Abstract
The bacterial pathogen Staphylococcus aureus controls many aspects of virulence by using the accessory gene regulator (agr) quorum sensing (QS) system. The agr system is activated by a macrocyclic peptide signal known as an autoinducing peptide (AIP). We sought to develop structurally simplified mimetics of AIPs for use as chemical tools to study QS in S. aureus. Herein, we report new peptidomimetic AgrC receptor inhibitors based on a tail-truncated AIP-II peptide that have almost analogous inhibitory activities to the parent peptide. Structural comparison of one of these peptidomimetics to the parent peptide and a highly potent, all-peptide-derived, S. aureus agr inhibitor (AIP-III D4A) revealed a conserved hydrophobic motif and overall amphipathic nature. Our results suggest that the AIP scaffold is amenable to structural mimicry and minimization for the development of synthetic agr inhibitors.
Keywords: accessory gene regulator, autoinducing peptides, peptidomimetics, quorum sensing, Staphylococcus aureus
Introduction
Quorum sensing (QS) allows bacteria to behave as a group at high cell densities and is closely connected to virulence in many common pathogens.[1] This intercellular communication system is mediated by small molecule or peptide signals that diffuse out of or are secreted by bacteria into the local environment. As the population grows, the concentration of QS signal likewise increases until a threshold is reached, whereupon the signal productively binds to its cognate receptor. The activated receptor then modifies gene expression levels to permit the bacteria to adopt a community lifestyle, whilst simultaneously increasing production of the QS circuitry, thus amplifying the QS response.[1a]
For many pathogens, QS allows bacteria to amass a sufficiently high population prior to initiating virulence phenotypes, thereby increasing their chances to successfully infect a host.[1a,2] The close connection between QS and bacterial infection, and perhaps more fundamentally, the chemical nature of QS signaling, has inspired considerable recent research into the design of chemical strategies to intercept QS pathways.[3] Many of these efforts have focused on the development of synthetic mimics of QS signals that can inhibit QS receptor:signal binding for use as chemical probes to block virulence phenotypes and to delineate basic QS mechanisms. Our laboratory[4] and others[1a,5] have contributed in this area, and we recently have focused on the common pathogen Staphylococcus aureus[4f–i,k,l,6] and related species.[4j]
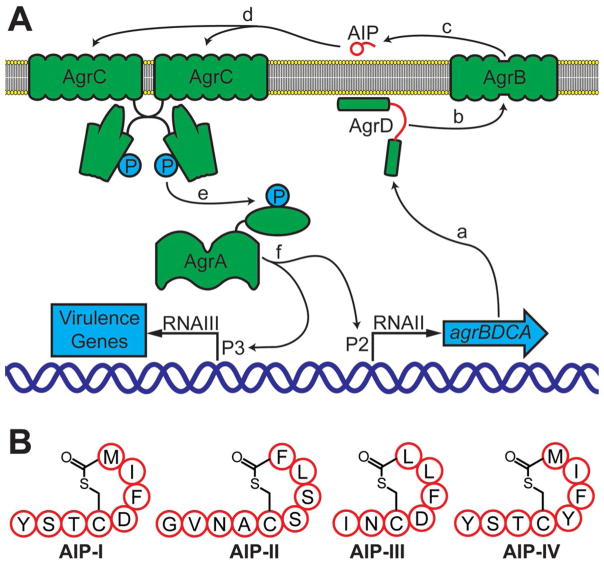
In staphylococci, QS is controlled by the accessory gene regulator (agr) system.[5t,7] In the case of S. aureus, this QS system controls the expression of over 100 virulence factors.[7b,8] The S. aureus agr machinery is composed of four proteins, AgrA–D, and a signaling molecule (derived from AgrD) termed an autoinducing peptide (or AIP; Figure 1 A).[9]
Figure 1.
Schematic representation of the agr QS circuit in S. aureus. A) a: The agr operon is expressed to produce the Agr proteins A–D. b and c: The AIP precursor AgrD is processed by AgrB and the mature AIP is transported out of the cell. d: The AIP signal binds to and activates AgrC, a transmembrane receptor and preformed dimer. e: AgrC phosphorylates and activates AgrA, the response regulator. f: AgrA binds promoters P2 and P3 and initiates transcription. See text. B) Structures of the four known S. aureus AIPs (I–IV). Single letter abbreviations used for amino acid residues.
AgrD is the precursor to the QS signal, which is processed by AgrB and secreted as the mature AIP (Figure 1 A).[5t,7,9a,10] The S. aureus AIP is a small macrocyclic peptide (7–9 residues), containing a short N-terminal tail and a thiolactone bridge between an internal cysteine side chain and the C terminus.[5j,9a] To date, four specificity subgroups of S. aureus have been characterized (groups I–IV), each defined by the unique peptide sequence of their AIPs (shown in Figure 1 B) and their target transmembrane receptor and histidine kinase, AgrC.[9a,11] When a threshold extracellular AIP concentration is reached, the peptide signal binds and activates AgrC. AgrC then phosphorylates and thereby activates its partner response regulator, the transcription factor AgrA.[5o] AgrA subsequently targets several promoters, including P2 and P3. P2 induces transcription of the agr operon and provides positive feedback for the autoinduction circuit.[9b] In turn, P3 drives transcription of RNA-III, which is a major regulator of virulence factor production in S. aureus.[12]
Blocking AgrC:AIP binding represents one strategy to attenuate QS signaling in S. aureus, and this approach has been the focus of considerable research over the past ~ 15 years.[4g,l,5t,7a,b] The initial discovery that AIP-I and AIP-II inhibit AgrC activity in non-self agr specificity groups was followed by studies of AIP derivatives with tail truncations, sequential alanine scans, and other amino acid substitutions to identify key structure–activity relationships (SARs) dictating ligand and receptor activity.[5j–l,q,7b] These early SAR studies suggested that the AIP ligand-binding site on AgrC was a hydrophobic cleft that could be a viable target for competitive inhibition with hydrophobic, peptidic ligands.[5l,m,13] Additional screening with tail-truncated AIPs resulted in further refinement of the SARs for AgrC activation and inhibition, thus suggesting that the presence of only two hydrophobic C-terminal groups within ligands was required for AgrC binding, whereas additional contacts on the N-terminal tail were essential for receptor activation.[5l,n] Tail-truncated AIP-II (t-AIP-II) was found to be quite potent amongst this class.[5l,n] Recently, we extended similar SAR studies to S. aureus AIP-III and found an analogous activity trend; namely, hydrophobic endocyclic residues were required for AgrC binding, whereas exocyclic tail contacts, along with the hydrophobic motifs, were required for activation.[4g–i,k,l] Our studies with AIP-III and mimetics thereof identified a number of highly potent, pan-group AgrC inhibitors, with AIP-III D4A being one of the most potent AgrC inhibitors reported to date.[4g]
Despite their potency, however, peptidic AgrC modulators possess several qualities that limit their utility as chemical tools. First, the AIP thiolactone bridges are hydrolytically unstable.[4l,6a,14] Second, although their macrocyclic framework renders AIPs more proteolytically stable than linear peptides, they are still susceptible to proteolysis.[4l,15] Third, AIP-type peptides have relatively low water solubilities due to their hydrophobic structures. Fourth, these ligands are typically prepared by using solid-phase synthesis techniques that do not lend themselves easily to large batch synthesis. We have begun to address some of these limitations through the development of lactam-bridged AIP-III mimetics.[4l] However, for the long term, we also seek to explore non-peptide, small molecule mimetics of AIPs that display enhanced stabilities and aqueous solubilities, reduced immunogenicity, and are amenable to larger scale synthesis relative to peptides. Indeed, identifying such compounds has recently been advanced as an important focus for the future[5t] and was the motivation for the current study.
Herein, we report our initial steps toward the development of structurally simplified AIP mimetics that inhibit AgrC activity in S. aureus. Starting with the scaffold of t-AIP-II, we designed and synthesized a series of minimized AIP-II peptidomimetics containing only three natural amino acid residues linked by an aliphatic spacer group. Screening of these compounds in the four groups of S. aureus for AgrC inhibition revealed three compounds that were pan-group inhibitors, with the most potent new inhibitor maintaining potency within a factor of seven of the parent peptide in each of the four groups. Structural analyses of one of these peptidomimetics by solution-phase NMR spectroscopy indicated that it possesses an overall amphipathic nature and a minimal hydrophobic motif similar to those of both the parent peptide t-AIP-II and our potent AgrC inhibitor, AIP-III D4A. Despite these similar features, we found this AIP mimetic significantly more soluble in aqueous media relative to AIP-III D4A and the other lead peptidomimetics reported herein. Together, this foundational work opens new avenues to the development of next-generation, small-molecule mimics of the AIPs.
Results and Discussion
Library design and synthesis
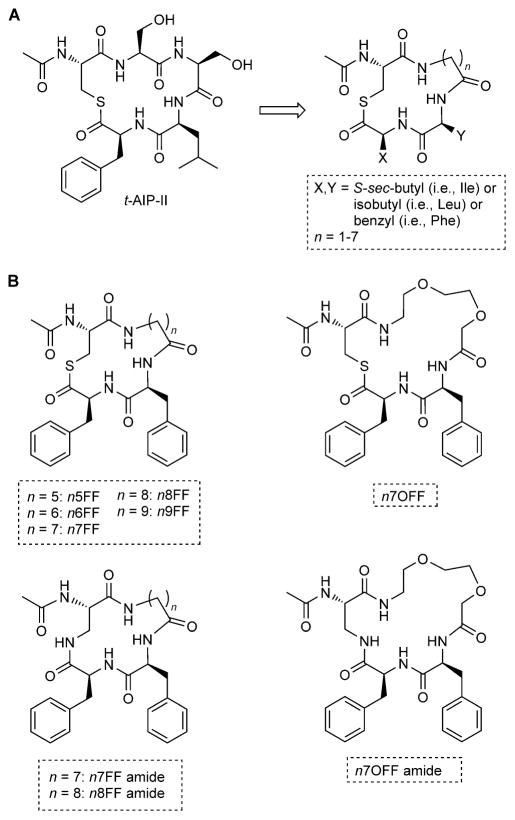
To develop our minimized AIP scaffold, we selected the truncated AIP, t-AIP-II, as our model peptide due to its small size and concomitant high potency (i.e., IC50 values for AgrC inhibition in the low nanomolar range for each S. aureus group).[5l] A differentiating feature of t-AIP-II (and its parent, AIP-II) is that it contains only two hydrophobic amino acids in the macrocycle (Scheme 1 A), as opposed to the three present in the other AIPs (Figure 1 B). We previously examined the solution-phase NMR structure of t-AIP-II and found that it adopts a rigid structure in solution with the Leu4 and Phe5 side chains pointing in the same direction and in close proximity to each other.[4k] This study, in conjunction with earlier mechanistic work,[5n] suggests that the presence and close positioning of these hydrophobic residues is critical for AgrC binding. We thus sought to retain hydrophobic functionality at these two positions in our minimized AIP-II scaffold, and systematically examined leucine, phenylalanine, and isoleucine in each of these positions. To streamline the scaffold thereafter, the two serine residues (Ser2 and Ser3) were replaced by a single aliphatic linker. For peptidic AgrC ligands, the macrocycle has been shown to require a defined size for AgrC recognition (16–17 atoms), with restrictions on the positions that can accept an extra atom.[5p] However, with our proposed aliphatic linker, which should be much more flexible than a peptidic amide backbone, we decided to explore different linker lengths, with a chain of methylenes varying between 1–7 carbons in length (yielding macrocycles of 13–19 atoms). Cys1 was retained to create the thiolactone moiety that closes the macrocycle and is known to be important for recognition by AgrC.[5l,n] Our overall modifications of t-AIP-II to create this new scaffold are summarized in Scheme 1 A.
Scheme 1.
A) Structure of t-AIP-II and its reduction to the minimized scaffold examined in this study. B) Selected t-AIP-II mimetics with potencies and/or structural features of interest.
We prepared a library of 63 peptidomimetics based on this simplified scaffold, with every permutation of the desired hydrophobic residues and linker lengths (Scheme 1 A), by using standard Fmoc solid-phase peptide synthesis (SPPS) methods.[16] Macrocyclization was performed according to our previously reported procedure,[4g] followed by routine HPLC purification and MS characterization (see the Experimental Section). This minimized AIP-II scaffold was anticipated to remedy several shortcomings of fully peptidic ligands. Because the number of natural amino acids was minimized, there was less opportunity for enzymatic degradation, thus improving ligand stability. In addition, fewer coupling steps were required to produce these peptidomimetics, thereby shortening synthesis times and increasing overall product yields (~25 % on average). The small size of these mimetics also opens up the possibility for larger scale production by using solution-phase synthesis.
Screening of peptidomimetic library
We screened the 63-compound library of AIP mimetics for AgrC modulatory activity in all four groups of S. aureus by using strains containing yfp reporter plasmids. These strains are summarized in the Experimental Section and include the multidrug-resistant strain USA300 LAC.[17] In these S. aureus reporters, binding of the native AIP to AgrC activates YFP production and can be measured by using fluorescence. Thus, compounds capable of inhibiting native AIP binding produce a reduction in fluorescence. Preliminary compound screening was performed at 10 μM in each strain; the full results of preliminary screening are available in the Supporting Information (Figure S1). We summarize key findings from these initial screens here. To simplify the discussion, the full library is described as three smaller sub-libraries. In the first sub-library (termed nxLX), there was a linker with n = x methylenes, residue 3 was leucine, and residue 4 (X) was leucine, isoleucine, or phenylalanine (Scheme 1 A). The second and third sub-libraries (nxIX and nxFX, respectively) were identical to the first sub-library except that residue 3 was an isoleucine or phenylalanine, respectively.
In sub-library nxLX, there were no compounds that completely abolished AgrC activity in group I S. aureus, but n3LF reduced activity to 17 % relative to a DMSO control. In group II, n6LF completely inhibited AgrC activity. In group III, n3LF, n5LF, n6LF, and n7LF all reduced AgrC activity to minimal levels. n3LF completely inhibited AgrC activity in group IV, highlighting the potential similarities between group I and group IV (which share almost identical native AIP ligands, Figure 1 B). Notably, all of the AgrC inhibitors identified in this sub-library contained Phe at residue 4.
In sub-library nxIX, the overall inhibitory activity observed was low. These results suggest that the presence of Ile at residue 4 on the minimized t-AIP-II scaffold is detrimental to AgrC binding. This sub-library was only active in the group III strain, with n5IF, n6IF, and n7IF reducing AgrC activity to minimal levels at a concentration of 10 μM. Interestingly, this set of active compounds contained the same Phe4 and had similar ring sizes as the group III inhibitors identified in the nxLX library.
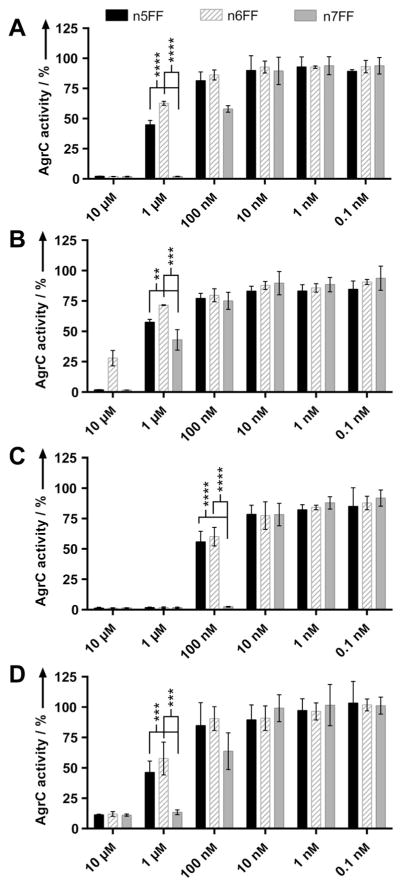
In sub-library nxFX, the mimetics n5FF, n6FF, and n7FF were found to reduce AgrC activity to minimal levels in all four groups of S. aureus, with n6FF displaying slightly less AgrC inhibition in group II than n5FF and n7FF. Conspicuously, these compounds all contained phenylalanine at residues 3 and 4.
In determining which compounds to advance in this study, mimetics with the noted double Phe3 and Phe4 motif and ring sizes of n =5–7 were deemed the most promising. Thus, we proceeded with n5FF, n6FF, and n7FF as our lead compounds from the peptidomimetic library (shown in Scheme 1B), and measured their AgrC inhibition over a range of concentrations to better gauge their relative activities using the four S. aureus reporter strains. The assay results, shown in Figure 2, identify n7FF as the top pan-group AgrC inhibitor, based on statistically significant differences in activity.
Figure 2.
Preliminary AgrC activity screening data for lead compounds. Compound activity data in A) group I, B) group II, C) group III, and D) group IV S. aureus strains at concentrations decreasing from 10 μM to 100 pM. Bar graphs indicate percent AgrC activity as measured by YFP fluorescence using the S. aureus reporter strains (see text). Error bars represent 95 % confidence intervals. The results of two-tailed t-tests indicate that at 1 μM in groups I, II, and IV, and at 100 nM in group III, there is at least a significant statistical difference in compound activity for n5FF and n6FF versus n7FF. ** p≤0.01, *** p≤0.001, and **** p≤0.0001.
Design of second-generation compounds and preliminary screening
We next scrutinized the structures of n5FF, n6FF, and n7FF to further delineate features that were important for their activity profiles against AgrC and design additional ligands with potentially enhanced activities. As the most active pan-group inhibitor (n7FF) had the largest macrocycle tested (19 atoms), we prepared the 1- and 2-carbon ring-expanded homologues, n8FF and n9FF, to examine how further expanding the ring size impacted activity (Scheme 1 B). We found that n8FF demonstrated very similar activity relative to n7FF and the parent peptide t-AIP-II, whereas n9FF was less active than either n7FF or n8FF in groups I–III, having estimated IC50 values in the micromolar range (Figure S2).[18] These results indicate that, at least for the nxFF series, the size of the flexible carbon linker can be varied only slightly without impacting the observed activity, and optimally, the number of atoms in the macrocycle should be three or four atoms greater than that in native AIP amide backbones (i.e., 16). Further, these data suggest that—assuming these minimized ligands target the same site on AgrC as native AIPs—all of the macrocycle amide contacts made by AIPs in the AgrC binding pocket might not be essential for binding, as long as the requisite space in the binding site is filled sufficiently.
Further study and manipulation of compounds n7FF and n8FF revealed that they had relatively low water solubilities, limiting their study at concentrations over 100 μM. We thus sought to explore related derivatives that were less hydrophobic to ease their routine handling; such a characteristic should also enhance their utilities as eventual chemical probes. Beginning with the lead compound n7FF, we envisaged that the addition of two oxygen atoms into the linker would be straightforward and increase its capacity to form hydrogen bonds with solvent. This new compound, n7OFF (n7 oxo-linker FF, Scheme 1 B), was readily prepared by incorporating the linker amino acid 8-(Fmoc-amino)-3,6-dioxaoctanoic acid (n7O) into our standard SPPS route.
Synthetic manipulation quickly revealed n7OFF to be approximately 100 times more soluble in water than n7FF, with a solubility near 10 mM. This difference in solubility was also apparent from calculated Clog P (n-octanol/water) values for these peptidomimetics (exact values in the Supporting Information). Indeed, the Clog P values for t-AIP-II and n7OFF are very similar and relatively low (indicative of lower hydrophobicity); the Clog P values for n7FF, n8FF, and AIP-III D4A are also quite similar, albeit larger than those for t-AIP-II and n7OFF (indicative of higher hydrophobicity; although in practice n7FF and n8FF are both less soluble than AIP-III D4A in water). In addition to possessing a desirable aqueous solubility profile, preliminary screening of n7OFF in the four AgrC reporter strains indicated that it was approximately as active as t-AIP-II, n7FF, and n8FF in groups I and II and modestly active in groups III and IV (Figures S3–S6).
Next, we examined nxFF derivatives that lacked the labile thioester bond, which is unstable in aqueous media at pH values above 7.4.[4l] Derivatives of this type could exhibit prolonged lifetimes and possibly enhanced activities in biologically relevant settings. Building on our recent study of amide-linked analogues of AIP-III D4A (in which the thiolactone was replaced with an amide), we prepared three amide-linked analogues of n7FF, n8FF, and n7OFF—termed n7FF amide, n8FF amide, and n7OFF amide (Scheme 1 B)—by substituting L-2,3-diaminopropionic acid (Dap) for Cys1 during our SPPS protocol and a final macrolactamization step (see the Experimental Section). These compounds were markedly more soluble in water than their parent thiolactones (also apparent in their Clog P values; Supporting Information), analogous to our observation for the AIP-III D4A amide.[41] However, preliminary biological screening indicated that they were significantly less active than their parent compounds (Figure S7). In all the S. aureus reporters, n7OFF amide showed no significant activity at 10 μM. Although the n7FF and n8FF amides both reduced AgrC activity to minimal levels in group II and group III at 10 μM, they were far less active in group I, and there was virtually no inhibition in group IV at 10 μM. These results suggest that the thioester bond is important for AgrC:ligand interactions in our minimized t-AIP-II system. These data also could indicate that, as this class of AIP mimetics becomes more hydrophilic, it becomes less effective at binding certain AgrC receptors.
Dose–response analyses of lead compounds
Screening of the 63-member library, followed by the second-generation design studies described above, identified n7FF, n8FF, and n7OFF as our most promising compounds for further follow-up. To facilitate a more quantitative comparison of their relative activities, we performed dose–response analyses on these three compounds by using the group I–IV S. aureus reporter strains and calculated IC50 values for AgrC inhibition. As a key control, we included the parent peptide, t-AIP-II, on which the mimetics were based. Dose–response data for n7FF amide, n8FF amide, and n7OFF amide were also collected for comparison to n7FF, n8FF, and n7OFF, respectively. The IC50 data are summarized in Table 1 (for a complete compilation of all trials, see Figures S3–S7).
Table 1.
IC50 values for selected peptidomimetics against S. aureus AgrC in groups I–IV.[a]
| Inhibitor | Group I | IC50 [nm][b] | Group IV | |
|---|---|---|---|---|
| Group II | Group III | |||
| t-AIP-II[c] | 260 (95–695) | 230 (190–270) | 4 (3–5) | 150 (90–260) |
| t-AIP-II[d] | 101 (88–117) | 97 (94–112) | 9.3 (7.7–11.4) | 140 (118–166) |
| n7FF | 340 (308–376) | 495 (435–564) | 34.8 (30.8–39.4) | 985 (825–1176) |
| n8FF | 468 (422–518) | 479 (410–559) | 125 (109–142) | 3486 (2301–5280) |
| n7OFF | 181 (154–213) | 583 (468–725) | 332 (280–395) | 5938 (4221–8352) |
| n7FF amide | 3935 (3593–4309) | 2200 (1727–2801) | 573 (472–696) | –[e] |
| n8FF amide | 4742 (4335–5187) | 1339 (1065–1683) | 2613 (2180–3131) | –[e] |
| n7OFF amide | –[f] | –[f] | –[f] | –[f] |
See Experimental Section for details of strains and assay protocols.
95 % confidence interval provided in parentheses.
Values reproduced from ref. [5l] with different S. aureus reporter strains.
Values differ slightly in groups III and IV from those previously reported in ref. [4k] due to day-to-day assay variations.
Data not shown due to non-sigmoidal curves in group IV, with maximal inhibition not reached at the highest concentration tested.
Inactive over the concentrations tested.
Dose–response analyses revealed that n7FF was the most consistent mimetic of t-AIP-II with regard to potency, as it maintained IC50 values within three- to sevenfold of those for t-AIP-II in all four groups of S. aureus. The homologue that was larger by one carbon, n8FF, was largely equipotent to n7FF in groups I and II but fourfold less potent in groups III and IV. The more soluble mimetic, n7OFF, was almost two times more potent than n7FF in group I, largely equipotent in group II, and far less potent in groups III and IV. Notably, the potency of n7OFF in group I matched that of the parent truncated AIP, t-AIP-II. This activity profile in groups I and II (the two more common S. aureus groups in human infection),[7b] coupled with its heightened aqueous solubility, suggest that the n7OFF scaffold could prove useful for QS probe development in S. aureus.
As we suspected from our single-concentration preliminary assays, the amide analogues of n7FF and n8FF were far less potent than their parents (i.e., an approximately tenfold increase in IC50 value; Table 1), and n7OFF amide was essentially inactive at all concentrations tested. Again, this loss in potency for the amide analogues relative to their thioester parents suggests that the thioester motif is critical for the binding of these simplified AIP-II mimics to AgrC receptors (assuming that they target AgrC directly).
NMR structural studies
We next sought to characterize the structure of n7OFF in aqueous solution to gain further insights into the origins of its biological activity. Such studies have been extremely valuable in our prior analyses of AIP-type derivatives that inhibit AgrC receptor activity.[4h, k, l] As n7OFF was based on the structure of t-AIP-II, a logical first step was to compare the structures of these two compounds in water. In addition, we wanted to compare the structure of n7OFF to that of our highly potent S. aureus AgrC inhibitor, AIP-III D4A (sequence=I-N-[C-A-F-L-L]), to gain insights into any structural similarities or differences that might affect the activity profile for n7OFF. We previously reported NMR structures for both t-AIP-II[4k] and AIP-III D4A,[4h] albeit in a mixed solvent system (acetonitrile/water). By using a higher field spectrometer equipped with a cryoprobe in the current study, we were able to obtain solution-phase NMR structures of t-AIP-II, AIP-III D4A, and n7OFF in water without any organic solvent added to improve solubility. The new structures were solved by using 2D ROESY spectra with excitation sculpting solvent suppression, with accompanying 1 D proton and 2 D TOCSY spectra to determine chemical shifts of proton resonances and aid in sequential assignments (a 1H,13C HSQC spectrum was also used for n7OFF; Tables S2–S4). Low-energy ensembles, created with MestReNova 10 NMR and Xplor-NIH[19] software, were used to find the most representative low-energy structures based on relative energy and RMS differences from the ensemble averages as determined by PyMOL (Figures S8–S13).
Not surprisingly, the structures of t-AIP-II in water and in the mixed solvent system were essentially identical, with a single amide bond contorted (Ser2 to Ser3) and the two serine side-chains in slightly different rotational conformations (Figure 3 A; see overlay in Figure S9). The relatively large number of ROE peaks observed in the ROESY spectrum and the intensities of the inter-residue peaks suggested that t-AIP-II adopted a highly rigidified conformation, as we had observed previously (ensemble average RMSD for the amide backbone atoms of only 0.05 Å).[4k] As before, the Leu4 and Phe5 side chains were positioned adjacently and pointed directly outwards away from the macrocycle. Similar to t-AIP-II, a comparison of the old and new solution structures for AIP-III D4A revealed only very subtle differences (Figure 3E; see overlay in Figure S13). The resolution of the newer NMR solution structure was similar to the original structure, with an ensemble average RMSD of 0.69 Å for the amide backbone atoms (Figures S11–S12). These results indicated that our previous conclusions drawn from the structures of t-AIP-II and AIP-III D4A in the mixed solvent system should also apply in pure water.
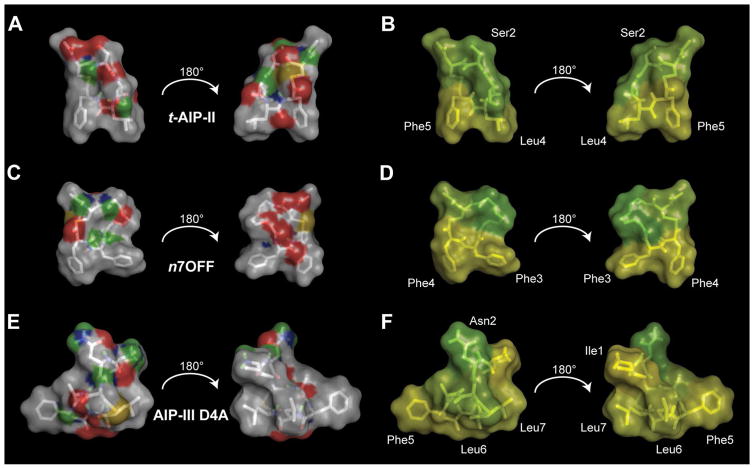
Figure 3.
Surface plots of t-AIP-II, n7OFF, and AIP-III D4A. A), C), and E) Hydrophobic atoms (i.e., carbons and hydrogens) are shown in light gray; the hydrophilic atoms are colored. Green represents hydrogen-bonding hydrogens, gold represents sulfur, blue represents nitrogen, and red represents oxygen. B), D), and F) Residue hydrophobicity is plotted on the surface of molecules by using the Eisenberg hydrophobicity scale. Green indicates less hydrophobicity, and yellow indicates more hydrophobicity. In n7OFF, the n7O linker is set to green, as it is hydrophilic. Selected residues are labeled for clarity. In (A) and (B), hydrophobic groups in t-AIP-II are oriented down, with hydrophilic moieties on either side oriented above, creating an overall amphipathic wedge. In (C) and (D), hydrophobic groups in n7OFF are oriented down, with hydrophilic moieties on either side oriented above, creating an analogous amphipathic wedge to that observed in t-AIP-II. In (E) and (F), the hydrophobic front face of AIP-III D4A is shown, and a subsequent rotation of 180° results in a back view with clustered hydrophobic groups.
Turning our attention to the n7OFF structure in water, we noted that the two endocyclic phenylalanine residues were well structured (Figures 3 C and S10), with multiple constraints indicated during analysis. Conversely, there were relatively few constraints indicated for the linker region; this suggests that movement of the linker atoms was sufficiently great to avoid the build-up of magnetization required for ROEs to be observed during the NMR experiment. The backbone atom RMSD from the ensemble was therefore relatively large compared to t-AIP-II and AIP-III D4A, at 0.96 Å.
We next compared the NMR structures of t-AIP-II, AIP-III D4A, and n7OFF in water. An initial examination of the placement of heteroatoms and hydrophobic amino acid residues suggested that all three structures were largely amphipathic (from observations using an Eisenberg hydrophobicity scale; Figures 3 B, D, and F).[20] Each structure had a hydrophilic face opposite a hydrophobic face, with aliphatic/aromatic groups clustered on the hydrophobic face of the peptide. It is interesting to note that previous SAR studies provided evidence that there is a hydrophobic binding pocket for AIP ligand recognition on AgrC receptors.[5m, t] We speculated that the amphipathic nature of this trio of ligands allows the hydrophobic face to insert in the proposed hydrophobic cleft of AgrC, while the hydrophilic face engages in favorable solvent interactions. Although we do not demonstrate conclusively here that the conformations adopted in aqueous solution are relevant to AgrC binding, there is a high probability that these macrocyclic, constrained compounds spend a majority of the time in a conformation in aqueous media that is favorable for AgrC binding. With this in mind, we analyzed overlays of t-AIP-II and n7OFF, and AIP-III D4A and n7OFF, using minimized RMS of similar atoms to gain further insights into the similarities and differences between these three structures (Figure 4).
Figure 4.
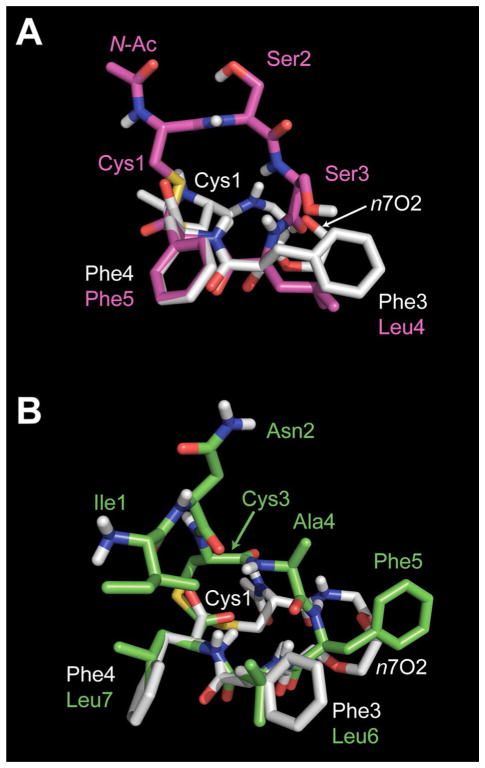
Overlaid NMR solution structures of t-AIP-II and n7OFF, and AIP-III D4A and n7OFF. A) t-AIP-II is shown in magenta; n7OFF is shown in white. B) AIP-III D4A is shown in green; n7OFF is shown in white. Residues are labeled and color coded according to the structure for clarity.
Overlays of the major solution conformations for t-AIP-II and n7OFF (Figure 4A), and AIP-III D4A and n7OFF (Figure 4 B), show that the two C-terminal residues and the bulk of the macrocycles on the hydrophobic faces of each compound occupy very similar positions. The RMS differences of the 19 atoms used for the fits of t-AIP-II with n7OFF and AIP-III D4A with n7OFF were 0.17 and 0.34 Å, respectively; note, these 19 atoms were all on the hydrophobic faces (additional views are available in Figure S14). Although the N-terminal acetyl group and flexible linker of n7OFF point away from the other macro-cyclic elements of t-AIP-II and AIP-III D4A, these two motifs both lie in the region suspected as being hydrophilic and therefore are likely to be positioned outside of the expected hydrophobic binding site on AgrC. When comparing the orientation of the macrocycle of t-AIP-II to that of n7OFF (Figure 4 A), we observed that the rigid macrocycle of t-AIP-II extended above the flexible linker in the n7OFF structure. In addition, when studying both overlays in Figure 4 in relation to n7OFF, the space occupied by the N-terminal acetyl group, Cys1, and Ser2 of the t-AIP-II macrocycle (Figure 4 A, magenta) appeared to be positioned similarly to Ile1 and Asn2 in the exocyclic tail of AIP-III D4A (Figure 4 B, green). In view of these collective structural features, it is possible that both t-AIP-II and n7OFF have reduced inhibitory activities relative to AIP-III D4A because, although they each can occupy a portion of space filled by AIP-III D4A, neither can completely fill the AgrC ligand-binding pocket quite as well as AIP-III D4A. To address this potential insufficient filling of space, exploring future AIP mimetics with a more rigidified bicyclic structure, complete with, for example, an aliphatic linker to mimic the girth of the AIP-III D4A Ile1 side chain, could prove fruitful. Of course, we acknowledge that the overlap of coinciding residues is not proof of an AgrC binding orientation, but the approximate similarities of the backbone conformations and hydrophobic side chains between n7OFF and these two peptides is certainly compelling and could prove to be important in understanding the mechanisms by which these ligands, and potentially other derivatives, interact with AgrC receptors.
Conclusion and Outlook
The goal of this study was to develop and characterize structurally simplified AIP mimetics as modulators of agr-type QS in the pathogen S. aureus. We have realized this objective. Herein, we report the design, synthesis, and biological characterization of simplified mimetics of the truncated native AIP, t-AIP-II. A focused library of 63 peptidomimetics was prepared by SPPS and evaluated for AgrC inhibition in the four agr groups of S. aureus by using reporter gene assays. Follow-up studies of lead compounds and second-generation mimetic design eventually revealed three peptidomimetics (n7FF, n8FF, and n7OFF) that were capable of inhibiting AgrC activity in the clinically relevant group I S. aureus strain with potencies similar to that of the parent peptide. In addition, n7FF maintained potencies in all four S. aureus groups largely similar to those of t-AIP-II, whereas n7OFF displayed enhanced water solubility relative to n7FF and n8FF.
Three structural features important for compound activity were identified through our analyses of these t-AIP-II mimetics. First, Phe3 and Phe4 were conserved in each of the most active peptidomimetics, thus suggesting that AgrC binding was optimized for this compound set when such hydrophobic, aromatic residues were present on the macrocycle. Second, the thioester moiety appeared important for ligand recognition, as modification to an amide largely obliterated the activities of our lead compounds. Third, an aliphatic chain of seven carbons in the mimetic macrocycle (n =7) appeared optimal for inhibitory activity against AgrC receptors.
We performed a series of solution-phase NMR experiments to gain insights into the structure of a lead mimetic, n7OFF, in aqueous solution. Structural comparisons of n7OFF with its parent peptide, t-AIP-II, and our previously reported potent AgrC inhibitor, AIP-III D4A, revealed a conserved amphipathic motif, with structural overlays emphasizing significant similarity in the respective hydrophobic regions of each molecule. These similarities in the hydrophobic region, which we posit to be important for AgrC binding, could prove useful in the design of future AIP mimetics.
Overall, the biological and structural results reported herein for peptidomimetics n7FF, n8FF, and n7OFF show that such a minimized AIP scaffold retains activity as a viable AgrC inhibitor. Ongoing studies are directed at further streamlining the AIP structure, using insights garnered here, to generate small-molecule AgrC modulators that are highly soluble, physically robust, and amenable to larger-scale synthesis relative to peptidic variants. Molecules of this type would represent valuable tools to study fundamental aspects of QS in S. aureus and its viability as a therapeutic target. For example, we are considering simple tail mimetics to replace the acetyl cap of the N terminus in n7OFF, as well as exploration of alternative amino acids and aromatic/hydrophobic groups beyond phenylalanine. In addition, the role of electronics in ligand activity has yet to be investigated systematically and could prove quite interesting. Lastly, we suspect that the simplified mimetics identified here might also be active against other staphylococcal species. These and related studies will be reported in due course.
Experimental Section
Reagents, instrumentation, and general methods
All chemical reagents and solvents were purchased from commercial sources (Chem-Impex International or Sigma–Aldrich) and used without further purification, except for dichloromethane (CH2Cl2), which was distilled and dried over activated molecular sieves. Water (18 MΩ) was purified by using a Thermo Scientific Barnstead Nanopure system. Solid-phase resin was purchased from NovaBiochem. t-AIP-II and AIP-III D4A were sourced from our in-house stocks.[4g, k]
All standard biological reagents were purchased from Sigma–Aldrich and used according to enclosed instructions. Brain heart infusion (BHI) was purchased from Teknova and prepared as instructed at pH 7.35.
Reversed-phase high performance liquid chromatography (RP-HPLC) was performed on a Shimadzu system equipped with an SCL-10Avp system controller, a DGU-20A5 degasser, an LC-20AT solvent delivery unit, an SIL-10AF autosampler, a CTO-20A column oven equipped with a manual injector, an SPD-M20A UV/Vis diode array detector, and an FRC-10A fraction collector. Solvent A was 18 MΩ water containing 0.1 % trifluoroacetic acid (TFA), and solvent B was HPLC grade acetonitrile (AcN) with 0.1 % TFA. For purification, a semi-preparative Kromasil Eternity C18 column (10 mm × 250 mm, 5 μm particle size with 100 Å pore size) was used with a linear gradient of 38–48 % solvent B at a flow rate of 5 mL per min for 30 min. For analytical samples, an analytical Kromasil Eternity C18 column (4.6 mm × 250 mm, 5 μm particle size with 100 Å pore size) was used to determine purity with a linear gradient of 10–95 % solvent B at a flow rate of 1 mL per min for 27 min. Peptide purities were assessed by integration of peaks detected at 220 nm.
MALDI-TOF mass spectrometry (MS) data were obtained on a Bruker Relex II spectrometer equipped with a 337 nm laser and a reflectron. In positive ion mode, the acceleration voltage was 25 kV. Exact mass (EM) data were obtained on either a Waters (Micromass) LCT ESI-TOF mass spectrometer or a Thermo Q Exactive Plus ESI-Q-IT (orbitrap) mass spectrometer.
Nuclear magnetic resonance (NMR) spectroscopy experiments were performed on either a 750 MHz Bruker Avance III instrument equipped with a TCI cryogenic probe, or a 600 MHz Varian Inova instrument equipped with a Bruker cryogenic probe.
Solid-phase peptide synthesis
The peptidomimetics were prepared on Dawson 3-(Fmoc-amino)-4-aminobenzoyl (Dbz) AM resin (100–200 mesh) according to standard Fmoc SPPS methods described by Chan and White.[16] All SPPS reactions were mixed by agitation on a shaker table. Briefly, the resin (0.050 mmol equiv) was swelled in CH2Cl2 for 30 min, and the solvent was exchanged for dimethylformamide (DMF). Piperidine (20 % in DMF, 2 mL, 5 min × 3) was used to effect standard Fmoc group deprotections. For each amino acid coupling, DMF (2 mL) containing the Fmoc-protected amino acid (0.2 mmol), N-[(dimethylamino)-1H-1,2,3-tri-azolo-[4,5-b]pyridin-1-ylmethylene]-N-methylmethanaminium hexafluorophosphate N-oxide (HATU, 0.2 mmol), and diisopropylethylamine (DIPEA, 0.4 mmol) were preactivated for 2.5 min, then added to the resin for 30 min. To acetylate the N-terminus, acetic anhydride (0.5 mmol) and DIPEA (0.35 mmol) were dissolved in DMF (2 mL) and added to the resin for 15 min. All couplings and deprotections were preceded by washing three times with an appropriate solvent, and reaction completion was monitored by Kaiser test. To install the n7 oxo linker into n7FF, 8-(Fmoc-amino)-3,6-dioxaoctanoic acid was incorporated during SPPS. To install the amide linkages into n7FF amide, n8FF amide, and n7OFF amide, L-2,3-diaminopropionic acid (Dap) was incorporated instead of cysteine during SPPS.
To activate the Dbz group prior to peptidomimetic cleavage, the resin was exchanged into CH2Cl2 and treated twice with 4-nitrophenylchloroformate (0.25 mmol) in CH2Cl2 (2.5 mL) for 30 min. The resin was then exchanged into DMF, and cyclic urea was formed by twice adding DIPEA (216 μL, 1.24 mmol) in DMF (2 mL) for 10 min. The resin was washed sequentially with DMF (2 × 2 mL), CH2Cl2 (2 × 2 mL), and Et2O (2 × 2 mL), dried under N2, and reduced for 18 h under vacuum. To cleave the linear peptidomimetic, the resin was treated with a cleavage cocktail of TFA/CH2Cl2/H2O/triisopropyl silane (36:2:1:1; 3 mL) for 2 h. The mixture was filtered into a round-bottomed flask, and the resin was washed with cleavage cocktail (2 × 3 mL).
The resulting solution was concentrated by rotary evaporation and precipitated in Et2O at −20 °C overnight. The solid precipitate was dissolved in AcN/H2O (1:1), frozen by using iPrOH/dry ice, and lyophilized. The lyophilized linear peptidomimetic was purified by semipreparative RP-HPLC according to the method outlined above. Collected fractions were analyzed by MALDI-MS to isolate the desired product, frozen by using iPrOH/dry ice, and lyophilized.
Macrocyclization and final product isolation
Macrocyclization of the linear peptidomimetics (to give thiolactones) was performed according to our previously reported method,[4k] with minor modifications. Cyclization buffer was prepared by dissolving guanidinium chloride (18.32 g, 192 mmol) to a final volume of 32 mL in 0.1 M Na2HPO4, adding AcN (8 mL), and adjusting the pH to 6.8. The linear peptidomimetic was dissolved in cyclization buffer (4 mL) in a 15 mL conical tube and agitated on a shaker table at 50°C for 2 h. Macrocyclization of the linear peptidomimetics (to give amides) was performed according to our previously reported method,[4l] with minor modifications. The deprotected linear peptide was dissolved in anhydrous tetrahydrofuran (6 mL) with (7-azabenzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (PyAOP, 2 equiv) and DIPEA (4 equiv). Reaction progress was monitored by RP-HPLC, and reaction completion was observed within 6 h. The macrocyclic peptidomimetics were then purified by RP-HPLC, and their masses were evaluated by MALDI-MS.
To verify product identity, exact mass determinations were obtained for final products by using ESI-MS (Table S1). Peptidomimetic purities were checked by analytical RP-HPLC by using the methods described above. All samples displayed purities in excess of 90% by peak integration, with >95 % purity for n7FF, n8FF, and n7OFF (traces available in the Supporting Information). Isolated peptidomimetics were stored as 1 mM stock solutions in DMSO at 4°C, with the exception of n7OFF, which was stored as a 4 mM stock DMSO solution (used specifically for screening in group IV S. aureus).
Reporter gene assays
The peptidomimetics were assayed for AgrC I–IV inhibition with the four YFP reporter strains listed in Table 2. Peptidomimetic stock solutions were serially diluted with DMSO, and aliquots (2 μL) of the diluted solution were added to each of the wells in a black, clear-bottom 96-well microtiter plate (Costar). An overnight culture of S. aureus strain was diluted 1:50 with fresh BHI (pH 7.35). A 198 μL portion of diluted culture was added to each well of the microtiter plate containing peptide (resulting in a 1% DMSO solution). Plates were incubated at 37°C with shaking at 200 rpm for 24 h. Fluorescence (λex = 500 nm/λem = 540 nm) and OD600 of each well were then recorded by using a BioTek Synergy 2 plate reader. IC50 values and 95% confidence intervals were determined by using GraphPad Prism 6 software with four-parameter variable slope dose–response curves. For full dose–response curves for active compounds, see the Supporting Information.
Table 2.
Bacterial reporter strains used for AgrC inhibition assays.
NMR experimental protocols
NMR experiments on t-AIP-II, n7OFF, and AIP-III D4A were conducted at ambient temperature in H2O/D2O (95:5). The t-AIP-II sample had a concentration ≥3 mM and a pH ≈6, and NMR data were collected on a 600 MHz spectrometer. The n7OFF sample had a concentration ≥1.5 mM and a pH ≈7, and NMR data were collected on a 750 MHz spectrometer. The AIP-III D4A sample had a concentration ≥700 μM and a pH ≈6.5, and NMR data were collected on a 750 MHz spectrometer. Chemical shifts were referenced to H2O at 4.79 ppm. Dilutions were prepared to check for differences in proton chemical shifts that would indicate aggregation issues; no such differences were observed.
Four standard NMR experiments were used. A proton NMR experiment was performed to check for lock and shim quality. A 1D proton NMR experiment with excitation sculpting (Bruker pulse sequence zgesgp) was performed to check the signal-to-noise ratio and aid in sequential assignments. Parameters included a sweep width of 9 ppm, 4 s acquisition time, 3 s relaxation delay, 32 scans, 27 026 real points, and full spectral size of 65 536 points. A 2D TOCSY experiment (Bruker pulse sequence mlevesgpph) was used to identify the proton resonances associated with each amino acid residue. The spin-lock mixing time was set to 80 ms for n7OFF and 120 ms for t-AIP-II and AIP-III D4A. TOCSY parameters included a sweep width of 9 ppm, a 3 s relaxation delay, 256 points in the indirect dimension, 1024 (n7OFF) or 2048 (t-AIP-II and AIP-III D4A) points in the direct dimension, and 2 scans. To obtain internuclear proton distances through dipolar couplings, 2D ROESY experiments were performed (Bruker pulse sequence roesyesgpph). The ROESY spin-lock mixing time was varied, with final experiments performed at 300 ms. Additional ROESY parameters included a sweep width of 9 ppm, a 3 s relaxation delay, 1024 points in the indirect dimension, 8192 points in the direct dimension, and 16 scans.
A 1H,13C HSQC experiment (Bruker pulse sequence hsqcetgpsisp2.2) was used to differentiate the chemical shifts of several methylene protons in n7OFF. Parameters included a sweep width of 7 ppm in the direct dimension and 165 ppm in the indirect dimension, centered at 4.7 ppm in the direct dimension and centered at 90.0 ppm in the indirect dimension, 2048 points in the direct dimension and 512 points in the indirect dimension, 1H,13C coupling detection of 145 Hz, a 3 s relaxation delay, and 2 scans.
NMR spectra were analyzed by using MestReNova 10 NMR processing software. Representative spectra are shown in the Supporting Information. Resonance assignments were determined by using standard sequential methodology.[23] The ROESY crosspeak volumes were integrated, and integral values were entered in a spreadsheet. The values were calibrated to provide estimated distances for each crosspeak with a uniform ±20% constraint allowance to account for spin diffusion. The obtained distances were formatted for constraint files compatible with the Xplor-NIH software suite (v2.31).[19] Summaries of the constraint files are provided in the Supporting Information. Three-dimensional structure calculations and refinements made use of the torsion angle molecular dynamics and the internal variable dynamics modules[24] of Xplor-NIH, with patches for the thioester bridge and ring closure.[4h] The minimized target function was composed of the experimental NMR constraints (ROE-derived interproton distances and torsion angles), a repulsive van der Waals potential for the non-bonded contacts,[25] a torsion angle database potential of mean force,[26] and a gyration volume potential.[27]
The structure RMSD determinations and image files were prepared by using PyMOL.[28] To produce the topology and parameter files for the non-natural n7O amino acid, the structure was inputted into the PRODRG online server to create topology and parameter files, and the atoms in the output files were renamed to standard convention.[29] Ensembles of 20% of the lowest-energy structures were averaged for each molecule, and a structure with a low RMS difference relative to the average and low relative energy was selected for each ensemble as a representative structure.
Acknowledgments
This work was supported by the Office of Naval Research (N00014-16-1-2185), Kimberly-Clark Corporation, and the Burroughs Welcome Fund. J.K.V. was supported in part by the UW–Madison NIH Chemistry–Biology Interface Training Program (T32 GM008505 and the UW–Madison Office of the Vice Chancellor for Research and Graduate Education (with funding from the Wisconsin Alumni Research Foundation)), and a National Science Foundation (NSF) Graduate Research Fellowship (DGE-1256259). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by the National Institutes of Health (NIH; P41 GM103399, P41 GM66326, RR02781, and RR08438) and the NSF (DMB-8415048, OIA-9977486, and BIR-9214394). MS instrumentation in the UW–Madison Department of Chemistry was supported by the NIH (1S10 OD020022-1) and NSF (CHE-9974839). NMR instrumentation in the UW–Madison Department of Chemistry was supported by the NIH (S10 OD012245). We thank Prof. Alexander Horswill for generously providing the S. aureus reporter strains.
Footnotes
Supporting information and ORCID identification numbers for the authors for this article can be found under http://dx.doi.org/10.1002/cbic.201600516.
References
- 1.a) Rutherford ST, Bassler BL. Cold Spring Harbor Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bassler BL. Daedalus. 2012;141:67–76. [Google Scholar]; c) Fuqua C, Greenberg EP. Nat Rev Mol Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 2.a) Parker CT, Sperandio V. Cell Microbiol. 2009;11:363–369. doi: 10.1111/j.1462-5822.2008.01272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Waters CM, Bassler BL. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 3.a) Welsh MA, Blackwell HE. FEMS Microbiol Rev. 2016;40:774–794. doi: 10.1093/femsre/fuw009. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Galloway WRJD, Hodgkinson JT, Bowden S, Welch M, Spring DR. Trends Microbiol. 2012;20:449–458. doi: 10.1016/j.tim.2012.06.003. [DOI] [PubMed] [Google Scholar]; c) Galloway WRJD, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Chem Rev. 2011;111:28–67. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]; d) Amara N, Krom BP, Kaufmann GF, Meijler MM. Chem Rev. 2011;111:195–208. doi: 10.1021/cr100101c. [DOI] [PubMed] [Google Scholar]; e) Njoroge J, Sperandio V. EMBO Mol Med. 2009;1:201–210. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Welsh MA, Blackwell HE. Cell Chem Biol. 2016;23:361–369. doi: 10.1016/j.chembiol.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) O’Reilly MC, Blackwell HE. ACS Infect Dis. 2016;2:32–38. doi: 10.1021/acsinfecdis.5b00112. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Welsh MA, Eibergen NR, Moore JD, Blackwell HE. J Am Chem Soc. 2015;137:1510–1519. doi: 10.1021/ja5110798. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Moore JD, Rossi FM, Welsh MA, Nyffeler KE, Blackwell HE. J Am Chem Soc. 2015;137:14626–14639. doi: 10.1021/jacs.5b06728. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Gorske BC, Blackwell HE. Org Biomol Chem. 2006;4:1441–1445. doi: 10.1039/b517681f. [DOI] [PubMed] [Google Scholar]; f) Fowler SA, Stacy DM, Blackwell HE. Org Lett. 2008;10:2329–2332. doi: 10.1021/ol800908h. [DOI] [PubMed] [Google Scholar]; g) Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. J Am Chem Soc. 2013;135:7869–7882. doi: 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]; h) Tal-Gan Y, Ivancic M, Cornilescu G, Cornilescu CC, Blackwell HE. J Am Chem Soc. 2013;135:18436–18444. doi: 10.1021/ja407533e. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Tal-Gan Y, Stacy DM, Blackwell HE. Chem Commun. 2014;50:3000–3003. doi: 10.1039/c4cc00117f. [DOI] [PubMed] [Google Scholar]; j) Yang T, Tal-Gan Y, Paharik AE, Horswill AR, Blackwell HE. ACS Chem Biol. 2016;11:1982–1991. doi: 10.1021/acschembio.6b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Tal-Gan Y, Ivancic M, Cornilescu G, Blackwell HE. Org Biomol Chem. 2016;14:113–121. doi: 10.1039/c5ob01735a. [DOI] [PMC free article] [PubMed] [Google Scholar]; l) Tal-Gan Y, Ivancic M, Cornilescu G, Yang T, Blackwell HE. Angew Chem Int Ed. 2016;55:8913–8917. doi: 10.1002/anie.201602974. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2016;128:9059–9063. [Google Scholar]
- 5.a) O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. Proc Natl Acad Sci USA. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chen G, Swem LR, Swem DL, Stauff DL, O’Loughlin CT, Jeffrey PD, Bassler BL, Hughson FM. Mol Cell. 2011;42:199–209. doi: 10.1016/j.molcel.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Müh U, Hare BJ, Duerkop BA, Schuster M, Hanzelka BL, Heim R, Olson ER, Greenberg EP. Proc Natl Acad Sci USA. 2006;103:16948–16952. doi: 10.1073/pnas.0608348103. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Müh U, Schuster M, Heim R, Singh A, Olson ER, Green-berg EP. Antimicrob Agents Chemother. 2006;50:3674–3679. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Morkunas B, Galloway WRJD, Wright M, Ibbeson BM, Hodgkinson JT, O’Connell KMG, Bartolucci N, Della Valle M, Welch M, Spring DR. Org Biomol Chem. 2012;10:8452–8464. doi: 10.1039/c2ob26501j. [DOI] [PubMed] [Google Scholar]; f) Morkunas B, Gal B, Galloway WRJD, Hodgkinson JT, Ibbeson BM, Tan YS, Welch M, Spring DR. Beilstein J Org Chem. 2016;12:1428–1433. doi: 10.3762/bjoc.12.137. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Amara N, Gregor R, Rayo J, Dandela R, Daniel E, Liubin N, Willems HME, Ben-Zvi A, Krom BP, Meijler MM. ChemBioChem. 2016;17:825–835. doi: 10.1002/cbic.201500676. [DOI] [PubMed] [Google Scholar]; h) Delago A, Mandabi A, Meijler MM. Isr J Chem. 2016;56:310–320. [Google Scholar]; i) Amara N, Mashiach R, Amar D, Krief P, Spieser SAH, Bottomley MJ, Aharoni A, Meijler MM. J Am Chem Soc. 2009;131:10610–10619. doi: 10.1021/ja903292v. [DOI] [PubMed] [Google Scholar]; j) Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Lyon GJ, Mayville P, Muir TW, Novick RP. Proc Natl Acad Sci USA. 2000;97:13330–13335. doi: 10.1073/pnas.97.24.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]; l) Lyon GJ, Wright JS, Muir TW, Novick RP. Biochemistry. 2002;41:10095–10104. doi: 10.1021/bi026049u. [DOI] [PubMed] [Google Scholar]; m) Wright JS, Lyon GJ, George EA, Muir TW, Novick RP. Proc Natl Acad Sci USA. 2004;101:16168–16173. doi: 10.1073/pnas.0404039101. [DOI] [PMC free article] [PubMed] [Google Scholar]; n) George EA, Novick RP, Muir TW. J Am Chem Soc. 2008;130:4914–4924. doi: 10.1021/ja711126e. [DOI] [PubMed] [Google Scholar]; o) Wang B, Zhao A, Novick RP, Muir TW. Mol Cell. 2014;53:929–940. doi: 10.1016/j.molcel.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; p) Johnson JG, Wang B, Debelouchina GT, Novick RP, Muir TW. ChemBioChem. 2015;16:1093–1100. doi: 10.1002/cbic.201500006. [DOI] [PMC free article] [PubMed] [Google Scholar]; q) McDowell P, Affas Z, Reynolds C, Holden MT, Wood SJ, Saint S, Cockayne A, Hill PJ, Dodd CE, Bycroft BW, Chan WC, Williams P. Mol Microbiol. 2001;41:503–512. doi: 10.1046/j.1365-2958.2001.02539.x. [DOI] [PubMed] [Google Scholar]; r) Scott RJ, Lian LY, Muharram SH, Cockayne A, Wood SJ, Bycroft BW, Williams P, Chan WC. Bioorg Med Chem Lett. 2003;13:2449–2453. doi: 10.1016/s0960-894x(03)00497-9. [DOI] [PubMed] [Google Scholar]; s) Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. J Microbiol Methods. 2009;77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; t) Wang B, Muir TW. Cell Chem Biol. 2016;23:214–224. doi: 10.1016/j.chembiol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; u) Ciardiello JJ, Galloway WRJD, O’Connor CJ, Sore HF, Stokes JE, Wu YT, Spring DR. Tetrahedron. 2016;72:3567–3578. [Google Scholar]
- 6.a) Kratochvil MJ, Tal-Gan Y, Yang T, Blackwell HE, Lynn DM. ACS Biomater Sci Eng. 2015;1:1039–1049. doi: 10.1021/acsbiomaterials.5b00313. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Broderick AH, Stacy DM, Tal-Gan Y, Kratochvil MJ, Blackwell HE, Lynn DM. Adv Healthcare Mater. 2014;3:97–105. doi: 10.1002/adhm.201300119. [DOI] [PubMed] [Google Scholar]
- 7.a) Le KY, Otto M. Front Microbiol. 2015;6:1174. doi: 10.3389/fmicb.2015.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Novick RP, Geisinger E. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 8.a) Thammavongsa V, Kim HK, Missiakas D, Schneewind O. Nat Rev Microbiol. 2015;13:529–543. doi: 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Queck SY, Jameson-Lee M, Villaruz AE, Bach THL, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. Mol Cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Ji G, Beavis R, Novick RP. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]; b) Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 10.a) Zhang L, Lin J, Ji G. J Biol Chem. 2004;279:19448–19456. doi: 10.1074/jbc.M311349200. [DOI] [PubMed] [Google Scholar]; b) Qiu RD, Pei WH, Zhang LS, Lin JQ, Ji GY. J Biol Chem. 2005;280:16695–16704. doi: 10.1074/jbc.M411372200. [DOI] [PubMed] [Google Scholar]; c) Kavanaugh JS, Thoendel M, Horswill AR. Mol Microbiol. 2007;65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- 11.Jarraud S, Lyon GJ, Figueiredo AMS, Gérard L, Vandenesch F, Etienne J, Muir TW, Novick RP. J Bacteriol. 2000;182:6517–6522. doi: 10.1128/jb.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Gupta RK, Luong TT, Lee CY. Proc Natl Acad Sci USA. 2015;112:14036–14041. doi: 10.1073/pnas.1509251112. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyon GJ, Wright JS, Christopoulos A, Novick RP, Muir TW. J Biol Chem. 2002;277:6247–6253. doi: 10.1074/jbc.M109989200. [DOI] [PubMed] [Google Scholar]
- 14.Wright JS, Jin R, Novick RP. Proc Natl Acad Sci USA. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson MM, Mack JL, Hall PR, Alsup AA, Alexander SM, Sully EK, Sawires YS, Cheung AL, Otto M, Gresham HD. Cell Host Microbe. 2008;4:555–566. doi: 10.1016/j.chom.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan W, White P. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. OUP; Oxford: 2000. [Google Scholar]
- 17.a) Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS. J Infect Dis. 2008;198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]; b) CDC. Morb Mortal Weekly Rep. 2003;52:793–795. [PubMed] [Google Scholar]; c) CDC. Morb Mortal Weekly Rep. 2003;52:992–996. [PubMed] [Google Scholar]
- 18.Compound n9FF was not tested in the group IV S. aureus reporter, as the one-carbon-smaller homologue n8FF was found to be less active in this group relative to n7FF and t-AIP-II.
- 19.a) Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]; b) Schwieters CD, Kuszewski JJ, Clore GM. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. [Google Scholar]
- 20.Eisenberg D, Schwarz E, Komaromy M, Wall R. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 21.a) Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. J Bacteriol. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kirchdoerfer RN, Garner AL, Flack CE, Mee JM, Horswill AR, Janda KD, Kaufmann GF, Wilson IA. J Biol Chem. 2011;286:17351–17358. doi: 10.1074/jbc.M111.231258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malone CL, Boles BR, Horswill AR. Appl Environ Microbiol. 2007;73:6036–6044. doi: 10.1128/AEM.00912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wüthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- 24.Schwieters CD, Clore GM. J Magn Reson. 2001;152:288–302. doi: 10.1006/jmre.2001.2413. [DOI] [PubMed] [Google Scholar]
- 25.Nilges M, Clore GM, Gronenborn AM. FEBS Lett. 1988;229:317–324. doi: 10.1016/0014-5793(88)81148-7. [DOI] [PubMed] [Google Scholar]
- 26.Clore GM, Kuszewski J. J Am Chem Soc. 2002;124:2866–2867. doi: 10.1021/ja017712p. [DOI] [PubMed] [Google Scholar]
- 27.Schwieters CD, Clore GM. J Phys Chem B. 2008;112:6070–6073. doi: 10.1021/jp076244o. [DOI] [PubMed] [Google Scholar]
- 28.The PyMOL Molecular Graphics System, Version 1.7. Schrödinger, LLC; 2015. [Google Scholar]
- 29.Schüttelkopf AW, van Aalten DMF. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]