Abstract
Chloroquine (CQ) and amodiaquine (AQ) have been used for treating or preventing malaria for decades, and their application has expanded into treating inflammatory disease in humans. CQ and AQ are applicable for controlling rheumatoid arthritis, but their molecular mechanisms of anti-inflammatory activity remain to be elucidated. In this study, we examined the effects of CQ and AQ on T cell activation and T cell-mediated immune response. CQ had no significant effect on T cell numbers, but decreased the population of T cells with a high division rate. However, AQ treatment significantly increased the number of cells with low division rates and eliminated cells with high division rates, resulting in the inhibition of T cell proliferation triggered by T cell receptor stimulation, of which inhibition occurred in developing effector T helper and regulatory T cells, regardless of the different exogenous cytokines. Interestingly, the cyclin-dependent kinase inhibitor p21 was significantly and dose-dependently increased by CQ, and more potently by AQ, while other cell cycle regulators were unchanged. Both CQ and AQ elevated the transcription level of p21 though the activation of p53, but also blocked p21 protein degradation in the presence of cycloheximide, causing p21 protein accumulation mainly in the nucleus. Sustained treatment of developing T cells with either CQ or AQ suppressed IFN-γ production in a dose dependent manner and potently inhibited the differentiation of IFN-γ-producing Th1 cells. These results demonstrate that CQ and AQ increase the expression level of p21 and inhibit T cell proliferation and the development of IFN-γ-producing Th1 cells, thereby revealing beneficial roles in treating a wide range of chronic inflammatory diseases mediated by inflammatory T cells.
Keywords: Amodiaquine, Chloroquine, p21, Proliferation, IFN-γ
1. Introduction
Malaria is a life-threatening infectious disease caused by parasites, mainly Plasmodium falciparum and Plasmodium vivax [1]. Chloroquine (CQ) is one of the antimalarial agents and this is extensively used in the treatment or prevention of malaria from P. vivax, Plasmodium ovale, and Plasmodium malariae [2,3]. The anti-malarial activity of CQ is explained by the inhibition of heme degradation in lysosomes, which occurs in red blood cells infected with malaria parasites [4–6]. The appearance of a CQ-resistant strain, P. falciparum sparked the development of more potent and efficient antimalarial drugs, which produced amodiaquine (AQ) by modifying the core structure of CQ and for treating CQ-resistant P. falciparum malaria infections [7,8]. In addition, CQ and AQ have beneficial roles in controlling cancer through the activation of p53 and the inhibition of autophagy [9–12], as well as chronic inflammatory diseases such rheumatoid arthritis, colitis, and multiple sclerosis via its immune suppression activity [13–18]. However, the detailed molecular mechanisms underlying the immunesuppressive effects of CQ and AQ have yet to be characterized.
CD4+ T helper (Th) cells play key roles in the regulation of immune responses against pathogenic infections including bacteria, viruses, and malaria parasites [19,20] by differentiating into effector T helper (Th) cells such as Th1, Th2 and Th17 cells [21]. In particular, dysregulated overproduction of IFN-γ by Th1 cells is responsible for the development of inflammatory diseases. As IFN-γ deficiency inhibits the development of bleomycin-induced lung inflammation and DSS-induced colitis [22,23], suppression of pro-inflammatory IFN-γ may be crucial for treating inflammatory diseases.
In this study, we have investigated whether anti-malarial agents CQ and AQ could affect the proliferation and differentiation of CD4+ T cells. Our results indicate that CQ and AQ suppresses T cell proliferation by increasing the p21 expression level, which is controlled at the level of transcriptional activation and post-translational protein stabilization, and thus inhibits the production of pro-inflammatory IFN-γ by Th1 cells.
2. Materials and methods
2.1. Materials and mice
Chloroquine (CQ, MW = 515.86) and amodiaquine (AQ, MW = 464.81) were purchased from Sigma–Aldrich (St. Louis, MO). Chloroquine was dissolved in PBS and amodiaquine was dissolved in DMSO. All cytokines were purchased from BD Biosciences (San Diego, CA). C57BL6 mice were housed under specific pathogen-free condition and sacrificed for primary T cell culture in accordance with IACUC guidelines at Ewha Womans University (IACUC No. 2012-01-071, 2014-01-011).
2.2. Primary CD4+ T cell culture and proliferation assay
CD4+ T cells were isolated from lymph node and spleen using CD4 mini MACS beads (Miltenyi Biotec, Auburn, CA) and stimulated with plate-bound anti-CD3/anti-CD28 Ab (2 μg/ml, BD Biosciences) in the presence of recombinant human IL-2 (rhIL-2, 10 U/mL) for 24 h. Cells were additionally treated with either CQ or AQ for an additional 24 h and 48 h. For cytotoxic assay, cells were incubated with water soluble tetrazolium salt reagent in EZ-CYTOX assay kit (DOGEN, Seoul, Korea) for 3 h and subjected to measure the absorbance at 450 nm. For proliferation assay, cells were labeled with 5 μM carboxyfluorescein diacetate, succinimidyl ester (CFSE, Sigma–Aldrich) before TCR stimulation, followed by flow cytometry. In addition, IL-12 (2 ng/mL), IL-4 (10 ng/mL), TGF-β (5 ng/ml) and IL-6 (10 ng/ml), or TGF-β (5 ng/ml) was added to the activated cells.
2.3. Cytokine measurement
Culture supernatants were harvested at 48 h for cytokine measurement using ELISA. EIA plate was coated with IFN-γ antibody (BD Biosciences) and incubated with culture supernatants, followed by incubation with biotinylated anti–IFN–γ antibody and measurement using an ELISA reader (Molecular Devices, Sunnyvale, CA). For intracellular cytokine staining, developing CD4+ T cells were stimulated with anti-CD3 antibody and incubated with monensin (4 μM) for 4 h before harvest. Cells were incubated with PE-conjugated IFN-γ (2 μg/ml, BD Biosciences) and then analyzed by FACS Calibur and a CellQuest software (BD Biosciences).
2.4. Real-time PCR
Cells were harvested at 48 h after TCR stimulation and subjected to total RNA preparation using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA was synthesized from mRNA using reverse transcriptase (Promega, Madison, WI) and used for quantitative real time PCR analysis (Applied Biosystems Inc., Foster City, CA). Specific primers were as follows; p21-FWD 5′-cgagaacggtggaactttgac-3′ and p21-REV 5′-tcccagacgaagttgccct-3′; IFN-γ-FWD 5′-agcaa-cagcaaggcgaaaa-3′ and IFN-γ-REV 5′-ctggacctgtgggttgttga-3′; actin-FWD 5′-agagggaaatcgtgcgtgac-3′ and actin-REV 5′-caa-tagtgatgacctggccgt-3′. Relative expression level was calculated after normalization to the level of actin.
2.5. Immunoblotting
Whole cell extracts were prepared from either CQ- or AQ-treated cells and resolved by SDS-PAGE. Expression of cell cycle-and autophagy-related proteins was analyzed using antibodies against p21, p27, PCNA, p53, LC3-I, LC3-II, ATG5, and actin (Santa Cruz Biotech, Santa Cruz, CA)
2.6. Statistical analysis
All experiments were performed at least three times. A set of data is given as mean ± SD. The statistical significance of results was calculated using a one-way analysis of variance (ANOVA) and was set at p < 0.05. *, p < 0.05; **, p < 0.005; ***, p < 0.0005.
3. Results
3.1. Attenuation of T cell division by CQ and AQ
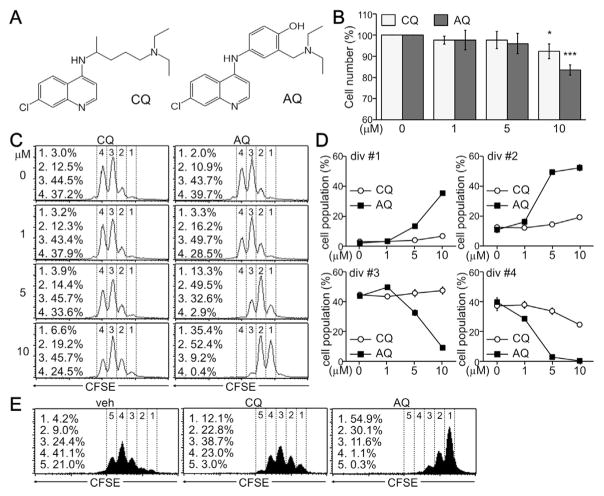
In order to determine the immunosuppressive effect of CQ and AQ, we treated TCR-triggered CD4+ T cells with either CQ or AQ for 24 h and analyzed its effect on CD4+ T cell activation and proliferation. CQ and AQ were similar in chemical structure to 4-amino-7-chloroquine, except for the side chain linked to an amine group (Fig. 1A). Although CQ and AQ had no significant cytotoxic effect, the number of CD4+ T cells was decreased by treatment with 10 μM CQ, and was more potently decreased by AQ at the same concentration (Fig. 1B). Further analysis using a CFSE-based assay confirmed that 10 μM CQ attenuated cell division rates. Interestingly, the number of developing T cells in divisions 1 and 2 was drastically elevated by AQ, whereas cell populations in divisions 3 and 4 were decreased by treatment with AQ (Fig. 1C). Quantitative analysis demonstrated that AQ increased the population of cells with low division rates in a dose-dependent manner and also decreased the number of cells with high division rates (Fig. 1D). Sustained treatment with CQ decreased cell populations at divisions 4 and 5, and AQ treatment more drastically inhibited the cell cycle progression of developing T cells (Fig. 1E).
Fig. 1.
Suppression of T cell division by CQ and AQ. (A) Structure of CQ and AQ. (B) CD4+ T cells were activated with anti-TCR antibodies for 24 h and treated with CQ and AQ for an additional 24 h. Cell viability was assayed using an EZ-CYTOX assay kit. Data are expressed as the mean ± SD of three independent experiments. *, p < 0.05; ***, p < 0.0005. (C) CD4+ T cells were labeled with CFSE and treated with CQ or AQ for 24 h under non-skewing conditions. Cells were assayed at day 2 after TCR stimulation by flow cytometry analysis. (D) The cell population at divisions #1, #2, #3 and #4 was quantitatively determined using the CellQuest software. Data are expressed as the mean ± SD of four independent experiments. (E) CFSE-labeled cells were treated with CQ and AQ for 48 h and subjected to flow cytometry analysis.
3.2. Anti-proliferative activity of CQ and AQ in effector Th and Treg cells
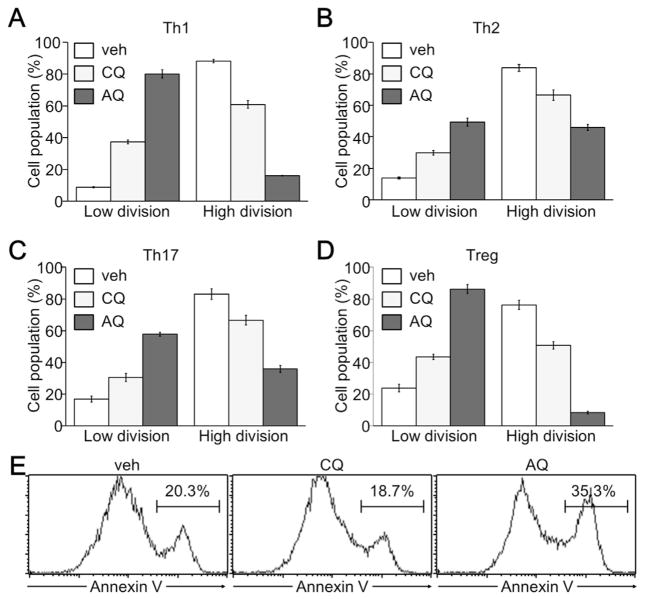
Since the suppression of Th cell proliferation by CQ and AQ was observed under non-skewing conditions, we thus examined the anti-proliferative effect of CQ or AQ on developing Th1, Th2, Th17, and Treg cells in response to different cytokines. The addition of IL-12 and IL-4 induced highly proliferative Th1 and Th2 cell development respectively. Strong cell division in developing Th1 and Th2 cells was inhibited by treatment with CQ, and more drastically by AQ (Fig. 2A and B). Furthermore, TGF-β with IL-6 or TGF-β alone promoted activated CD4+ T cells to differentiate into Th17 or Treg cells. Under Th17 and Treg-skewing conditions, CQ and AQ also increased the population of cells with lower division rates, but decreased the population of cells with higher division rates (Fig. 2C and D). AQ was more potent than CQ in the suppression of cell proliferation of developing Th1, Th2, Th17, and Treg cells. AQ treatment also resulted in an increase in apoptotic cell death (Fig. 2E).
Fig. 2.
Anti-proliferative activity of CQ and AQ in developing effector Th and Treg cells. CD4+ T cells were isolated using MiniMACS CD4 microbeads and stimulated with anti-CD3 (2 μg/mL) and anti-CD28 (2 μg/mL) antibodies for 24 h. Cells were then treated with either 10 μM CQ or 10 μM AQ for 48 h in the presence of IL-12 (2 ng/ml) and anti-IL-4 (5 μg/mL) for Th1 (A), IL-4 (10 ng/ml) and anti–IFN–γ (5 μg/mL) for Th2 (B), TGF-β (5 ng/ml), IL-6 (20 ng/ml), anti–IFN–γ (5 μg/mL), and anti-IL-4 (5 μg/mL) for Th17 (C), or TGF-β (10 ng/ml), anti–IFN–γ (5 μg/mL), and anti-IL-4 (5 μg/mL) for Treg cells (D). The cell population at low divisions (division 1 through 3) and high divisions (division 4 through 6) was determined by flow cytometry and the CellQuest software. Data are expressed as the mean ± SD of three independent experiments. (E) Developing Th1 cells in the presence of either 10 μM CQ or 10 μM AQ were subjected to annexin V staining and flow cytometry analysis. Annexin V-positive apoptotic cell populations were determined and a representative image is shown from three independent experiments.
3.3. Induction of nuclear p21 protein by CQ and AQ
We then investigated the molecular mechanisms underlying the anti-proliferative functions of CQ and AQ. We first checked the expression level of cell cycle regulators including p21, p27, and PCNA. While p27 and PCNA were not changed at all by treatment with CQ or AQ, the p21 expression level was significantly increased by AQ and moderately increased by CQ, as evidenced by the densitometry data for band intensities in different sets (Fig. 3A and B). We next examined whether p21 induction was mediated by the inhibition of autophagy-induced protein degradation after treatment with CQ or AQ [24–26]. As previously reported [27,28], CQ treatment increased the level of the autophagosome membrane-bound form of LC3B. AQ also strongly increased the level of LC3B degradation via autophagy but had no effect on ATG5 expression (Fig. 3A). In addition, an increased expression of p21 was mostly observed in the nucleus of developing T cells, not in the lysosomal compartment, which was evidenced by the staining of lysosome associated membrane protein-1 (LAMP-1) (Fig. 3C). CQ and AQ sustained the p21 protein level in the presence of a protein synthesis inhibitor, cycloheximide (Fig. 3D), indicating p21 protein stabilization by CQ and AQ. We have also found that p21 expression was remarkably increased at the transcriptional level after treatment with CQ or AQ (Fig. 3E). The p53-induced p21 promoter activity was significantly enhanced by treatment with CQ and AQ, although the expression level of p53, a transcriptional activator of p21, was unchanged (Fig. 3A and F). These results suggest that CQ and AQ increased p21 expression at both the transcriptional and post-translational level.
Fig. 3.
Induction of p21 protein expression by CQ and more prominently by AQ. (A–B) TCR-triggered CD4+ T cells were incubated with different amounts of CQ or AQ for 24 h. Whole cell extracts were resolved by SDS-PAGE and subjected to immunoblotting with antibody against p21, p27, PCNA, p53, LC3B, ATG5, or actin (A). The intensity of the p21 protein band was quantitatively analyzed by a densitometry scan (B). (C) CQ- or AQ-treated T cells were fixed in 4% paraformaldehyde and stained with antibody against p21 and LAMP1, followed by subsequent incubation with Alexa Fluor 594- and Alexa Fluor 488-conjugated secondary antibody. The nuclei were blue counterstained with DAPI. (D) Activated CD4+ T cells were treated with cycloheximide (+CHX) in the presence of either CQ or AQ and harvested for immunoblotting analysis. (E) Total RNA was obtained from CQ- or AQ-treated T cells and subjected to reverse transcription and real time-PCR analysis. The relative level of p21 mRNA was calculated after normalization to the actin level. *, p < 0.05; **, p < 0.005; ***, p < 0.0005. Three independent experiments were performed and data are given as the mean ± SD. (F) Highly transfectable 293T cells were transfected with the p21 promoter reporter gene with or without p53 expression vector and additionally treated with CQ (50 μM) or AQ (50 μM) for 24 h. Reporter activity was measured using luciferase assay kit and the relative luciferase unit was calculated after normalization. The activity under vehicle-treated conditions was set to 1. *, p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. CQ or AQ suppressed IFN-γ production and Th1 cell differentiation
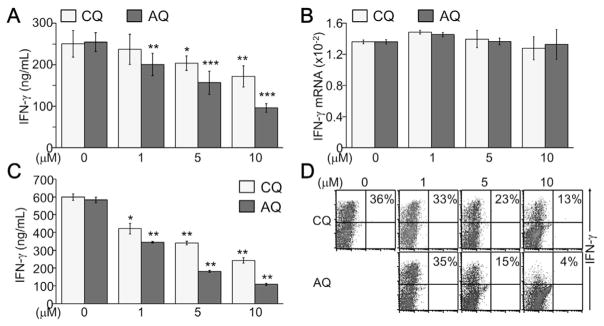
We lastly analyzed the effect of CQ and AQ on inflammatory cytokine IFN-γ production by developing T cells. IFN-γ production was dose-dependently suppressed by both CQ and AQ, even at concentrations lower than 10 μM (Fig. 4A). However, the mRNA level of the IFN-γ gene was not changed by treatment with either CQ or AQ (Fig. 4B). Sustained treatment with CQ and AQ substantially decreased IFN-γ production by differentiated T cells (Fig. 4C). Under Th1-skewing conditions, CQ and AQ suppressed IFN-γ-producing Th1 cell development as evidenced by analysis of intracellular cytokine staining of IFN-γ. Both CQ and AQ dose-dependently suppressed Th1 cell differentiation, but AQ was more likely to suppress it than CQ (Fig. 4D), implying an anti-inflammatory activity of CQ and AQ.
Fig. 4.
Inhibition of IFN-γ production and Th1 cell development by CQ and AQ. (A) Activated CD4+ T cells were treated with CQ or AQ under non-skewing conditions. Cell supernatants were collected at day 2 and subjected to ELISA for determining IFN-γ production. *, p < 0.05; **, p < 0.005; ***, p < 0.0005. (B) Total RNA was harvested from the cells and subjected to quantitative real-time PCR analysis. (C) CD3+ T cells were induced to differentiate into Th1 cells and incubated with CQ or AQ for an additional 4 days. Cells were re-stimulated with an anti-CD3 antibody (1 μg/ml) for 24 h and cell supernatants were collected for measuring IFN-γ by ELISA. Data are given as the mean ± SD of at least four independent experiments. *, p < 0.05; **, p < 0.005. (D) Differentiated Th1 cells in the presence of CQ or AQ were re-stimulated with PMA and ionomycin for 6 h and incubated with PE-conjugated anti–IFN–γ antibody, followed by flow cytometry analysis. Representative images are shown from four independent experiments.
4. Discussion
Our results demonstrated that CQ and AQ suppressed T cell proliferation through the induction of cyclin-dependent kinase inhibitor p21, which was controlled at the transcriptional level by CQ- and AQ-induced p53 activation and also at the post-translational level by CQ- and AQ-mediated inhibition of protein degradation. Suppression of T cell proliferation resulted in a significant IFN-γ reduction and an inhibition of Th1 cell differentiation. These results support that CQ and AQ play beneficial roles in suppressing chronic inflammatory and autoimmune disease [13–18], as well as identified their molecular mechanisms of p21-dependent inhibition of inflammatory Th1 cell development.
AQ was more potent than CQ in the induction of p21 expression and in the suppression of cell proliferation. While CQ substitutes 2-amino-5-diethylaminopentate in the 4-amine position of 4-aminoquinoline, AQ contains 2-[(diethylamino)methyl]phenol; this substitution reveals stronger anti-inflammatory functions, which provides an informative basis for the further development of promising anti-inflammatory drugs. Although CQ and AQ activate the p53-induced transcription of p21, it still needs to be clarified how these small molecules stimulate p53 activity and whether they directly bind to the p53 protein. Very recently, AQ has been shown to function as a direct agonist of nuclear receptor 4 A 1 (NR4A1, also referred to as Nurr1) and suppress macrophage activation, thereby suppressing Parkinson’s disease [29]. It would be interesting to identify the proteins that directly interact with CQ or AQ and control the inflammatory disease. In addition, CQ and AQ maintain p21 protein level in the presence of cycloheximide, since CQ and AQ function as inhibitors of a lysosomal degradation pathway that controls the quality of the cytoplasm by eliminating protein aggregates and damaged organelles [30].
Suppression of T cell proliferation by CQ and AQ resulted in the suppression of IFN-γ production by T cells and the inhibition of Th1 cell differentiation in vitro. T cell proliferation is essential for their development into effector Th cell, as well as Treg cells, and CQ and AQ suppress the early proliferation of CD4+ T cells, suggesting the potential roles of CQ and AQ in the regulation of effector Th- and Treg-mediated immune responses. Although this study reveals immune suppression by CQ and AQ treatment through the inhibition of Th1 cell development restraining cell proliferation, more detailed analysis of CQ and AQ on immune modulation is required for the proper application of CQ and AQ in human diseases [16].
Acknowledgments
This work was supported by Mid-career Researcher Program through NRF grant funded by the MEST (NRF-2013R1A2A2A01068302 for ESH and NRF-2015R1D1A1A01056936 for WHY).
Footnotes
Conflict of interest
We have no financial conflict of interest.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2016.04.105.
References
- 1.Buffet PA, Safeukui I, Deplaine G, Brousse V, Prendki V, Thellier M, Turner GD, Mercereau-Puijalon O. The pathogenesis of Plasmodium falciparum malaria in humans: insights from splenic physiology. Blood. 2011;117:381–392. doi: 10.1182/blood-2010-04-202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achan J, Talisuna AO, Erhart A, Yeka A, Tibenderana JK, Baliraine FN, Rosenthal PJ, D’Alessandro U. Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malar J. 2011;10:144. doi: 10.1186/1475-2875-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mushtaque M, Shahjahan Reemergence of chloroquine (CQ) analogs as multi-targeting antimalarial agents: a review. Eur J Med Chem. 2015;90:280–295. doi: 10.1016/j.ejmech.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 4.Pandey AV, Bisht H, Babbarwal VK, Srivastava J, Pandey KC, Chauhan VS. Mechanism of malarial haem detoxification inhibition by chloroquine. Biochem J. 2001;355:333–338. doi: 10.1042/0264-6021:3550333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguiar AC, de Santos RM, Figueiredo FJ, Cortopassi WA, Pimentel AS, Franca TC, Meneghetti MR, Krettli AU. Antimalarial activity and mechanisms of action of two novel 4-aminoquinolines against chloroquine-resistant parasites. PLoS One. 2012;7:e37259. doi: 10.1371/journal.pone.0037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olafson KN, Ketchum MA, Rimer JD, Vekilov PG. Mechanisms of hematin crystallization and inhibition by the antimalarial drug chloroquine. Proc Natl Acad Sci U S A. 2015;112:4946–4951. doi: 10.1073/pnas.1501023112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olliaro P, Mussano P. Amodiaquine for treating malaria. Cochrane Database Syst Rev. 2003:CD000016. doi: 10.1002/14651858.CD000016. [DOI] [PubMed] [Google Scholar]
- 8.Andrews KT, Fisher G, Skinner-Adams TS. Drug repurposing and human parasitic protozoan diseases. Int J Parasitol Drugs Drug Resist. 2014;4:95–111. doi: 10.1016/j.ijpddr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim EL, Wustenberg R, Rubsam A, Schmitz-Salue C, Warnecke G, Bucker EM, Pettkus N, Speidel D, Rohde V, Schulz-Schaeffer W, Deppert W, Giese A. Chloroquine activates the p53 pathway and induces apoptosis in human glioma cells. Neuro Oncol. 2010;12:389–400. doi: 10.1093/neuonc/nop046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon YH, Cho KS, Hwang JJ, Lee SJ, Choi JA, Koh JY. Induction of lysosomal dilatation, arrested autophagy, and cell death by chloroquine in cultured ARPE-19 cells. Invest Ophthalmol Vis Sci. 2010;51:6030–6037. doi: 10.1167/iovs.10-5278. [DOI] [PubMed] [Google Scholar]
- 11.Qiao S, Tao S, Rojo de la Vega M, Park SL, Vonderfecht AA, Jacobs SL, Zhang DD, Wondrak GT. The antimalarial amodiaquine causes autophagic-lysosomal and proliferative blockade sensitizing human melanoma cells to starvation- and chemotherapy-induced cell death. Autophagy. 2013;9:2087–2102. doi: 10.4161/auto.26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang MC, Wu MY, Hwang MH, Chang YT, Huang HJ, Lin AM, Yang JC. Chloroquine enhances gefitinib cytotoxicity in gefitinib-resistant nonsmall cell lung cancer cells. PLoS One. 2015;10:e0119135. doi: 10.1371/journal.pone.0119135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landewe RB, Miltenburg AM, Breedveld FC, Daha MR, Dijkmans BA. Cyclosporine and chloroquine synergistically inhibit the interferon-gamma production by CD4 positive and CD8 positive synovial T cell clones derived from a patient with rheumatoid arthritis. J Rheumatol. 1992;19:1353–1357. [PubMed] [Google Scholar]
- 14.Landewe RB, Miltenburg AM, Verdonk MJ, Verweij CL, Breedveld FC, Daha MR, Dijkmans BA. Chloroquine inhibits T cell proliferation by interfering with IL-2 production and responsiveness. Clin Exp Immunol. 1995;102:144–151. doi: 10.1111/j.1365-2249.1995.tb06648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimura M, Hidaka N, Akaza T, Tadokoro K, Juji T. Immunosuppressive effects of chloroquine: potential effectiveness for treatment of post-transfusion graft-versus-host disease. Transfus Med. 1998;8:209–214. doi: 10.1046/j.1365-3148.1998.00160.x. [DOI] [PubMed] [Google Scholar]
- 16.Hugosson E, Bjorkman A, Troye-Blomberg M. Chloroquine enhances the number of IL-10 producing cells and the expression of B7-2 and ICAM-1 in in vitro-cultured PBMC. Scand J Immunol. 2002;55:399–408. doi: 10.1046/j.1365-3083.2002.01051.x. [DOI] [PubMed] [Google Scholar]
- 17.He Y, Xu Y, Zhang C, Gao X, Dykema KJ, Martin KR, Ke J, Hudson EA, Khoo SK, Resau JH, Alberts AS, MacKeigan JP, Furge KA, Xu HE. Identification of a lysosomal pathway that modulates glucocorticoid signaling and the inflammatory response. Sci Signal. 2011;4:ra44. doi: 10.1126/scisignal.2001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagar J, Ranade S, Kamath V, Singh S, Karunanithi P, Subramani S, Venkatesh K, Srivastava R, Dudhgaonkar S, Vikramadithyan RK. Therapeutic potential of chloroquine in a murine model of inflammatory bowel disease. Int Immunopharmacol. 2014;21:328–335. doi: 10.1016/j.intimp.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Perlmann P, Troye-Blomberg M. Malaria and the immune system in humans. Chem Immunol. 2002;80:229–242. doi: 10.1159/000058846. [DOI] [PubMed] [Google Scholar]
- 20.Haskins K, Cooke A. CD4 T cells and their antigens in the pathogenesis of autoimmune diabetes. Curr Opin Immunol. 2011;23:739–745. doi: 10.1016/j.coi.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen ES, Greenlee BM, Wills-Karp M, Moller DR. Attenuation of lung inflammation and fibrosis in interferon-gamma-deficient mice after intra-tracheal bleomycin. Am J Respir Cell Mol Biol. 2001;24:545–555. doi: 10.1165/ajrcmb.24.5.4064. [DOI] [PubMed] [Google Scholar]
- 23.Ito R, Shin-Ya M, Kishida T, Urano A, Takada R, Sakagami J, Imanishi J, Kita M, Ueda Y, Iwakura Y, Kataoka K, Okanoue T, Mazda O. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin Exp Immunol. 2006;146:330–338. doi: 10.1111/j.1365-2249.2006.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan ZW, Hou JK, He W, Fan L, Huang Y. Chloroquine enhances cobalt chloride-induced leukemic cell differentiation via the suppression of autophagy at the late phase. Biochem Biophys Res Commun. 2013;430:926–932. doi: 10.1016/j.bbrc.2012.12.052. [DOI] [PubMed] [Google Scholar]
- 25.Liang X, Tang J, Liang Y, Jin R, Cai X. Suppression of autophagy by chloroquine sensitizes 5-fluorouracil-mediated cell death in gallbladder carcinoma cells. Cell Biosci. 2014;4:10. doi: 10.1186/2045-3701-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seo BR, Lee SJ, Cho KS, Yoon YH, Koh JY. The zinc ionophore clioquinol reverses autophagy arrest in chloroquine-treated ARPE-19 cells and in APP/ mutant presenilin-1-transfected Chinese hamster ovary cells. Neurobiol Aging. 2015;36:3228–3238. doi: 10.1016/j.neurobiolaging.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Kimura N, Kumamoto T, Kawamura Y, Himeno T, Nakamura KI, Ueyama H, Arakawa R. Expression of autophagy-associated genes in skeletal muscle: an experimental model of chloroquine-induced myopathy. Pathobiology. 2007;74:169–176. doi: 10.1159/000103376. [DOI] [PubMed] [Google Scholar]
- 28.Modernelli A, Naponelli V, Giovanna Troglio M, Bonacini M, Ramazzina I, Bettuzzi S, Rizzi F. EGCG antagonizes Bortezomib cytotoxicity in prostate cancer cells by an autophagic mechanism. Sci Rep. 2015;5:15270. doi: 10.1038/srep15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim CH, Han BS, Moon J, Kim DJ, Shin J, Rajan S, Nguyen QT, Sohn M, Kim WG, Han M, Jeong I, Kim KS, Lee EH, Tu Y, Naffin-Olivos JL, Park CH, Ringe D, Yoon HS, Petsko GA. Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson’s disease. Proc Natl Acad Sci U S A. 2015;112:8756–8761. doi: 10.1073/pnas.1509742112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]