Abstract
CD38 is an activation marker that is present on recently activated T cells, but absent on resting memory T cells. In this study, we show that CD45RO+CD38+ beta cell antigen-specific CD4+ T cells were present at higher frequencies in T1D subjects compared to those in healthy subjects. These results imply an ongoing beta cell immunity years after onset of diabetes and suggest these activated T cells have an active role in the disease process. The antigen specificities of these activated T cells were determined by a novel CD154 T cell epitope mapping assay. While each patient usually had a unique set of epitopes recognized by these T cells, two epitopes, DR0401 restricted modified preproinsulin peptide (PPI78-90K88S) and ZnT266-285, were repeatedly identified in multiple subjects. Identifying these T cells and their specific antigenic epitopes might provide immunotherapeutic targets for personalized therapies.
Introduction
Type 1 diabetes (T1D) is a chronic inflammatory autoimmune disease in which islet beta cells are selectively destroyed [1, 2]). Autoreactive T cells that recognize islet beta-cell antigens (βCAs) [3–5] play a major role in disease pathogenesis [6–9]. Susceptibility to disease incidence is highly associated with HLA class II genes, particularly the DR4-DQ8 and DR3-DQ2 haplotypes [10, 11]. CD4 T cells specific to βCAs are speculated to be the major players in initiating the pathogenic process [6, 12]. Many studies have demonstrated that CD4+ T-cells, isolated from T1D patients or from the islets and pancreata of NOD mice, recognize antigenic peptides derived from βCAs. These include preproinsulin [13, 14], glutamic acid decarboxylase (GAD) 65 and 67 isotypes [15], islet glucose-phosphatase catalytic subunit related protein (IGRP) [16, 17], chromogranin A (ChgA) [18, 19], and zinc transporter 8 (ZnT 8) [20, 21]. Numerous antigenic epitopes derived from these antigens have been identified [3, 4]. Experiments in NOD mice, which carry a single MHC class II allele and unique genetic background, demonstrated that the Insulin B9-23 epitope is the major auto-antigenic epitope in T1D pathogenesis [14]. However, in humans who carry multiple class II alleles and a diverse genetic background, antigens and epitopes critical to the disease process are not fully defined.
We hypothesized that each individual T1D subject has a unique repertoire of βCA-specific T cells. Identification of the T cell epitopes of these antigens and detection of these epitope specific T cells could facilitate the understanding of T1D pathogenesis and subsequent development of antigen specific immunomodulation therapies.
We also hypothesized that destruction of beta cells in islets is an active process that continues even after onset of T1D, and that T1D subjects years after disease onset could have recently activated βCA-specific T cells. We investigated the use of biomarkers that track activated βCA-specific T cells for identification of target antigens and T cell epitopes critical to the disease process. CD38 is a type II glycoprotein that is expressed on surface naïve T cells and recently activated memory T cells [22–24], but is not expressed in resting memory T cells [23, 24]. Thus, the presence of antigen specific CD38+ memory T cells for a given antigen is an indication of active immunity directed against that antigen [23, 24].
We exploited the specificity of CD38 expression on memory T cells to identify βCA-specific CD4+ T cells from PBMC that were immune active in vivo. We used a CD154 up-regulation assay to track overall βCA-specific CD4+ T cells ex vivo. With the combination of CD38 and CD45RA/RO markers, we were able to detect, enumerate and compare the frequency of immune active (CD45RO+CD38+) βCA-specific T cells in T1D subjects and healthy controls. Furthermore, we developed a novel CD154 up-regulation epitope mapping assay to define the epitope specificity of these βCA-specific T cells. This study provides a novel strategy to investigate the role of CD4 T cells in T1D pathogenesis. In the future, this approach can potentially be used to tailor personalized antigen specific immunomodulation therapy.
Materials and methods
Human subjects
The healthy donors and T1D patients were recruited locally after approval by the Benaroya Research Institute Institutional Review Board and with the written consent of participants. A total of 15 subjects were recruited for each group. Age, duration of disease and serum C-peptide status for the T1D patients is shown in Supplemental Table 1. The healthy control group was age-matched with the T1D group. All subjects in both the T1D group and healthy control group had the T1D disease susceptible HLA-DRB1*0401-DQB1*0302 haplotype.
Peptides
For T1D associated auto-antigens, peptide libraries containing peptides of 20 aa in-length with 12 aa overlapping of adjacent peptides to cover each protein were synthesized (Mimotopes, Clayton, Victoria, Australia). Glutamate decarboxylase isoform 65 (GAD65) protein (Access#: NP_000809) had 72 peptides; zinc transporter 8 (ZnT8) protein (Access#: NP_776250.2) had 39 peptides; islet specific glucose-6-phosphatase catalytic subunit-related protein (IGRP) (Access#: NP_066999) had 43 peptides; chromogranin-A (ChgA) protein (Access#: AAP36245.1) had 56 peptides; and preproinsulin protein (Access#: NP_001008996) had 2 modified peptides, PPI78-90K88S[25] and PPI35-47R46E (InsB:11-23R22E) [26]. Peptide library for influenza A matrix protein (MP) (Access#: ACP41108) had 30 peptides.
Ex vivo T cell activation, CD154 enrichment and T cell sorting
A modified CD154 up-regulation assay was used to identify βCA -specific T cells ex vivo. Briefly, 20 million freshly isolated PBMC were stimulated in vitro for 3 h with a set of βCA peptide libraries containing GAD65, IGRP, ZnT8, ChgA and two modified preproinsulin peptides, a total of 212 peptides, at the final concentration of 0.5 µg/mL for each peptide in the presence of anti-CD40 (1 µg/mL) (clone HB-14, Miltenyi Biotec, San Diego, CA). Anti-CD40 antibody was added during the in vitro stimulation to prevent the down regulation of CD154 molecules through CD40/CD154 interaction on newly activated T cells [27]. After stimulation, PBMC were collected and stained with anti-CD154-PE antibody (clone 5C8, Miltenyi Biotec, San Diego, CA) followed by labeling with anti-PE microbeads (clone PE4-14D10, Miltenyi Biotec, San Diego, CA). The antigen responsive T cells with up-regulated CD154 were enriched on a magnetic bar according to the manufacture’s protocol. The enriched cells were further labeled with antibodies (all from BD Biosciences) including anti-CD3-V500 (clone SP34-2), anti-CD4-APC-H7 (clone RPA-T4), anti-CD45RA-PE-Cy7 (clone HI 100), anti-CD45RO-FITC (clone UCHL1), anti-CD38-V450 (clone HB7), anti-CD69-APC (clone L78), anti-CD14-PerCP (clone MΦ9) and anti-CD19-PerCP (clone Leu-12). Via-Probe™ (BD Biosciences), together with anti-CD14 and anti-CD19 were used to dump CD14+, CD19+ and dead cells. βCA responsive T cells were identified by up-regulation of CD154 and CD69 on CD4 T cells. CD154+CD69+ CD45RO+CD38+ cells were identified as in vivo immune active islet βCA-specific T cells. Anti-CD278-BV786 (clone DX29) was used to evaluate ICOS expression. In samples with significant numbers of CD45RO+CD38+ T cells, the T cells were sorted out and expanded as oligo-clones.
Gating strategy to determine CD38 positivity on CD154+ cells
In order to set the flow cytometric gate to define positivity of CD45RO and CD38 markers, total CD4 T cells collected from a pre-enrichment tube were used. The gate was set at a position such that the sum of CD45RO-CD38+ and CD45RO+CD38- populations was at a maximum, and the CD45RO+CD38+ population was at a minimum. Using this strategy, the activated memory T cells were set at the lowest possible level to avoid an overestimation of the activated memory T cell population.
Oligocloning and expansion of T cells
Sorted antigen specific T cells were seeded into round bottom 96-well plate at 10 cells/well, with 1.5X105 irradiated PBMC as feeder cells, in 200 µL of T cell culture medium (TCM: RPMI1640 containing 10% (v/v) of pooled human serum, 1% (v/v) of L-Glutamine, sodium pyruvate, and Penicillin/Streptomycin, all Invitrogen, CA) and 1 µg/mL of PHA. On the next day, each well was supplemented with 40 IU (in 10 µL of TCM) of recombinant human IL-2 (Roche, IL). After 7–10 days culture at 37°C, 5% CO2, expanded T cells became visible pellets in the 96-well plate. These T cell pellets were transferred to the flat-bottom 96-well plate and fed with 100 µL of fresh TCM supplemented with 200 IU/mL of IL-2. When the T cells became confluent in the plate, the cells were split and fed with fresh TCM and IL-2, and eventually placed in a 48-well plate to obtain 5–10X106 of T cells for CD154 epitope mapping assay.
CD154 epitope mapping
Once the T cells were successfully expanded, T cells from each oligoclone were washed and suspended at 0.5X106/mL in TCM containing 1 µg/mL of CD40 blocking antibody. 105 T cells in 200 µL from each oligoclone were stimulated with 43 different pools of 5 T1D antigen peptides and with DMSO only as a negative control. The cells were stimulated for 3 h, and then stained with Abs against CD3-FITC, CD4-PerCP, CD69-APC and CD154-PE for 10 min. After washing, the upregulation of CD154 after antigen stimulation was analyzed by flow cytometry. If a well contained a subpopulation of T cells responding to the T1D antigen peptide pool stimulation, a second round of CD154 epitope mapping assay was performed with individual peptides contained in that T1D antigen peptide pool.
Tetramer staining
Ex vivo tetramer staining and phenotype analysis were performed as described previously [17].
TCR Vbeta sequencing
CD4+ T cells from PBMC were sorted into three fractions: CD45RO-(5.6X106 naïve T cells), CD45RO+CD38- (3.2X106 resting memory T cells), and CD45RO+CD38+ (0.23X106 activated T cells). Genomic DNA from each fraction was isolated and subjected to TCR Vbeta CDR3 region deep sequencing by Adaptive Biotechnology (Seattle, WA)[28]. Preproinsulin specific T cell oligoclones were also subjected to survey sequencing by Adaptive Biotechnology.
Statistical analysis
We used Student’s t tests and Pearson correlation analyses in this study as described in each figure legend.
Results
CD38 expression on memory CD4+ T cells as an indication of recent antigen-specific activation
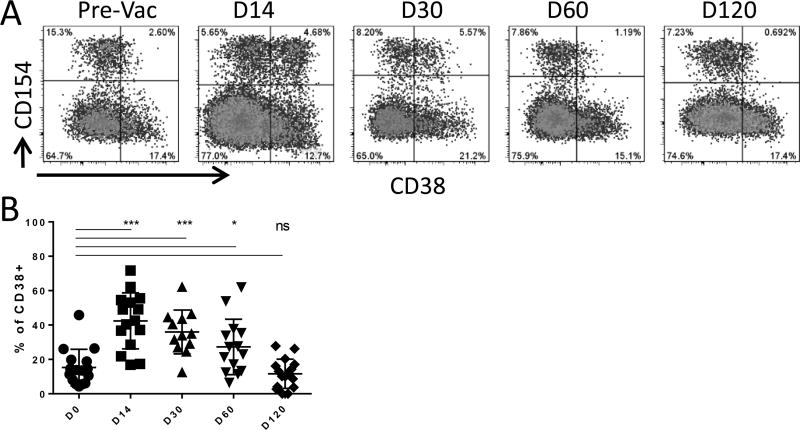
In our previous influenza vaccination and infection study, we noticed that the expression of CD38 on memory T cells was dynamically regulated [24]. To confirm that CD38 could be a useful marker in identifying recent in vivo activated T cells, the kinetics of CD38 expression on influenza A MP specific T cells from subject pre- and post- FLUZONE vaccination were examined. For these experiments, PBMC were stimulated with a peptide library covering MP for 3h in vitro. MP peptide-activated T cells in total PBMC were stained with anti-CD154-PE and enriched using anti-PE magnetic beads. The enriched cells were stained with anti-CD4, anti-CD45RA, anti-CD38, and anti- CD14/CD19/via-Probe. Cells were gated on CD4+CD45RA- memory T cells, and CD38 expression on CD154+ T cells was examined at different time points (Figure 1 A). MP-specific memory CD4+ T cells were CD38- or very dim before vaccination. This result also suggested that CD38 was not upregulated by 3 hrs. of peptide stimulation. A substantial proportion of these cells expressed CD38 at day 14 post-vaccination. The proportion of MP-specific cells that expressed CD38 declined at 4 weeks after vaccination, and gradually returned to the pre-vaccination level after 2–3 months (Figure 1 A and B). This result implies that CD38 expression on memory T cells can be used as a marker for recently activated T cells and suggests that the duration of CD38 expression on Flu specific memory CD4+ T cells lasted for 2–3 months after activation (Figure 1 A and B).
Figure 1. Dynamics of CD38 expression on memory CD4+ T cells after activation.
A. Dynamics of CD38 expression on MP-specific memory CD4+ T cells during the course of vaccination. PBMC from pre-, day 14, 30, 60 and 120 post-FLUZONE vaccination were stimulated with Flu MP peptide library for 3 hrs in vitro. Analyzed cells were only gated on CD4+CD45RO+ memory T cells. The CD154+CD38+ population marked the recently activated MP specific T cells. Data are from 1 representative subject.
B. Summary of CD38 expression dynamics during the course of vaccination for n = 12 subjects. Student’s t test was used to compare % of CD38+ MP specific memory T cells before and after defined days of vaccination. Data were compared using two-tail paired Student’s t test, *: p<0.05, ***: p<0.0001.
Beta cell antigen (βCA) specific T cells from subsets of T1D patients contained recently activated T cells
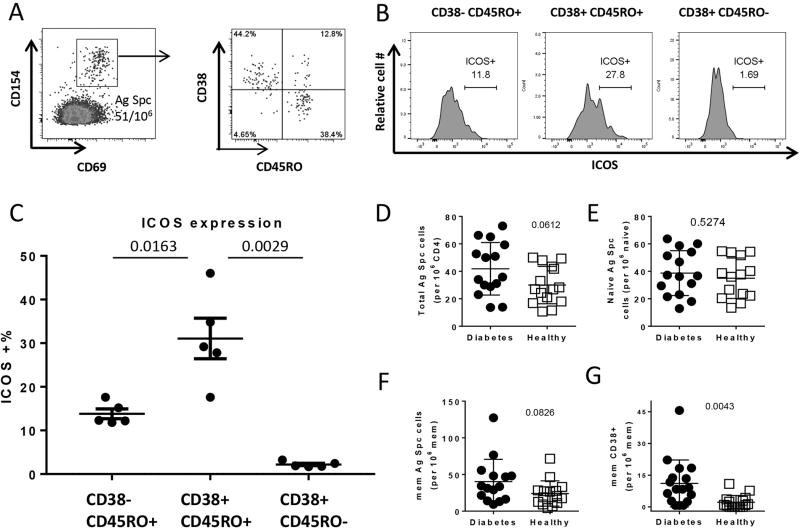
For further evaluation of whether CD38 expression could be used to identify recently activated βCA-specific CD4+ T cells, PBMC from T1D subjects were stimulated with 210 overlapping peptides derived from 4 different βCAs: GAD65, IGRP, ZnT8, ChgA with the addition of two modified preproinsulin peptides: DR0401 restricted preproinsulin 78-90K88S (PPI78-90K88S) [25] and DQ8 restricted PPI35-47R46E (InsB11-23R22E) [26]. βCA-specific CD4 T cells were identified by the co-expression of CD154 and CD69 surface markers on CD4+ T cells. Figure 2A shows the representative staining profiles from a T1D subject. The CD45RO and CD38 phenotypes of these CD154+CD69+ βCA-specific T cells were analyzed (right panels of Figure 2A). These panels show three major subsets, the naïve CD45RO-CD38+ subset, and the CD45RO+CD38- and CD45RO+CD38+ memory subsets of βCA-specific CD4 T cells. Phenotype of islet specific cells within these different CD38 populations in T1D subjects was further evaluated by examining another T cell activation marker ICOS. Example of ICOS expression in one subject is show in Figure 2B, and results from 5 T1D subjects are shown in Figure 2C. We observed that significant higher percentage of islet specific cells within the CD45RO+CD38+ subset expressed ICOS compared to CD45RO+CD38- and CD45RO-CD38+ subsets. This data support the premise that the CD45RO+CD38+ as the activated memory subset and the CD45RO+CD38- as the less activated or resting memory subset.
Figure 2. Frequency and Phenotype analysis of βCA-specific CD4+ T cells in T1D and healthy subjects.
A. Detection of antigen-specific CD4+ T cells by CD154 and CD69 surface markers and expression of CD38 on memory cells in a T1D subject.
B. ICOS expression on different CD154+CD69+CD38+/- subsets in a T1D subject.
C. ICOS expression on different CD154+CD69+CD38+/- subsets in 5 T1D subjects.
D. Comparison of total βCA-specific CD4+ T cells between T1D subjects and healthy controls.
E. Comparison of naïve (CD4+CD45RO-) βCA-specific T cells between T1D subjects and healthy controls.
F. Comparison of memory (CD4+CD45RO+) βCA-specific T cells between T1D subjects and healthy controls.
G. Comparison of immune active (CD4+CD45RO+CD38+) memory βCA specific T cells between T1D subjects and healthy controls.
Data were compared using two-tail unpaired Student’s t test.
Frequencies of total (CD154+CD69+), naïve (CD45RO-), memory (CD45RO+) and immune active memory (CD45RO+CD38+) βCA-specific T cells in both T1D and healthy subjects were then examined (Figure 2D–G). Statistically significant differences were observed for immune-active memory antigen specific T cells between T1D and healthy subjects (Student’s t test: p=0.0043), but not for total (Student’s t test, p=0.0612), naïve (Student’s t test: p=0.5274) or memory (Student’s t test: p=0.0826) antigen specific T cells (Figure 2 B–E). Percentages of CD45RO+ βCA-specific T cells which were CD38+ were also higher in T1D subjects compared to controls (data not shown). Correlation between frequency of islet specific cells and duration of disease was not observed within the T1D group.
Active immune responses were focused on a unique set of antigenic epitopes in each patient
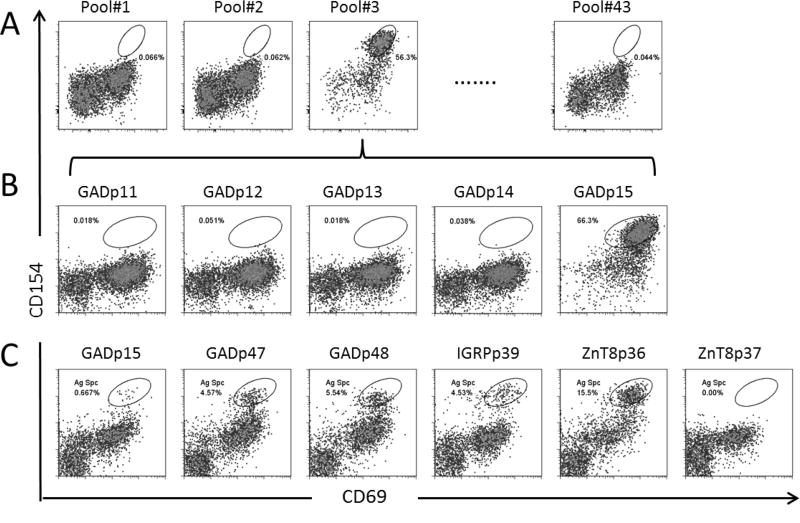
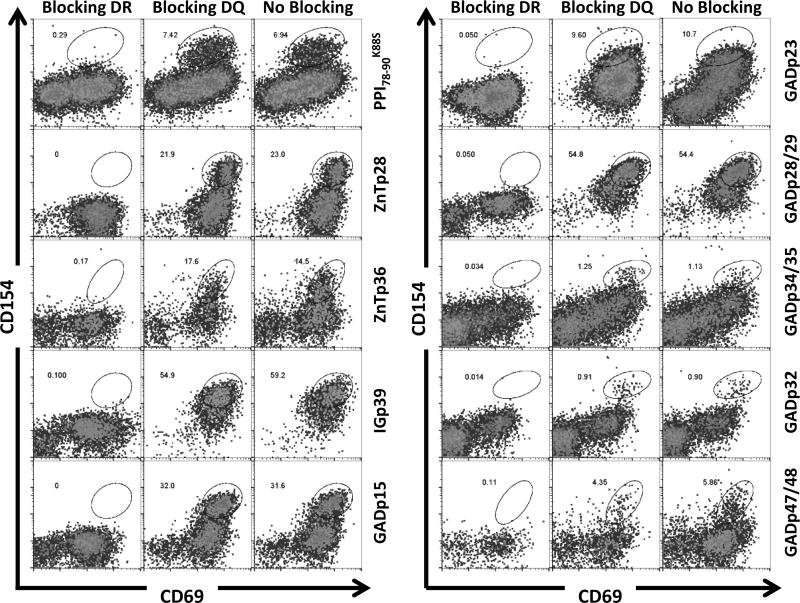
Next we sought to identify the antigenic epitopes recognized by the CD38+ βCA-specific cells. To achieve this, CD154+CD69+CD38+CD45RO+ T cells were sorted into a 96 well plate with 10 cells per well and expanded as oligoclones with PHA. After 1 month of expansion, each oligoclone was aliquoted into 43 different wells in a 96-well plate and stimulated with 43 pools of peptides (5 peptides per pool for a total of 212 peptides). Antigen specific T cells upregulated CD154 and CD69 upon peptide stimulation (Figure 3A). When a positive response was identified, the responding oligoclone was re-stimulated with individual peptides from the positive peptide pool to define the antigenic peptide (Figure 3B). In this way, specific epitopes of the T cell lines were identified. Figure 3C shows multiple antigenic peptides that were identified from one oligoclone. Table 1 lists peptides that contain antigenic epitopes to elicit active immune responses in four T1D patients. Supplemental Table 2 lists the frequency of peptide identified in all CD38+CD45RO+ oligoclones. Interestingly, CD154+CD69+CD38+CD45RO+ T cells from each patient recognized a unique set of antigenic peptides. The responses to the GAD65 and IGRP were diverse, with no identical epitopes identified across patients. In contrast, responses to PPI78-90K88S and ZnT8266-285 were detected in all 4 subjects tested. Response to ChgA was not observed. To determine whether the epitopes recognized by these CD38+ T cells were presented by HLA-DR or HLA-DQ, we analyzed HLA restrictions for 10 oligoclones that we had cells available (Figure 4). We found that the reactivities of all these oligoclones were blocked by anti-HLA-DR antibody but not by anti-HLA-DQ antibody, including the reactivity to GADp32-specific peptide, which has been shown to contain a HLA-DQ restricted epitope [29] (Figure 4). These results suggest that all these oligoclones recognizes peptides presented on HLA-DR.
Figure 3. Identification of T cell epitopes by CD154 epitope mapping assay.
A. Pooled epitope mapping on T cell oligoclone #3. T cell oligoclone #3 from subject K787T1D was stimulated with 43 peptide pools, respectively, each contained 5 peptides with 212 peptides in total. Peptide pool#3 stimulated oligoclone #3 to upregulate CD154 and CD69.
B. Individual peptide epitope mapping. T cell oligoclone #3 was stimulated with individual peptides contained in peptide pool #3 and GADp15 was identified as the antigenic epitope
C. Multiple antigenic T cell epitopes were identified in T cell oligoclone #4 from subject K787T1D.
Table 1.
Antigenic epitopes identified in 4 T1D subjects
| T1D Subject | Protein | Peptide # | Peptide Sequence |
|---|---|---|---|
|
| |||
| K787T1D | Insulin | PPI78-90K88S | QPLALEGSLQSRG |
|
| |||
| GAD65 | GADp15 | DVMNILLQYVVKSFDRSTKV | |
| GADp45 | ICKKYKIWMHVDAAWGGGLL | ||
| GADp47 | GGLLMSRKHKWKLSGVERAN | ||
| GADp48 | HKWKLSGVERANSVTWNPHK | ||
| GADp55 | YDLSYDTGDKALQCGRHVDV | ||
|
| |||
| ZnT8 | ZnT8p28 | ILKDFSILLMEGVPKSLNYS | |
| ZnT8p36 | VRREIAKALSKSFTMHSLTI | ||
|
| |||
| IGRP | IGp39 | QLYHFLQIPTHEEHLFYVLS | |
|
| |||
| ChgA | NEI | ||
|
| |||
| K276T1D | Insulin | PPI78-90K88S | QPLALEGSLQSRG |
|
| |||
| GAD65 | GADp23 | RYFNQLSTGLDMVGLAADWL | |
|
| |||
| ZnT8 | ZnT8p28 | ILKDFSILLMEGVPKSLNYS | |
|
| |||
| IGRP | IGp31 | KWCANPDWIHIDTTPFAGLV | |
|
| |||
| ChgA | NEI | ||
|
| |||
| K743T1D | Insulin | PPI78-90K88S | QPLALEGSLQSRG |
|
| |||
| GAD65 | GADp28 | EYVTLKKMREIIGWPGGSGD | |
| GADp29 | REIIGWPGGSGDGIFSPGGA | ||
| GADp32 | YAMMIARFKMFPEVKEKGMA | ||
| GADp34 | KGMAALPRLIAFTSEHSHFS | ||
| GADp35 | LIAFTSEHSHFSLKKGAAAL | ||
|
| |||
| ZnT8 | ZnT8p28 | ILKDFSILLMEGVPKSLNYS | |
|
| |||
| IGRP | IGp23 | HQVILGVIGGMLVAEAFEHT | |
|
| |||
| ChgA | NEI | ||
|
| |||
| K877T1D # | GAD65 | GADp70 | KVNFFRMVISNPAATHQDID |
|
| |||
| K877T1D* | Insulin | PPI78-90K88S | QPLALEGSLQSRG |
|
| |||
| GAD65 | GADp10 | CACDQKPCSCSKVDVNYAFL | |
| GADp11 | SCSKVDVNYAFLHATDLLPA | ||
|
| |||
| ZnT8 | ZnT8p28 | ILKDFSILLMEGVPKSLNYS | |
|
| |||
| IGRP | NEI | ||
|
| |||
| ChgA | NEI | ||
NEI: No Epitope Identified
For subject K877T1D, only four CD38+ memory T cells were obtained for oligoclonal expansion and epitope mapping.
Since only four CD38+ memory T cells were obtained from subject K877T1D for oligoclonal expansion and epitope mapping, oligoclones expanded from resting memory CD4 T cells were used to identify the epitopes for this subject.
Figure 4. Determine HLA restriction of antigenic peptide.
Ten Oligoclones were stimulated with corresponding peptides in the presence or absence of HLA-DR (L243) or –DQ8 (SPVL3) blocking antibody, respectively. The HLA restriction of antigen specific T cells was determined by blocking of expression of activation markers CD154 and CD69.
Persistence of TCR of expanded clonotype in the periphery
Since CD38+ PPI78-90K88S T cells were prevalent in the T1D subjects, we evaluated whether PPI-reactive T cells of a specific clonotype could be maintained in the periphery. For that purpose, CD38+ oligoclones from subject K276T1D (isolated as described above) were stimulated with PPI78-90K88S. CD154+CD69+ cells were sorted and expanded. Specificity of these cells to PPI78-90K88S was confirmed by upregulation of CD154 upon reactivation. Deep TCR sequencing was carried out with these oligoclonal populations. (Supplemental Table 3 shows the top 10 functional Vbeta sequences).
To determine whether PPI78-90K88S specific T cells of identical clonotype were being retained in the periphery, the subject K276T1D was recalled 4 months after the oligocloning assay was performed. PBMC were sorted into naïve (CD45RO-, 5.6X106 cells), resting memory (CD45RO+CD38-, 3.2X106 cells) and immune-active memory (CD45RO+CD38+, 0.23X106 cells) CD4+ T cells. The TCR repertoire of each population was subjected to genomic TCR Vbeta deep sequencing analysis and 163518, 110264 and 66484 functionally rearranged TCR Vbeta sequences were obtained from each population, respectively. When the DNA sequences of the top 10 rearranged Vbeta VDJ regions of PPI78-90K88S specific T cells were compared to the TCR repertoire from the above 3 populations, the PPI78-90K88S specific TCR VDJ sequence (TGTGCCAGCAGTTACGGAATAGGGAGGGCAGATACGCAGTATTTT) was detected as one of the productive TCR VDJ regions in the memory population (1 out of 110264 unique TCR Vbeta). This Vbeta sequence is equivalent to 3.7 cells per million [30] (Supplemental Figure 1). This suggests that PPI specific T cells can divide and persist in the periphery for at least 4 months.
Discussion
This study demonstrates the presence of in vivo immune-active βCA-specific T cells in T1D subjects. While activation markers like CD69 and CD25 can be used to define T cell activation status, their upregulation is relatively transient [31, 32]. Expression of CD38 in memory cells after activation can last for roughly 2 to 3 months [23, 24], making it an ideal biomarker to track T cells that are actively involved in autoimmune responses. Our results show that activated βCA-specific T cells are present at a much higher frequency in T1D subjects compared to healthy controls. These data suggest that βCA-specific T cells were likely activated by islet beta cell antigens in vivo, and might be involved in T1D pathogenesis. Indeed C-peptide could also be detected in serum of our cohort (Supplementary Table 1). Presence of C-peptide in the serum indicates the presence of viable beta cells, and these beta cells act as a source of autoantigen for the activation of βCA-specific T cells.
To identify the epitope specificity of activated βCA-specific T cells, we developed a CD154 up-regulation epitope mapping assay. We analyzed samples from 4 T1D subjects, and we detected a unique population of βCA-specific T cells with different epitope specificities in each subject. Among the 5 βCA we tested, immune active T cells recognizing preproinsulin, ZnT8, GAD65 and IGRP were detected; T cells that recognized ChgA were not detected. The epitope specificity of GAD65 and IGRP specific T cells among the 4 subjects tested were quite diverse. T cells from these subjects did not share the same epitope specificity towards these two proteins. However, responses to modified preproinsulin epitope (PPI78-90K88S) and ZnT8 epitope (ZnT266-285) were shared by all 4 subjects tested. Furthermore, T cells that responded to these two epitopes were frequently contained in multiple oligoclones (Supplemental Table 2), which suggests a high frequency of these T cells in the immune-active T cell fraction. These results indicate that the PPI78-90K88S epitope and the ZnT266-285 epitope are dominant antigenic epitopes which could potentially be targets for antigen-specific therapies. The relative abundance of PPI78-90K88S reactive T cells in the periphery was also supported by the longitudinal detection of PPI memory T cells of identical clonotype in the periphery.
The CD154 upregulation epitope mapping approach also led to the discovery of new epitopes, including ZnT8266-285 epitopes, which have not been previously reported. Although T cell epitopes for GAD65 and IGRP have been studied extensively, our current study also identified novel epitopes of these two proteins. These results illustrated the robustness of this mapping approach, and emphasize the high degree of diversity among individual autoimmune repertoires.
It is clear that T1D incidence is highly associated with HLA-DQ8. Thus there is great interest in identifying DQ8 restricting T cells. To our surprise, the responses of all the oligoclones to their corresponding peptides we tested were blocked by HLA-DR, not HLA-DQ blocking antibody, including an oligoclone specific to GADp32 peptide which contains HLA-DQ8 eptitope by tetramer staining [29]. One interpretation of this finding is that in this subject, HLA-DR and DQ8 restricted T cells may play different roles in the pathogenesis: DQ8 restricted T cells may be contributing to disease susceptibility or initiation [14], while DR restricted T cells may become dominant during disease progression and after disease onset.
In summary, we demonstrate that a small fraction of beta cell antigen specific memory T cells show evidence of recent antigen stimulation and persist in the circulation, even after onset of T1D. This type of targeted detection method can be used to study the roles of antigen specific T cells in the pathogenesis of type 1 diabetes, but also has potential application in development of personalized antigen specific vaccines.
Supplementary Material
Acknowledgments
We wish to thank Cynthia Cousens-Jacobs for her assistance in the preparation of this manuscript.
Footnotes
This project was funded in part by NIH Grant #1 DP3 DK106909-01 entitled “Phenotypic analysis of islet antigen-specific effector T cells in pre-diabetic subjects”. Dr. William Kwok is the PI.
References
- 1.Anderson MS, Bluestone JA. The NOD mouse: a model of immune dysregulation. Annu Rev Immunol. 2005;23:447–85. doi: 10.1146/annurev.immunol.23.021704.115643. [DOI] [PubMed] [Google Scholar]
- 2.Tisch R, McDevitt H. Insulin-dependent diabetes mellitus. Cell. 1996;85(3):291–7. doi: 10.1016/s0092-8674(00)81106-x. [DOI] [PubMed] [Google Scholar]
- 3.Bach JF. Autoimmunity and type I diabetes. Trends Endocrinol Metab. 1997;8(2):71–4. doi: 10.1016/s1043-2760(96)00271-8. [DOI] [PubMed] [Google Scholar]
- 4.Roep BO, Peakman M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb Perspect Med. 2012;2(4):a007781. doi: 10.1101/cshperspect.a007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Lorenzo TP, Peakman M, Roep BO. Translational mini-review series on type 1 diabetes: Systematic analysis of T cell epitopes in autoimmune diabetes. Clin Exp Immunol. 2007;148(1):1–16. doi: 10.1111/j.1365-2249.2006.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Pontesilli O, Gill RG, La Rosa FG, Lafferty KJ. The role of CD4+ and CD8+ T cells in the destruction of islet grafts by spontaneously diabetic mice. Proc Natl Acad Sci U S A. 1991;88(2):527–31. doi: 10.1073/pnas.88.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christianson SW, Shultz LD, Leiter EH. Adoptive transfer of diabetes into immunodeficient NOD-scid/scid mice. Relative contributions of CD4+ and CD8+ T-cells from diabetic versus prediabetic NOD.NON-Thy-1a donors. Diabetes. 1993;42(1):44–55. doi: 10.2337/diab.42.1.44. [DOI] [PubMed] [Google Scholar]
- 8.DiLorenzo TP, Graser RT, Ono T, Christianson GJ, Chapman HD, Roopenian DC, Nathenson SG, Serreze DV. Major histocompatibility complex class I-restricted T cells are required for all but the end stages of diabetes development in nonobese diabetic mice and use a prevalent T cell receptor alpha chain gene rearrangement. Proc Natl Acad Sci U S A. 1998;95(21):12538–43. doi: 10.1073/pnas.95.21.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinkse GG, Tysma OH, Bergen CA, Kester MG, Ossendorp F, van Veelen PA, Keymeulen B, Pipeleers D, Drijfhout JW, Roep BO. Autoreactive CD8 T cells associated with beta cell destruction in type 1 diabetes. Proc Natl Acad Sci U S A. 2005;102(51):18425–30. doi: 10.1073/pnas.0508621102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aly TA, Ide A, Jahromi MM, Barker JM, Fernando MS, Babu SR, Yu L, Miao D, Erlich HA, Fain PR, Barriga KJ, Norris JM, Rewers MJ, Eisenbarth GS. Extreme genetic risk for type 1A diabetes. Proc Natl Acad Sci U S A. 2006;103(38):14074–9. doi: 10.1073/pnas.0606349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castano L, Eisenbarth GS. Type-I diabetes: a chronic autoimmune disease of human, mouse, and rat. Annu Rev Immunol. 1990;8:647–79. doi: 10.1146/annurev.iy.08.040190.003243. [DOI] [PubMed] [Google Scholar]
- 12.Bradley BJ, Haskins K, La Rosa FG, Lafferty KJ. CD8 T cells are not required for islet destruction induced by a CD4+ islet-specific T-cell clone. Diabetes. 1992;41(12):1603–8. doi: 10.2337/diab.41.12.1603. [DOI] [PubMed] [Google Scholar]
- 13.Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, Hering BJ, Hafler DA. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature. 2005;435(7039):224–8. doi: 10.1038/nature03625. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, Wegmann DR, Hutton JC, Elliott JF, Eisenbarth GS. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005;435(7039):220–3. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarbell KV, Lee M, Ranheim E, Chao CC, Sanna M, Kim SK, Dickie P, Teyton L, Davis M, McDevitt H. CD4(+) T cells from glutamic acid decarboxylase (GAD)65-specific T cell receptor transgenic mice are not diabetogenic and can delay diabetes transfer. J Exp Med. 2002;196(4):481–92. doi: 10.1084/jem.20011845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee R, Wagar D, Stephens TA, Lee-Chan E, Singh B. Identification of CD4+ T cell-specific epitopes of islet-specific glucose-6-phosphatase catalytic subunit-related protein: a novel beta cell autoantigen in type 1 diabetes. J Immunol. 2005;174(9):5306–15. doi: 10.4049/jimmunol.174.9.5306. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Danke NA, Berger D, Reichstetter S, Reijonen H, Greenbaum C, Pihoker C, James EA, Kwok WW. Islet-specific glucose-6-phosphatase catalytic subunit-related protein-reactive CD4+ T cells in human subjects. J Immunol. 2006;176(5):2781–9. doi: 10.4049/jimmunol.176.5.2781. [DOI] [PubMed] [Google Scholar]
- 18.Stadinski BD, Delong T, Reisdorph N, Reisdorph R, Powell RL, Armstrong M, Piganelli JD, Barbour G, Bradley B, Crawford F, Marrack P, Mahata SK, Kappler JW, Haskins K. Chromogranin A is an autoantigen in type 1 diabetes. Nat Immunol. 2010;11(3):225–31. doi: 10.1038/ni.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stadinski B, Kappler J, Eisenbarth GS. Molecular targeting of islet autoantigens. Immunity. 2010;32(4):446–56. doi: 10.1016/j.immuni.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Chimienti F, Devergnas S, Favier A, Seve M. Identification and cloning of a beta-cell-specific zinc transporter, ZnT-8, localized into insulin secretory granules. Diabetes. 2004;53(9):2330–7. doi: 10.2337/diabetes.53.9.2330. [DOI] [PubMed] [Google Scholar]
- 21.Wenzlau JM, Juhl K, Yu L, Moua O, Sarkar SA, Gottlieb P, Rewers M, Eisenbarth GS, Jensen J, Davidson HW, Hutton JC. The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc Natl Acad Sci U S A. 2007;104(43):17040–5. doi: 10.1073/pnas.0705894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dianzani U, Funaro A, DiFranco D, Garbarino G, Bragardo M, Redoglia V, Buonfiglio D, De Monte LB, Pileri A, Malavasi F. Interaction between endothelium and CD4+CD45RA+ lymphocytes. Role of the human CD38 molecule. J Immunol. 1994;153(3):952–9. [PubMed] [Google Scholar]
- 23.Han A, Newell EW, Glanville J, Fernandez-Becker N, Khosla C, Chien YH, Davis MM. Dietary gluten triggers concomitant activation of CD4+ and CD8+ alphabeta T cells and gammadelta T cells in celiac disease. Proc Natl Acad Sci U S A. 2013;110(32):13073–8. doi: 10.1073/pnas.1311861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, James E, Gates TJ, DeLong JH, LaFond RE, Malhotra U, Kwok WW. CD4+ T cells recognize unique and conserved 2009 H1N1 influenza hemagglutinin epitopes after natural infection and vaccination. Int Immunol. 2013;25(8):447–57. doi: 10.1093/intimm/dxt005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Danke N, Roti M, Huston L, Greenbaum C, Pihoker C, James E, Kwok WW. CD4+ T cells from type 1 diabetic and healthy subjects exhibit different thresholds of activation to a naturally processed proinsulin epitope. J Autoimmun. 2008;31(1):30–41. doi: 10.1016/j.jaut.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Chow IT, Sosinowski T, Torres-Chinn N, Greenbaum CJ, James EA, Kappler JW, Davidson HW, Kwok WW. Autoreactive T cells specific for insulin B: 11–23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc Natl Acad Sci U S A. 2014;111(41):14840–5. doi: 10.1073/pnas.1416864111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frentsch M, Arbach O, Kirchhoff D, Moewes B, Worm M, Rothe M, Scheffold A, Thiel A. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat.Med. 2005;11(10):1118–1124. doi: 10.1038/nm1292. [DOI] [PubMed] [Google Scholar]
- 28.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114(19):4099–107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chow IT, Yang J, Gates TJ, James EA, Mai DT, Greenbaum C, Kwok WW. Assessment of CD4+ T cell responses to glutamic acid decarboxylase 65 using DQ8 tetramers reveals a pathogenic role of GAD65 121–140 and GAD65 250–266 in T1D development. PLoS One. 2014;9(11):e112882. doi: 10.1371/journal.pone.0112882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, Steen MS, LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, Wu D, Wood BL, Rieder MJ, Robins H. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 31.Caruso A, Licenziati S, Corulli M, Canaris AD, De Francesco MA, Fiorentini S, Peroni L, Fallacara F, Dima F, Balsari A, Turano A. Flow cytometric analysis of activation markers on stimulated T cells and their correlation with cell proliferation. Cytometry. 1997;27(1):71–76. doi: 10.1002/(sici)1097-0320(19970101)27:1<71::aid-cyto9>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 32.Ziegler SF, Levin SD, Johnson L, Copeland NG, Gilbert DJ, Jenkins NA, Baker E, Sutherland GR, Feldhaus AL, Ramsdell F. The mouse CD69 gene. Structure, expression, and mapping to the NK gene complex. J Immunol. 1994;152(3):1228–36. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.