Abstract
Blood biopsy has many advantages over tissue biopsy for diagnosing acquired T790M mutation in patients with non-small-cell lung cancer, such as being less risky and painful. New techniques with high sensitivity (eg, droplet digital PCR) show promising results during blood biopsy, but the positive rates of identification are still quite unclear. Whether there are other factors, except technology, affecting the results of blood biopsy is unclear. In this study, we used conventional amplification refractory mutation system to detect tumor tissue or blood for T790M mutation in patients clinically resistant to tyrosine kinase inhibitors. A total of 45 patients treated at West China Hospital between 2014 and 2016 were analyzed. The positive rate of T790M mutation was 70.8% based on tissue biopsy and 37.5% based on blood biopsy. Of the 24 patients whose epidermal growth factor receptor gene was genotyped through tissue and blood biopsy, 10 (41.7%) were concordant for T790M mutation status (κ=0.006). Of the 17 patients positive for T790M by tissue biopsy, 7 (41.2%) were positive for T790M by blood biopsy, and 3 of these 7 were only weakly positive. Of the 7 patients negative for T790M by tissue biopsy, 2 (28.6%) were positive by blood biopsy. Our T790M detection rate is higher than that reported by other studies using digital droplet PCR. These results suggest that other factors (eg, clinical features), intrinsically connected with circulating tumor DNA level, also affect the results of blood biopsy, and thus cannot be controlled through technological optimization.
Keywords: non-small-cell lung cancer, T790M mutation, re-biopsy, liquid biopsy
Introduction
Certain EGFR mutations in patients with non-small-cell lung cancer (NSCLC) are associated with acquired resistance to first-line epidermal growth factor receptor tyrosine kinase inhibitors (EGFR TKIs). The most common resistance-conferring mutation is T790M. Patients with this mutation often respond to the EGFR TKI known as AZD9291, so the mutation must first be confirmed through re-biopsy.1–3 This normally involves re-biopsy of the tumor tissue for subsequent sequencing, but biopsy is associated with safety risks and pain.
Blood biopsy has been proposed as a less risky and painful alternative. Blood contains circulating tumor DNA (ctDNA) within tumor cells as well as circulating free DNA.4 These DNA can then be sequenced. New techniques with higher sensitivity (eg, droplet digital PCR [ddPCR]), compared with conventional amplification refractory mutation system (ARMS), show promising results while testing ctDNA, but the positive rates are still quite varied and unclear.5–9 This forces blood biopsy by ctDNA to be an alternative approach,10 and tissue-based molecular analysis remains the recommended standard for evaluating resistance to TKIs.11 Finding out the factors affecting the performance and reliability of the tests is necessary for the optimization of blood biopsy by ctDNA. Other factors that affect the tests, except technology, are still unclear.
Here, we genotyped EGFR from either tissue or blood biopsy samples for detecting various EGFR mutations linked to TKI resistance in patients with NSCLC. In particular, we focused on the mutation T790M. DNA extracted from both types of biopsies was sequenced using the conventional ARMS. We chose this system for two reasons: first, it is still the standard system used for tissue biopsy at our hospital; second, we wanted to know whether there are other factors, except technology, affecting the results. The findings of this study may shed light on the optimization of the performance and reliability of blood biopsy, especially relative to tissue biopsy, for diagnosing acquired EGFR TKI resistance associated with T790M.
Materials and methods
Study design
This observational study was conducted on patients treated at West China Hospital (Chengdu, People’s Republic of China) between 2014 and 2016. Patients were eligible if they had advanced NSCLC involving EGFR mutation(s), were clinically resistant to EGFR TKIs (gefitinib, erlotinib, icotinib, or afatinib), and were undergoing re-biopsy for EGFR genotyping as part of their routine clinical care. The study was approved by the Biomedical Research Ethics Committee of West China Hospital of Sichuan University, and all patients signed the informed consent form.
Blood sampling and sequencing
Peripheral blood was collected within 2 weeks of re-biopsy into a 10-mL vacutainer containing ethylenediaminetetraacetic acid. Blood samples were transported to our laboratory within 2 h of being drawn, plasma was isolated by centrifugation at 2,000× g for 10 min, and the supernatant was further cleared at 8,000× g for an additional 10 min. The Circulating Nucleic Acid Kit (AmoyDx, Xiamen, People’s Republic of China) was used to isolate ctDNA according to the manufacturer’s protocol. EGFR genotyping was performed on tissue biopsies according to standard institutional procedures in a certified laboratory, with conventional ARMS sequencing carried out using the Human EGFR Gene Mutations Fluorescence PCR Diagnostic Kit (AmoyDx). The same kit was used to genotype EGFR from ctDNA. This kit has been approved for clinical use by the China Food and Drug Administration since 2010.
Statistical analysis
Diagnostic concordance between ARMS sequencing of tissue-derived DNA or ctDNA was assessed using Cohen’s κ, and the concordance was assessed for significance using the McNemar test. Percent agreement values were calculated based on the main diagonal in 2×2 tables. All statistical analyses were performed with SPSS 20.0 (IBM Corp., Armonk, NY, USA), and the significance threshold was set as P<0.05.
Results
Patient characteristics
From January 2014 to June 2016, 45 patients were enrolled in the study (Figure 1, Table 1). Of the 45 patients, 4 (8.9%) received two types of EGFR TKI. As the most recent treatment, 8 (17.8%) received chemotherapy, 4 (8.9%) received EGFR TKI plus chemotherapy, and 33 (73.3%) received EGFR TKI.
Figure 1.
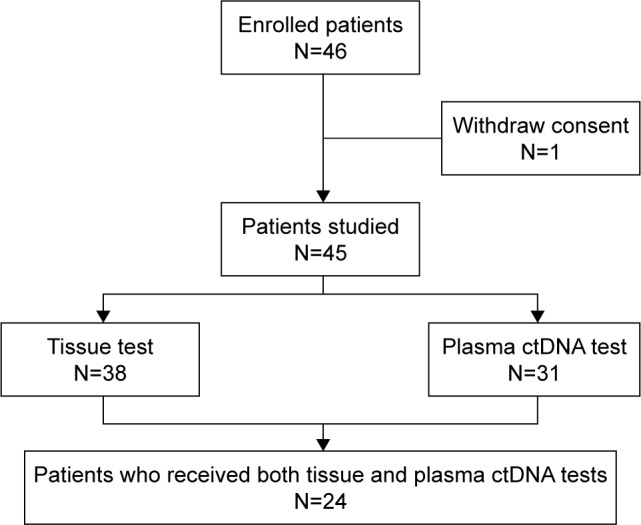
Patient flow diagram.
Abbreviation: ctDNA, circulating tumor DNA.
Table 1.
Patient characteristics
| Characteristics | N (%) |
|---|---|
| Age, years | |
| Median | 53.2 |
| Range | 36–75 |
| Sex | |
| Male | 23 (51.1) |
| Female | 22 (48.9) |
| Histology (initial biopsy) | |
| Adenocarcinoma | 42 (93.3) |
| Squamous carcinoma | 3 (6.7) |
| Other | 0 (0.0) |
| Stage before re-biopsy | |
| IIIB | 2 (4.4) |
| IV | 43 (95.6) |
| Type of initial EGFR mutation | |
| 19 Del | 30 (66.7) |
| L858R | 15 (33.3) |
| Other | 0 (0.0) |
| Type of initial EGFR TKI | |
| Gefitinib | 37 (82.2) |
| Erlotinib | 4 (8.9) |
| Icotinib | 4 (8.9) |
| Line of initial EGFR TKI | |
| First | 39 (86.7) |
| Second | 6 (13.3) |
| Response to initial EFGR TKI | |
| CR/PR/SD | 43 (93.3) |
| PD | 2 (6.7) |
| Second biopsy | |
| Tissue biopsy | 38 (84.4) |
| Blood biopsy | 31 (68.9) |
| Both | 24 (53.3) |
Abbreviations: EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; CR, complete response; PR, partial response; SD, stable disease; PD progressive disease.
EGFR genotyping in tissue biopsy and plasma ctDNA
Among patients whose tissue biopsy was sequenced, 25 showed two EGFR mutations, 12 showed one mutation, and 1 had three mutations (Table 2). Among patients whose plasma ctDNA was sequenced, 14 had two types of EGFR mutation, 12 had no mutation, and 5 showed one mutation.
Table 2.
EGFR genotyping of initial and secondary biopsy
| EGFR genotyping | Initial
|
Secondary
|
|
|---|---|---|---|
| Tissue N=45 (%) |
Tissue N=38 (%) |
ctDNA N=31 (%) |
|
| Exon 19 deletion | 30 (66.7) | 28 (73.7) | 12 (38.7) |
| L858R | 15 (33.3) | 11 (28.9) | 6 (19.4) |
| T790M | |||
| Weakly positivea | 0 (0.0) | 0 (0.0) | 4 (12.9) |
| Positive | 0 (0.0) | 26 (68.4) | 9 (29.0) |
| S768I | 0 (0.0) | 0 (0.0) | 2 (6.5) |
| Negative | 0 (0.0) | 0 (0.0) | 12 (38.7) |
Note:
The curve rises up, but does not reach the peak.
Abbreviation: ctDNA, circulating tumor DNA.
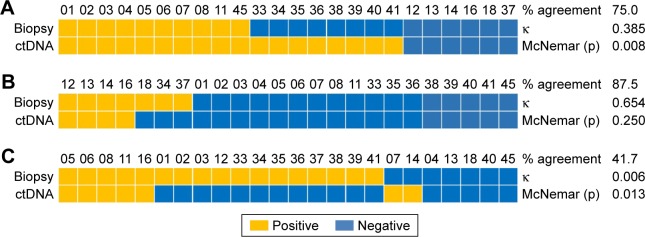
Of the 24 patients whose tissue and plasma ctDNA were sequenced (Table 3), 18 (75.0%) were concordant for exon 19 deletion (κ=0.385; Figure 2A), 21 (87.5%) were concordant for L858R (κ=0.654; Figure 2B), and 10 (41.7%) were concordant for T790M (κ=0.006; Figure 2C).
Table 3.
EGFR genotyping in patients who received both tissue and plasma ctDNA tests
| EGFR genotyping | Initial
|
Secondary
|
|
|---|---|---|---|
| Tissue N=24 (%) |
Tissue N=24 (%) |
ctDNA N=24 (%) |
|
| Exon 19 deletion | 16 (66.7) | 18 (75.0) | 10 (41.7) |
| L858R | 8 (33.3) | 7 (29.2) | 4 (16.7) |
| T790M | |||
| Weakly positivea | 0 (0.0) | 0 (0.0) | 3 (12.5) |
| Positive | 0 (0.0) | 17 (70.8) | 6 (25.0) |
| S768I | 0 (0.0) | 0 (0.0) | 2 (8.3) |
| Negative | 0 (0.0) | 0 (0.0) | 9 (37.5) |
Note:
The curve rises up, but does not reach the peak.
Abbreviation: ctDNA, circulating tumor DNA.
Figure 2.
Comparison of EGFR mutation status based on tissue biopsy or plasma ctDNA with respect to (A) exon 19 deletion, (B) L858R mutation, or (C) T790M mutation.
Abbreviation: ctDNA, circulating tumor DNA.
The positive rate for T790M mutation was 70.8% based on tissue sequencing and 37.5% based on plasma ctDNA sequencing (Table 3). Of the 24 patients for whom both tissue and plasma ctDNA sequencing results were available, 9 (37.5%) showed the same results, although the plasma ctDNA results for one-third of these patients were only weakly positive. Of the 17 patients who were positive for T790M based on the tissue test, only 7 (41.2%) were positive for the plasma ctDNA test, and 3 of those 7 were only weakly positive. Of the 7 patients negative for T790M based on tissue sequencing, 2 were positive based on plasma ctDNA sequencing (Table 4).
Table 4.
T790M mutation tested by both tumor tissue and plasma ctDNA levels
| Tissue biopsy | Plasma ctDNA
|
|
|---|---|---|
| Positive | Negative | |
| Positive | 7 | 10 |
| Negative | 2 | 5 |
Abbreviation: ctDNA, circulating tumor DNA.
Discussion
In this study, we used conventional ARMS to sequence the EGFR gene of patients with NSCLC for mutations associated with TKIs resistance from re-biopsied tumor DNA or from plasma ctDNA. Our results suggest that ctDNA can be a suitable template for EGFR genotyping in these patients, although the positive rate for T790M (37.5%) based on blood biopsy is lower than that based on tissue sequencing (70.8%). These findings support the usefulness of plasma ctDNA for mutation analysis as established in several other disease contexts9,12–17 and also suggest that there are other factors affecting the performance results of the test.
We sequenced the EGFR gene from tissue and blood using conventional ARMS despite its lower sensitivity,18 rather than using newer techniques such as ddPCR,12,19–21 because we wanted to analyze both types of biopsy in the same way to identify whether there are other factors, except technology, affecting the results. This should not threaten the validity of our results because studies using newer techniques also show a large variation in T790M-positive rates,6–9,12–15,19–22 and the same held true even in four prospective studies on NSCLC patients with acquired resistance.6–8,23 Indeed, the fact that our T790M-positive rate based on tissue sequencing (70.8%) was similar to that of several previous studies (63%, 66%, and 76.5%)6–8 suggests that our study population was representative of the target patient population.
Some important characteristics of the four prospective studies and this study are listed in Table S1. All these prospective studies used new techniques with higher sensitivity. Mok et al23 used allele-specific amplification, but the kit (cobas®, Roche Diagnostics Limited, Indianapolias, IN, USA) had as high a sensitivity and specificity as the ddPCR system.19 However, our T790M rate was not the lowest one, either in all the patient receiving blood biopsy or in patients confirmed by tissue biopsy. In contrast, a clinical trial7 reported the lowest T790M rate in both two situations. These results indicate that the less sensitive, conventional ARMS used in the present study does not necessarily perform worse than the sequencing techniques used in other studies. This implies that technology is not the primary reason why the T790M rate of ctDNA sequencing is highly varied.
While some authors have suggested the need to optimize the timing of blood biopsy,24 this seems unlikely to explain the higher T790M rate, since in the present study and four other prospective studies,6–8,23 both blood and tissue biopsies were performed at the same time after disease progression. It seems more likely that the higher T790M rate obtained with conventional ARMS is due to high ctDNA levels; in fact, high levels of circulating free DNA are associated with poor prognosis.25,26 This implies that the risk of false negatives is much higher with blood biopsy than tissue biopsy in patients with good prognosis. We do not know whether the poor prognosis causes the high ctDNA levels in the present study. However, we do know that the disease course of patients in the clinical trial (mentioned earlier),7 who only received first-line EGFR TKI treatment, is shorter compared to patients who received a multiline treatment in real-world studies. We believe the factors causing high ctDNA levels are intrinsically connected with clinical features, different from those factors relating to preanalytical plasma processing and ctDNA extraction or detection technique,10 and cannot be controlled through technological optimization.10 It is necessary to find out what these factors are, so that patients with high ctDNA levels can be identified to choose blood biopsy as preferred diagnostic approach. In the meantime, and even more importantly, it may be advisable to use tissue biopsy for patients with low ctDNA levels in order to avoid the risk of false-negative by blood biopsy.
Future work should attempt to identify the clinical features relating to ctDNA level as well as additional factors that may affect the sensitivity and specificity of blood biopsy in order to clarify for which patients and at what times in disease progression this type of biopsy can be reliably performed. At the same time, studies should continue to examine how to make re-biopsy as safe as possible, especially since some patients, such as those with low ctDNA levels, may not be suitable for blood biopsy.
Supplementary material
Table S1.
Characteristics of the studies
| Characteristics | Sundaresan et al1 | Yanagita et al2 | Takahama et al3 | Mok et al4 | Our research |
|---|---|---|---|---|---|
| Type | Prospective | Prospective | Prospective | Prospective | Prospective |
| Enrolling condition | Real world | Clinical trial | Real world | Real world | Real world |
| Patient feature | AR | AR | AR | AR | AR |
| Sequencing method | |||||
| Tissue | IS | ddPCR | ddPCR | Cobas® (ASA) | AmoyDx (ARMS) |
| Plasma ctDNA | AS PCR | ddPCR | ddPCR | Cobas® version 2 (ASA) | AmoyDx (ARMS) |
| Positive rate of T790M | |||||
| In all the patients (%) | 50.0 | 23.0 | 28.8 | 52.5 | 25.0–37.5 |
| In patients CTB (%) | 60.0 | 33.3 | 64.5 | 52.5 | 41.2 |
Abbreviations: AR, acquired resistance; IS, institution’s standard; ASA, allele-specific amplification; AS PCR, allele-specific PCR; ddPCR, droplet digital-PCR; ARMS, amplification refractory mutation system; CTB, confirmed by tissue biopsy.
References
- 1.Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res. 2016;22(5):1103–1110. doi: 10.1158/1078-0432.CCR-15-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yanagita M, Redig AJ, Paweletz CP, et al. A prospective evaluation of circulating tumor cells and cell-free DNA in EGFR mutant non-small cell lung cancer patients treated with erlotinib on a phase II trial. Clin Cancer Res. 2016;22(24):6010–6020. doi: 10.1158/1078-0432.CCR-16-0909. [DOI] [PubMed] [Google Scholar]
- 3.Takahama T, Sakai K, Takeda M, et al. Detection of the T790M muta tion of EGFR in plasma of advanced non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan oncology group 8014LTR study) Oncotarget. 2016;7(36):58492–58499. doi: 10.18632/oncotarget.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376(7):629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81472196). We also acknowledge Qian Zhao, Xiao-juan Zhou, Yong Xu, Jin Wang, and Li Ren for their contributions to this work.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372(18):1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 4.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 5.Sheikine Y, Rangachari D, McDonald DC, et al. EGFR testing in advanced non-small-cell lung cancer, a mini-review. Clin Lung Cancer. 2016;17(6):483–492. doi: 10.1016/j.cllc.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 6.Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res. 2016;22(5):1103–1110. doi: 10.1158/1078-0432.CCR-15-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanagita M, Redig AJ, Paweletz CP, et al. A prospective evaluation of circulating tumor cells and cell-free DNA in EGFR mutant non-small cell lung cancer patients treated with erlotinib on a phase II trial. Clin Cancer Res. 2016;22(24):6010–6020. doi: 10.1158/1078-0432.CCR-16-0909. [DOI] [PubMed] [Google Scholar]
- 8.Takahama T, Sakai K, Takeda M, et al. Detection of the T790M mutation of EGFR in plasma of advanced non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan oncology group 8014LTR study) Oncotarget. 2016;7(36):58492–58499. doi: 10.18632/oncotarget.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3375–3382. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Normanno N, Denis MG, Thress KS, Ratcliffe M, Reck M. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget. 2017;8(7):12501–12516. doi: 10.18632/oncotarget.13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan DS, Yom SS, Tsao MS, et al. The International Association for the Study of Lung Cancer Consensus Statement on Optimizing Management of EGFR Mutation-Positive Non-Small Cell Lung Cancer: Status in 2016. J Thorac Oncol. 2016;11(7):946–963. doi: 10.1016/j.jtho.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2(8):1014–1022. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6:20913. doi: 10.1038/srep20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JY, Qing X, Xiumin W, et al. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC-12-02) Oncotarget. 2016;7(6):6984–6993. doi: 10.18632/oncotarget.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sueoka-Aragane N, Katakami N, Satouchi M, et al. Monitoring EGFR T790M with plasma DNA from lung cancer patients in a prospective observational study. Cancer Sci. 2016;107(2):162–167. doi: 10.1111/cas.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous mechanisms of primary and acquired resistance to third-generation EGFR inhibitors. Clin Cancer Res. 2016;22(19):4837–4847. doi: 10.1158/1078-0432.CCR-15-1915. [DOI] [PubMed] [Google Scholar]
- 18.Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep. 2014;4:6269. doi: 10.1038/srep06269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90(3):509–515. doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Seki Y, Fujiwara Y, Kohno T, et al. Picoliter-droplet digital polymerase chain reaction-based analysis of cell-free plasma DNA to assess EGFR mutations in lung adenocarcinoma that confer resistance to tyrosine-kinase inhibitors. Oncologist. 2016;21(2):156–164. doi: 10.1634/theoncologist.2015-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chai X, Ren P, Wei B, et al. A comparative study of EGFR oncogenic mutations in matching tissue and plasma samples from patients with advanced non-small cell lung carcinoma. Clin Chim Acta. 2016;457:106–111. doi: 10.1016/j.cca.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Reck M, Hagiwara K, Han B, et al. ctDNA determination of EGFR mutation status in European and Japanese patients with advanced NSCLC: The ASSESS Study. J Thorac Oncol. 2016;11(10):1682–1689. doi: 10.1016/j.jtho.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 23.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376(7):629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chia PL, Do H, Morey A, Mitchell P, Dobrovic A, John T. Temporal changes of EGFR mutations and T790M levels in tumour and plasma DNA following AZD9291 treatment. Lung Cancer. 2016;98:29–32. doi: 10.1016/j.lungcan.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Ai B, Liu H, Huang Y, Peng P. Circulating cell-free DNA as a prognostic and predictive biomarker in non-small cell lung cancer. Oncotarget. 2016;7(28):44583–44595. doi: 10.18632/oncotarget.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu YJ, Zhang HB, Liu YH, et al. Estimation of cell-free circulating EGFR mutation concentration predicts outcomes in NSCLC patients treated with EGFR-TKIs. Oncotarget. 2017;8(8):13195–13205. doi: 10.18632/oncotarget.14490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Characteristics of the studies
| Characteristics | Sundaresan et al1 | Yanagita et al2 | Takahama et al3 | Mok et al4 | Our research |
|---|---|---|---|---|---|
| Type | Prospective | Prospective | Prospective | Prospective | Prospective |
| Enrolling condition | Real world | Clinical trial | Real world | Real world | Real world |
| Patient feature | AR | AR | AR | AR | AR |
| Sequencing method | |||||
| Tissue | IS | ddPCR | ddPCR | Cobas® (ASA) | AmoyDx (ARMS) |
| Plasma ctDNA | AS PCR | ddPCR | ddPCR | Cobas® version 2 (ASA) | AmoyDx (ARMS) |
| Positive rate of T790M | |||||
| In all the patients (%) | 50.0 | 23.0 | 28.8 | 52.5 | 25.0–37.5 |
| In patients CTB (%) | 60.0 | 33.3 | 64.5 | 52.5 | 41.2 |
Abbreviations: AR, acquired resistance; IS, institution’s standard; ASA, allele-specific amplification; AS PCR, allele-specific PCR; ddPCR, droplet digital-PCR; ARMS, amplification refractory mutation system; CTB, confirmed by tissue biopsy.