, the substantial biological insights afforded provide compelling justification to pursue such structural studies.
Fig. 1.
Basic workflow for the preparation, crystallization and structure determination of a chromatin complex.
2. Prepare Nucleosome Core Particles (NCP)
The preparation of recombinant nucleosome core particles appropriate for structural studies has been described in detail in previous publications (Luger, Rechsteiner, & Richmond, 1999a; 1999b; Luger, Rechsteiner, Flaus, Waye, & Richmond, 1997b), and these descriptions are still comprehensive and appropriate today. The reader is encouraged to follow closely the procedures described in these publications since the details can determine both the efficiency and quality of the resulting nucleosomes. We offer here some observations based on our experiences, which we hope will augment the descriptions in these publications.
We prefer to use anion-exchange high performance chromatography over preparative gel electrophoresis to purify the reconstituted nucleosome core particles for the scalability and throughput advantages. While preparative gel electrophoresis could offer higher resolution purification, we have not yet encountered situations that require such potential higher resolution.
We use GE Healthcare Source 15Q resin for the anion-exchange purification of reconstituted nucleosome core particles, and find this resin to be at least as good as the Tosoh TSK DEAE-5PW employed in the Luger et al publications (Luger et al., 1997b; Luger, Rechsteiner, & Richmond, 1999a; 1999b). Although we have not evaluated the GE Healthcare Mono Q resin for purifying reconstituted nucleosomes, we suspect this resin would also be appropriate based on our other comparisons of Source Q and Mono Q resins.
We routinely evaluate Source Q fractions of reconstituted nucleosome core particles by native polyacrylamide gel electrophoresis (PAGE), but one has to be careful in interpreting the results since the peak fractions will contain significant concentrations of salt, which promote dissociation of the nucleosome core particles during electrophoresis. For a more accurate assessment of nucleosome core particle quality, samples should be dialyzed against the storage low salt buffer before electrophoresis. Otherwise, the amount of free DNA and non-canonical nucleosome species might be overestimated.
The choice of the nucleosomal DNA to reconstitute with histones may be dependent on whether the associated chromatin factor or enzyme has any sequence preference. In the case that the chromatin factor or enzyme has no sequence preference, one could use the symmetric human alpha satellite nucleosome positioning sequence used successfully to crystallize the nucleosome core particle at 1.9 Å resolution (Davey, Sargent, Luger, Maeder, & Richmond, 2002; T. J. Richmond & Davey, 2003). However, one should be aware that preparing a symmetric nucleosome positioning sequence requires significantly more effort than preparing an asymmetric nucleosome positioning sequence (four preparative enzymatic steps versus just one). We have successfully used the asymmetric Widom 601 nucleosome positioning sequence used extensively in the chromatin field for our structural studies (Kim, Chatterjee, Jennings, Bartholomew, & Tan, 2015; Lowary & Widom, 1998; Makde et al., 2010; McGinty et al., 2014; Thåström et al., 1999), and we observe slightly higher nucleosome core particle reconstitution yields using this sequence. One potential disadvantage is a lower yield of plasmid containing multiple inserts of the Widom 601 DNA sequence: typically, a 12 liter preparation of pST55-16xNCP601a (a high copy number plasmid containing 16 copies of a 147 bp Widom DNA repeat) provides 50–100 mg of plasmid, which yields 20–50 mg of the 147 Widom DNA repeat. Comparable yields for other asymmetric nucleosome positioning sequences can be twice as high.
The plasmid containing multiple inserts of the nucleosome positioning sequence should be propagated in the HB101 E. coli strain for the recA- genotype to reduce recombination of the relatively unstable plasmid. Even when using HB101, one may notice a small fraction of plasmid with less than the full number of inserts, but inserts appear to be lost as individual units and not fractions of an insert.
For crystallizing chromatin complexes, it is helpful to have minimally 3 to 5 mg of purified nucleosome core particles at a concentration of ~10 mg/ml.
3. Prepare chromatin protein
It is difficult to provide general instructions on how to prepare chromatin proteins since each protein is highly individual. Seeing that one will need milligram quantities of the chromatin protein, recombinant methods are often used to overexpress the individual protein or protein complex in an appropriate heterologous system. Standard approaches should be used to express and purify the protein (Burgess & Deutscher, 2009; Curr Protoc Protein Sci, n.d.). We try to prepare at least 5 mg of protein at a final concentration of > 5 mg/ml in an appropriate buffer, with higher concentrations required for larger proteins and complexes and those requiring high salt for concentration. The concentration buffer conditions are optimized to reduce aggregation of the concentrated protein. We use dynamic light scattering to assess the aggregation state (we strive for a single peak with a polydispersity of < 20%). We also aim for a purity of > 95%, noting that the chromatin protein does not need to be absolutely pure since further purification of the chromatin protein/nucleosome complex will be carried out (see item 4 below).
4. Purify chromatin complex
4.1 Analytical purification
We prefer to use size exclusion chromatography to purify the chromatin protein or enzyme in complex with the nucleosome (the complex will be referred to as the “chromatin complex”) for several reasons. Size exclusion chromatography is a relatively robust and simple method, it is scalable (enabling both analytical and preparative uses), and the buffer exchange properties allow samples to be concentrated immediately after the run. Preparative electrophoresis of the chromatin complex is an alternative that may be particularly useful if overlapping chromatography peaks of different complexes can be resolved by electrophoresis. We do note that not all chromatin complexes stable to size exclusion chromatography will be stable to electrophoresis. Selective precipitation with polyethylene glycol is yet another alternative purification method for chromatin complexes (Arnaudo et al., 2013).
In order to ensure stoichiometric preparations of chromatin complexes, we first optimize reconstitution buffer, especially salt concentration, and chromatin protein/nucleosome ratios using analytical scale reconstitutions prior to proceeding to preparative scales. An example of this optimization is described here.
-
1
Equilibrate a Superdex 200 column in filtered MilliQ water at a flow rate of 0.2 to 0.4 ml/min. We have used both Superdex 200 HR 10/30 and Superdex 200 increase 10/300 columns with similar results though Superdex increase 10/300 columns can be run at higher flow rates.
-
2
Prepare 300 ml of appropriate initial size exclusion chromatography buffer filtered through a 0.45 μm filter. The precise buffer composition should be selected to stabilize the chromatin complex, a decision that may require iterative optimization. We use 5 mM Tris-Cl pH 7.6, 35–75 mM NaCl, 1 mM DTT, and any complex-specific additives for our initial attempts. When possible, our initial choice of salt concentration is based on quantitative binding experiments using fluorescence quenching HIFI binding experiments in varying conditions (Hieb, D’Arcy, Kramer, White, & Luger, 2012; Kim et al., 2015; McGinty et al., 2014; Winkler, Luger, & Hieb, 2012).
-
3
Equilibrate Superdex 200 column in same chromatography buffer.
-
4
Plan the recipe for the chromatin complex. For analytical reconstitutions we typically prepare 400 μl of complex with 0.5 – 1.0 mg/ml NCP and 2.2 – 2.5 molar equivalents of a chromatin protein anticipated to bind with a 2:1 chromatin protein:NCP stoichiometry. This corresponds to a slight excess of the chromatin protein to drive saturation. In cases where a 1:1 nucleosome binding is expected, the molar equivalents are halved. Depending on the size and shape of the chromatin protein, the excess chromatin protein can often be separated from the chromatin protein/NCP complex by size exclusion chromatography.
An example used to reconstitute the PRC1 ubiquitylation module/NCP complex is provided here.
-
Record the concentration of the available chromatin protein and nucleosome:
PRC1 ub module prep 1 10.0 mg/ml 235 μM NCP prep 196 22.3 mg/ml 109 μM -
Decide how much NCP you wish to use. Then calculate the amount in moles of chromatin protein you need to reconstitute the appropriate stoichiometric chromatin protein/NCP complex plus additional chromatin protein to drive formation of the chromatin complex. The final volume should be less than 500 μl.
18.0 μl 109 μM NCP prep 196 (final concentration 1 mg/ml) 20.8 μl 235 μM PRC1 ub module prep 1 (2.5 molar equivalents to NCP) 361 μl reconstitution buffer (10 mM Tris-Cl pH 7.6, 35 mM NaCl, 1 mM DTT, 10 μM ZnSO4) 400 μl total volume
-
-
5
Prepare the chromatin complex mixture by adding the components in order (buffer, then NCP, then PRC1 ub module), usually at room temperature. Add the chromatin protein in 2–5 aliquots with at least 5 minutes between each addition:
361 μl reconstitution buffer 18.0 μl 109 μM NCP prep 196 5.2 μl 235 μM PRC1 ub module prep 1 obs: precipitation, then immediate clearing 5.2 μl 235 μM PRC1 ub module prep 1 obs: precipitation, then immediate clearing 5.2 μl 235 μM PRC1 ub module prep 1 obs: precipitation, then immediate clearing 5.2 μl 235 μM PRC1 ub module prep 1 obs: precipitation, then immediate clearing -
6
Purify the chromatin complex by size exclusion chromatography over the equilibrated Superdex 200 column:
Load the sample through the sample loop using a 1 ml disposable syringe
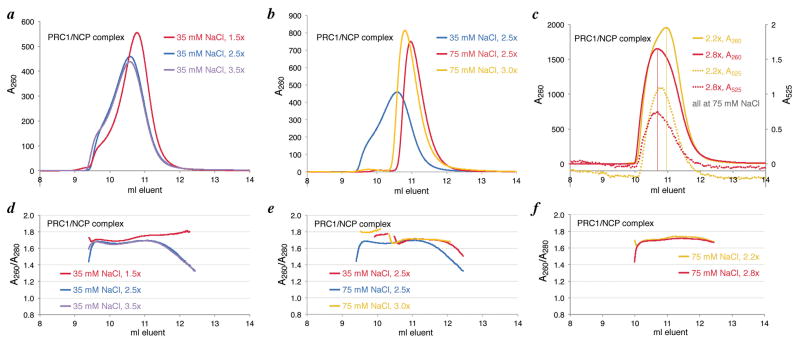
Elute the sample using a flow rate of 0.4 ml/min (or 0.75 ml/ml for Superdex 200 increase columns) collecting 1 min = 0.4 ml fractions and monitoring both A260 and A280 if possible. The PRC1 ub module/NCP complex usually elutes between 0.42 and 0.48 column volumes, while the free PRC1 ub module protein usually elutes around 0.58 column volumes (Fig. 2a, b).
-
7
Analyze peak fractions by SDS-PAGE using an 18% gel.
-
8
The quality of complex is assessed as follows:
Examine the width and symmetry of the chromatographic peak of the chromatin complex. A front shoulder often suggests super-stoichiometric non-specific binding to the nucleosome as observed initially with the reconstitution described above (Figure 2a). Meanwhile a back shoulder may suggest unsaturated complex or free nucleosome. The presence of a peak for the chromatin protein implies unbound protein resulting from too stringent of reconstitution conditions or too much protein added in the reconstitution mixture. We also plot and examine the A260/A280 ratio that acts as a surrogate for the NCP to chromatin protein ratio (Figure 2d). A flat A260/A280 ratio across the chromatin complex peak indicates a homogenous and saturated complex (Figure 2e, f). One can also use the absorbance of a fluorophore attached to the chromatin protein to further interrogate the quality of the complex based on alignment of the fluorophore absorbance and A260 peaks (Figure 2f, and see more below).
Examine the stoichiometry of the chromatin protein and histones in peak fractions via SDS-PAGE. Homogenous, saturated complexes show similar stoichiometry across the entire peak as opposed to higher levels of the chromatin protein relative to histones at the front edge of the peak versus the back edge.
-
9
Based on the above analysis, adjust the reconstitution buffer conditions until a high-quality complex is obtained. For the PRC1 ubiquitylation module-NCP complex described above, the NaCl concentration in the reconstitution buffer was systematically increased to 75 mM to eliminate the observed front shoulder indicative of super-stoichiometric binding of PRC1 to the NCP (Figures 2a and 2b).
-
10
With optimized buffer conditions established, we then optimize the ratio of chromatin protein to nucleosome by repeating analytical reconstitutions followed by size exclusion chromatography, titrating the chromatin protein into the nucleosome. We first use a broad titration series, for example 1.1:1, 2.2:1, 3.3:1, etc., to determine preferred stoichiometry and to further rule out super-stoichiometric binding by examining peak profile and retention time shift upon increasing chromatin protein (Figures 2a and 2d). This broad titration series can then be refined using a more targeted titration (Figures 2b and 2e). Again fluorescently labeled chromatin factor can aid in the analysis. Such optimization led us to use 2.8–3.0:1 PRC1 ub module:NCP for preparative scale reconstitutions (Figures 2c and 2f). Note that at a ratio of 2.8:1 PRC1 ub module:NCP, the A260 peak (detecting mostly nucleosomal DNA) and the A525 peak (detecting the fluorescently labeled chromatin enzyme) align, whereas at a ratio of 2.2:1, the two peaks do not align (Figure 2f).
-
8
If the Superdex 200 column will not be used for additional runs, wash the column in water at 0.4 ml/min, then store in 25% ethanol at 0.2 ml/min flow rate.
Fig. 2.
Size exclusion chromatography for the purification and analysis of a reconstituted chromatin complex. a) and b) A260 chromatograms for reconstituted chromatin complexes on an analytical scale using specified salt concentrations and molar equivalents of chromatin protein relative to NCP. c) A260 and A525 for fluorescently labeled chromatin complex reconstituted on a preparative scale. d–f) A260/A280 ratio for chromatograms in top panels.
4.2 Preparative purification
After optimizing the reconstitution on analytical scale, we reconstitute and purify the chromatin complex on a preparative scale. Here we describe the procedure used in our laboratory to purify chromatin complexes by size exclusion chromatography on preparative scale for crystallization.
-
1
Equilibrate a Superdex 200 column in optimized buffer as described above for analytical scale experiments.
-
2
Plan the recipe for the chromatin complex. This might be the single most important step of the procedure and due care is warranted.
An example used to reconstitute the PRC1 ub module/NCP complex is provided here.
-
Record the concentration of the available chromatin protein and nucleosome:
PRC1 ub module prep 3 8.54 mg/ml 120 μM NCP prep 222 12.9 mg/ml 62.9 μM -
Decide how much NCP you wish to use. Then calculate the amount in moles of chromatin protein you need to reconstitute the appropriate stoichiometric chromatin protein/NCP complex. Determine the amount of salt contributed by the chromatin protein buffer and the amount of salt additional salt needed to raise the overall reconstitution salt concentration to the desired level. Finally, calculate the volume of 1 M Tris-Cl pH 7.6 to add an additional 25 mM Tris-Cl (to counteract the relatively low pH 6.0 in the NCP storage solution). The final volume should be less than 500 μl.
121 μl 62.9 μM NCP prep 222 ~1.55 mg 7.5 μl 1 M Tris-Cl pH 7.6 1.9 μl 5 M NaCl (75 mM final concentration) 110 μl 120 μM PRC1 ub module prep 3 (2.8:1 ratio) 59.6 μl MilliQ water 300 μl total volume
-
-
5
Prepare the chromatin complex mixture by adding the components in order (water, NCP, then Tris, then salt, then PRC1 ub module), usually at room temperature. Add the chromatin protein in 2–5 aliquots with at least 5 minutes between each addition and record observations. It is not uncommon to observe some precipitation upon addition of the chromatin protein that clears upon mixing, especially at lower salt concentrations.
59.6 μl MilliQ water 121 μl 62.9 μM NCP prep 222 7.5 μl 1 M Tris-Cl pH 7.6 1.9 μl 5 M NaCl 27.6 μl 120 μM PRC1 ub module prep 3 obs: precipitation, then immediate clearing 27.6 μl 120 μM PRC1 ub module prep 3 obs: precipitation, then immediate clearing 27.6 μl 120 μM PRC1 ub module prep 3 obs: precipitation, then immediate clearing 27.6 μl 120 μM PRC1 ub module prep 3 obs: precipitation, with some cloudiness remaining 300 μl total volume -
6
Purify the chromatin complex by size exclusion chromatography over the equilibrated Superdex 200 column as described above for analytical reconstitutions (Figures 2c and 2f).
-
7
Pool the chromatin complex (PRC1 ub module/NCP here) Superdex fractions (typically 3–5 fractions of 0.4 ml/fraction, or 1.2–2 ml) in Eppendorf tubes. We often add PMSF to 0.1 mM to minimize proteolysis of the complex.
5. Concentrate chromatin complex
-
Concentrate the chromatin complex pool using a Vivaspin 500, 10 kD cutoff centrifuge concentration device to a final concentration of ~10 mg/ml. Use a microcentrifuge speed of 10 K rpm or manufacturer’s recommended centrifugation speed for all concentration centrifugation steps. For ill-behaved samples, consider concentrating an aliquot to first confirm that the chromatin complex can be concentrated to high concentrations without aggregation before committing the entire sample.
Wash concentration device twice with size exclusion chromatography buffer, discard filtrates.
Concentrate chromatin complex pool through successive spins of 400–500 μl until the final volume is less than 150 μl. 5–10 minutes is usually sufficient to concentrate 500 μl per spin. Substantially longer centrifugation times may indicate aggregation of the sample in the concentration buffer.
Transfer concentrated chromatin complex sample to a new Eppendorf tube.
Add 500 μl of size exclusion chromatography buffer to concentration device and allow to stand for 5 min at room temperature. Then centrifuge at 10 K rpm for 5–10 minutes to concentrate this wash to less than 10 μl.
Combine wash with concentrate chromatin complex pool. Mix gently.
Centrifuge chromatin complex sample for 5 minutes at 20°C and then transfer supernatant to new Eppendorf tube. Avoid any particulate matter at the bottom of the tube.
Analyze aggregation state of concentrated sample by dynamic light scattering (DLS) or similar appropriate method.
-
Quantitate the chromatin complex. We typically quantitate using the UV absorbance of the nucleic acid component of the NCP within the chromatin complex under denaturing basic conditions to dissociate the chromatin and histone proteins from the nucleosomal DNA. With some complexes, we do not observe a clear peak at 260 nm. Rather the absorbance continues to increase at lower wavelengths. In such cases, quantitation using this protocol can lead to overestimation of complex concentration. Additionally, when using chromatin proteins with significant absorption at 260 nm, this protocol will need to be adjusted accordingly.
For samples with an expected concentration of 1–25 mg/ml, prepare 1:200 dilution of chromatin complex in 0.2 N NaOH: 497.5 μl 0.2 N NaOH + 2.5 μl sample.
Blank UV spectrophotometer with quartz cuvette containing 0.2 N NaOH
Record UV spectrum of chromatin complex in 0.2 N NaOH from 320 to 220 nm. Record A320 and A260 of sample.
-
Calculate concentration of chromatin complex. For example, for the PRC1 ub module/NCP complex in the size exclusion chromatography example, the molecular weight of the PRC1 ub module/NCP was 290,234 Da (2:1 complex), and the molecular weight of the 147 bp double stranded DNA in the complex was 97,020:
adjusted A260 of 1:200 dilution in 0.2 N NaOH = A260 – A320 = 0.564
A260 of stock = 200 × 0.564 = 112.8
concentration of DNA in stock = 112.8 × 40 μg/ml = 4.51 mg/ml
concentration of chromatin complex = 4.51 mg/ml × 290,234/97,020 = 13.5 mg/ml
The extinction coefficient was adjusted from that of DNA in neutral pH conditions based on empirical measurements. We have observed that this calculation can occasionally overestimate the chromatin complex concentration, so it is best to consider the calculated chromatin complex concentration as an estimate and not a firm measurement.
-
Calculate the purification yield of the chromatin complex based on the limiting reagent when reconstituting the chromatin complex (in this example, the NCP). For example:
started with 1.55 mg of NCP complex, equivalent to 7.59 nmol for the 204,921 Da NCP
ended with 100 μl of PRC1 ub module/NCP complex @ 13.5 mg/ml, or 4.65 nmol PRC1 ub module/NCP complex.
purification yield = 4.65/7.59 = 61.3% (yields are typically between 50 and 80%)
Use the chromatin complex immediately for crystallization trials, or store complex on ice in cold room, or where appropriate, add glycerol to 20%, flash freeze in liquid nitrogen and store at −80°C.
6. Crystallize chromatin complex
6.1 Crystallization
Standard crystallization approaches can be used to crystallize the chromatin complex. As with purification of chromatin proteins, the process of crystallizing a chromatin complex is highly variable and dependent on the properties of the individual complex. Nevertheless, basic principles and concepts will apply to the crystallization of many chromatin complexes, and we share such ideas here:
As with most any crystallization trial, one should survey a range of pH, salt and precipitating conditions. Since many chromatin complexes are sensitive to salt, it may be prudent to bias towards low salt conditions and near neutral pH. These conditions can be fulfilled by commercial low ionic strength crystallization screens and screens for protein-nucleic acid complexes (Hampton Natrix screens, Hollis lab Protein-Nucleic Acid Complex Crystal Screen, (Pryor, Wozniak, & Hollis, 2012)).
For initial crystallization screens, we use 96 well microbatch under oil trials using 1 μl of sample + 1 μl of the precipitating solution. Robotics can be helpful for setting up such trials or ones using even smaller volumes, but robotics are not necessary.
Divalent ions can play different roles in chromatin complex crystallization. Magnesium ions (Mg2+) are often used in protein/nucleic acid cocrystallization. However, Mg2+ is also used to condense chromatin (Fredericq, Hacha, Colson, & Houssier, 1991), and nucleosome core particles are crystallized at the salting in transition balancing Mn2+ as a precipitating agent and KCl as a solubilizing agent (Hanson, Alexander, Harp, & Bunick, 2004; Luger, Mäder, Richmond, Sargent, & Richmond, 1997a; Rhodes, Brown, & Klug, 1989). As such, use of millimolar concentrations of Mg2+ in the crystallization trials could favor precipitating the chromatin complex instead of controlled crystallization. It should be noted, however, that the Sir3 silencing protein was crystallized with nucleosome core particles in the presence of 10 mM MgCl2 (Armache et al., 2011).
As with crystallizing any protein, the use of orthologous proteins can be a fruitful strategy. In the case of the RCC1/nucleosome complex, initial crystals were grown using human RCC1 but the crystals were hollow and produced very poor diffraction. Substituting yeast RCC1 (Srm1) produced morphologically more perfect crystals, but nonisotropic diffraction of X-rays limited to about 7 Å in the worse directions. On the other hand, Drosophila RCC1 (Bj1) complexed with the nucleosome produced crystals with both external and internal order, permitting diffraction data collection to 2.9Å (Makde et al., 2010; Makde & Tan, 2013)
6.2 Fluorescent labeling of chromatin protein for cocrystallization with the nucleosome
One recurrent challenge we observe in the crystallization of chromatin complexes is the undesired crystallization of subcomplexes. This is especially true when working with chromatin proteins bound to the NCP, where the nucleosome particle often crystallizes without the chromatin protein bound. This can lead to much time wasted optimizing false leads to obtain crystals large enough to verify crystal contents by SDS-PAGE. To short circuit this problem and streamline crystal screening, we use fluorescently labeled chromatin proteins during screening. This allows us to verify the presence of the chromatin protein in any crystallization hits in situ, following procedures developed more generally for protein crystallization (Meyer, Betzel, & Pusey, 2015; Pusey et al., 2015).
We label our chromatin proteins through the N-α amino group using (5-(and-6)-Carboxyrhodamine 6G, succinimidyl ester using pH conditions that favor conjugation specifically at the N-terminus. This fluorophore is small and inexpensive and has stable spectrophotometric properties over a wide range of pHs often used in crystallization screens. The N-terminal labeling is convenient and unlikely to interfere with crystallization as terminal regions of proteins are often unstructured in macromolecular crystals. Of note, it is possible that some non-specific labeling of lysine side chain amines may occur in this pH-controlled reaction. However, at labeling percentages under 2% of the total chromatin protein pool, any resulting heterogeneity is minimal. Moreover, if undesired side chain modification of the chromatin protein prevents nucleosome binding, the improperly labeled protein is often purified away from the chromatin complex during size exclusion chromatography and at trace levels causes little to no change of the overall chromatin protein to nucleosome ratio. This methodology can also be applied to other fluorophores and conjugation chemistries.
An example of fluorescent labeling of a chromatin protein and incorporation into a chromatin complex is shown here.
Prepare your chromatin protein for labeling. In order to achieve efficient labeling of the chromatin protein, reactive nucleophiles (especially primary amines) that can consume the fluorophore must be removed or minimized. Buffer exchange can be performed by dialysis, size exclusion chromatography, or through a desalting column. Additionally, if the chromatin protein is sufficiently concentrated, dilution into labeling buffer may be adequate given the low levels of labeling required. In anticipation of labeling requirements, the chromatin protein can often be concentrated in a suitable buffer for labeling.
-
We typically label 2–5 mg of chromatin protein at 1–4 mg/ml. An example of labeling the PRC1 ubiquitylation module is shown here.
Dissolve (5-(and-6)-Carboxyrhodamine 6G, succinimidyl ester in DMF (dimethyl formamide) at 10 mM.
-
Decide on the quantity of chromatin protein to label and prepare labeling mixture
250 μl 235 μM PRC1 ub module prep 1 (2.5 mg) 738 μl labeling buffer (10 mM HEPES pH 7.5, 300 mM NaCl) 12 μl 10 mM carboxyrhodamine succinimidyl ester 1000 μl total volume (2.5 mg/ml chromatin protein) Mix at room temperature for 1 hour.
Purify the labeled chromatin protein. We prefer to use size exclusion chromatography as it further purifies the chromatin protein and adds an extra level of quality control, but a desalting column is a suitable alternative. We perform size exclusion chromatography in a buffer optimized for concentration of the chromatin protein after clarification of the labeling reaction mixture in a tabletop microcentrifuge. We note that two successive spins are often required to remove all particulate matter.
Concentrate the labeled chromatin protein as described above for the unlabeled protein.
-
Quantitate the labeled protein and labeling efficiency. Because the fluorophore absorbs at 280 nm, the A280,total for the labeled chromatin protein is the sum of the A280,protein and the A280,fluor. Therefore, the protein concentration and labeling percentage can be determined by subtracting A280,fluor. from the A280,total. This A280,fluor. can be calculated by measuring the absorbance of the fluorophore at its peak absorbance (A532,fluor. in this example) and 280 nm. In our typical chromatin protein buffers, the A532,fluor./A280,fluor. ratio for carboxyrhodamine is approximately 3. We refer to this ratio as the correction factor (CF). Importantly, most chromatin proteins have little to no absorption above 300 nm. Thus, by measuring the absorbance of the labeled protein at 280 nm and 532 nm, we can calculate the concentration of the protein and the fluorophore in our labeled protein samples. An example for the PRC1 ub module is shown here.
Determine the CF for carboxyrhodamine in chromatin protein concentration buffer = A532,fluor./A280,fluor.. For other fluorophores replace A532 with Amax. A280,fluor = A532,total./CF
-
Measure A280,total, A532,total and A600,total at appropriate dilution for labeled chromatin protein
adjusted A280,total of 1:21 dilution = A280,total – A600,total = 0.304
A280,total of stock = 21 × 0.304 = 6.38
adjusted A532,total of 1:21 dilution = A532,fluor. = A524,total – A600,total = 0.064
A532,total of stock = 21 × 0.064 = 1.34
[Chromatin protein] = (A280,total -A280,fluor)/εprotein = (6.38 – 1.34/3)/0.924 ml/mg = 6.42 mg/ml = 151 μM
Percent labeling = (A532,total/εfluor.)/[Chromatin protein] = 1.34/92,000 M−1/0.000151 M = 9.6%
-
Reconstitute the chromatin complex with the fluorescently labeled chromatin protein. We typically include 0.5–2% labeled chromatin protein in our reconstitutions. An example using PRC1 ub module/NCP is shown below.
Prepare 0.5% labeled PRC1 ub module
95.9 μl 234.7 μM unlabeled PRC1 ub module 10.2 μl 120.5 μM PRC1 ub module (9.6 % labeled) Prepare reconstitution mixture
69.9 μl MilliQ water 124 μl 61.9 μM NCP prep 201 7.5 μl 1 M Tris-Cl pH 7.6 2.2 μl 5 M NaCl 24.1 μl 0.5 % labeled PRC1 ub module obs: precipitation, then immediate clearing 24.1 μl 0.5 % labeled PRC1 ub module obs: precipitation, then immediate clearing 24.1 μl 0.5 % labeled PRC1 ub module obs: precipitation, then immediate clearing 24.1 μl 0.5 % labeled PRC1 ub module obs: precipitation, then immediate clearing 300 μl total volume The chromatin complex can be purified, concentrated, quantitated and used in crystallization screens as described above for the unlabeled complex. We record the A525 along with the A260 and A280 during size exclusion chromatography. This allows us to visualize the alignment of the chromatin protein (A525) and NCP (A260) independently to confirm saturation of the chromatin complex (Fig. 2c).
Observe fluorescence of crystals. We observed crystals grown with fluorescently labeled chromatin complexes using a Nikon SM7 1500 stereoscope equipped with a Nikon Intensilight C-HGFI light source, a DsRed filter (Ex 545/30 nm, DM 570 nm, BA 620/60 nm), and a CoolSnapEZ Turbo 1394 camera.
7. Post-crystallization soaks to improve diffraction
Post-crystallization soaks of chromatin complex crystals appear to be particularly important for obtaining high resolution diffraction. This is consistent with previous observations that the order within nucleosome core particle crystals could be improved by post-crystallization soaks in methylpentanediol (MPD) or other alcohols (T. J. Richmond, Finch, Rushton, Rhodes, & Klug, 1984; Struck, Klug, & Richmond, 1992). Therefore, it is highly advisable to spend significant efforts exploring post-crystallization soaking conditions once chromatin complex crystals have been grown, assuming of course that sufficient numbers of such crystals can be grown.
The underlying approach is to find a base soaking solution in which the crystal is stable, and then to gradually increase the concentration of the additive, usually a dehydration agent. We usually perform this procedure using loops to transfer crystals between drops of different compositions, but one can imagine other procedures using dialysis buttons or diffusing volatile reagents. One should also consider using dehydration equipment such as the HC-1 device developed at EMBL Grenoble which may provide a more reproducible and direct way of manipulating the hydration state of macromolecular crystals (M. W. Bowler et al., 2015; Sanchez-Weatherby et al., 2009; Wheeler, Russi, Bowler, & Bowler, 2012).
-
Determine a crystallization base soaking solution in which the crystal is stable. We usually start with the precipitant used in the relevant crystallization trial and increase the precipitant concentration by 1.25 fold (though older crystallization drops may need higher precipitant concentrations). So for example, if the precipitant contained 20 mM Tris-Cl pH 7.6, 50 mM NaAc and 6% PEG2000-MME, we would use 20 mM Tris-Cl pH 7.6, 50 mM NaAc and 7.5% PEG2000-MME as the trial crystallization base soaking solution.
Transfer a small crystal into the trial crystallization base soaking solution and observe the crystal under a microscope. If the crystal dissolves, it is likely that the precipitant concentration needs to be increased, although the pH or the salt concentration might have to be manipulated as well. If the crystal cracks, it is likely the precipitant concentration needs to be decreased. The trial crystallization base soaking solution is ideally one in which the crystals is stable for at least 15 min at the appropriate temperature. We usually perform soaking experiments at room temperature, but we did observe that soaking RCC1/nucleosome crystals at 4°C was important for obtaining high resolution diffraction (Makde & Tan, 2013).
-
Determine a soaking protocol that permits the additive to be soaked into the crystal without damaging the external morphology of the crystal.
First decide what soaking additive and what final concentration of this additive you wish to examine. Possible soaking additives are MPD, PEG400, PEG2000 and PEG550-MME. Then decide what size steps of the additive concentration to employ. For example, you might wish to soak MPD into the crystal to a final concentration of 24% in 4% steps. In this case, you would prepare the crystallization base soaking solution with 0, 4, 8, 12, 16, 20, 24% MPD.
Use a crystal loop to transfer a crystal into 10 μl of the crystallization base soaking solution with no added MPD. Incubate at room temperature for desired length of time (e.g. 5 min). Observe the crystal under a microscope for possible changes in external morphology.
Transfer crystal into 10 μl of the crystallization base soaking solution with 4% MPD. Incubate and observe as before.
Repeat step (c) until crystal is transferred to the final additive concentration (24% MPD in this example). If cracking or defects are observed during the process, continue with the procedure for at least two more additive additions. It is not uncommon to observe cracking or defects followed by annealing of these cracks or defects upon further soaks, reflecting repacking of the molecules in the crystal.
Where appropriate, flash cool the crystal by plunging into liquid nitrogen before storage in liquid nitrogen.
Notes
Unfortunately, there is no simple soaking protocol that will handle all cases. The appropriate protocol needs to be determined empirically. Potentially important variables include the identity of the soaking additive, the final concentration of the soaking additive, the number of steps used to reach the final concentration of the soaking additive, the method used to achieve such steps (e.g. transfer via loops), the soaking temperature and the soaking time per step.
Since the ultimate criteria of judging success of a soaking protocol is the quality of X-ray diffraction and since one will want to examine many soaking protocols, one approach is to process many crystals through the various soaking protocols and to then examine the diffraction in succession at a home or synchrotron source.
It is advisable to examine a variety of dehydration soaking agents including but not necessarily limited to MPD, PEG400, PEG2000 and PEG550-MME.
You can assess the maximum concentration for an additive by increasing the concentration of the additive until crystals show defects that do not anneal with further additions of the additive.
The soaking protocol can manipulate more than one additive, perhaps to introduce a cryoprotectant as well as a dehydrating agent. For example, you could increase both glycerol and PEG400 conditions simultaneously or in succession.
Make sure to allow enough time for the crystal to equilibrate against the soaking solution, particularly for the final soak. This may be particularly important for the reproducibility of the soaking protocol.
We have observed that post-crystallization protocols optimized for crystals harvested from fresh drops do not necessarily work with crystals harvested from older crystallization drops.
One may wish to decrease the incremental step of the dehydrating agent or additive in the soaking solutions to achieve more gradual dehydration. For example, we used 2% steps of PEG400 when dehydrating RCC1/nucleosome crystals (Makde et al., 2010; Makde & Tan, 2013).
Bibliography
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science (New York, NY) 2011;334(6058):977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaudo N, Fernandez IS, McLaughlin SH, Peak-Chew SY, Rhodes D, Martino F. The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle. Nature Structural & Molecular Biology. 2013;20(9):1119–1121. doi: 10.1038/nsmb.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science (New York, NY) 2006;311(5762):856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- Bowler MW, Mueller U, Weiss MS, Sanchez-Weatherby J, Sorensen TLM, Thunnissen MMGM, et al. Automation and Experience of Controlled Crystal Dehydration: Results from the European Synchrotron HC1 Collaboration. Crystal Growth & Design. 2015;15(3):1043–1054. [Google Scholar]
- Burgess RR, Deutscher MP, editors. Methods in Enzymoloogy vol. 463: Guide to Protein Purification. Vol. 463. Elsevier/Academic Press; 2009. pp. 1–851. [Google Scholar]
- Coligan John E, Dunn Ben M, Speicher David W, Wingfield Paul T., editors. Current protocols in protein science. [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. Journal of Molecular Biology. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- Fredericq E, Hacha R, Colson P, Houssier C. Condensation and precipitation of chromatin by multivalent cations. Journal of Biomolecular Structure & Dynamics. 1991;8(4):847–865. doi: 10.1080/07391102.1991.10507849. [DOI] [PubMed] [Google Scholar]
- Hanson BL, Alexander C, Harp JM, Bunick GJ. Preparation and crystallization of nucleosome core particle. Methods in Enzymology. 2004;375:44–62. doi: 10.1016/s0076-6879(03)75003-4. [DOI] [PubMed] [Google Scholar]
- Hieb AR, D’Arcy S, Kramer MA, White AE, Luger K. Fluorescence strategies for high-throughput quantification of protein interactions. Nucleic Acids Research. 2012;40(5):e33. doi: 10.1093/nar/gkr1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, et al. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science (New York, NY) 2013;340(6136):1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-A, Chatterjee N, Jennings MJ, Bartholomew B, Tan S. Extranucleosomal DNA enhances the activity of the LSD1/CoREST histone demethylase complex. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of Molecular Biology. 1998;276(1):19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997a;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods in Molecular Biology (Clifton, NJ) 1999a;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods in Enzymology. 1999b;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. Journal of Molecular Biology. 1997b;272(3):301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- Makde RD, Tan S. Strategies for crystallizing a chromatin protein in complex with the nucleosome core particle. Analytical Biochemistry. 2013;442:138–145. doi: 10.1016/j.ab.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. 2010;467(7315):562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, et al. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523(7560):366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. 2014;514(7524):591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Makde RD, Tan S. Recognition of the nucleosome by chromatin factors and enzymes. Current Opinion in Structural Biology. 2016:37. doi: 10.1016/j.sbi.2015.11.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Betzel C, Pusey M. Latest methods of fluorescence-based protein crystal identification. Acta Crystallographica. Section F, Structural Biology Communications. 2015;71(Pt 2):121–131. doi: 10.1107/S2053230X15000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor EE, Wozniak DJ, Hollis T. Crystallization of Pseudomonas aeruginosa AmrZ protein: development of a comprehensive method for obtaining and optimization of protein-DNA crystals. Acta Crystallographica Section F, Structural Biology and Crystallization Communications. 2012;68(Pt 8):985–993. doi: 10.1107/S1744309112025316. http://doi.org/10.1107/S1744309112025316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusey M, Barcena J, Morris M, Singhal A, Yuan Q, Ng J. Trace fluorescent labeling for protein crystallization. Acta Crystallographica. Section F, Structural Biology Communications. 2015;71(Pt 7):806–814. doi: 10.1107/S2053230X15008626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Brown RS, Klug A. Crystallization of nucleosome core particles. Methods in Enzymology. 1989;170:420–428. doi: 10.1016/0076-6879(89)70060-4. [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. 2003;423(6936):145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 A resolution. 1984;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Sanchez-Weatherby J, Bowler MW, Huet J, Gobbo A, Felisaz F, Lavault B, et al. Improving diffraction by humidity control: a novel device compatible with X-ray beamlines. Acta Crystallographica Section D Biological Crystallography. 2009;65(Pt 12):1237–1246. doi: 10.1107/S0907444909037822. [DOI] [PubMed] [Google Scholar]
- Struck MM, Klug A, Richmond TJ. Comparison of X-ray structures of the nucleosome core particle in two different hydration states. Journal of Molecular Biology. 1992;224(1):253–264. doi: 10.1016/0022-2836(92)90588-b. [DOI] [PubMed] [Google Scholar]
- Thåström A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. Journal of Molecular Biology. 1999;288(2):213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- Wheeler MJ, Russi S, Bowler MG, Bowler MW. Measurement of the equilibrium relative humidity for common precipitant concentrations: facilitating controlled dehydration experiments. Acta Crystallographica Section F, Structural Biology and Crystallization Communications. 2012;68(Pt 1):111–114. doi: 10.1107/S1744309111054029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler DD, Luger K, Hieb AR. Quantifying Chromatin-Associated Interactions: The HI-FI System. Methods in Enzymology. 2012;512:243–274. doi: 10.1016/B978-0-12-391940-3.00011-1. [DOI] [PubMed] [Google Scholar]
- Zhou B-R, Jiang J, Feng H, Ghirlando R, Xiao TS, Bai Y. Structural Mechanisms of Nucleosome Recognition by Linker Histones. Molecular Cell. 2015 doi: 10.1016/j.molcel.2015.06.025. http://doi.org/10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed]